
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MLPH-SHARPIN (FusionGDB2 ID:54339) |
Fusion Gene Summary for MLPH-SHARPIN |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MLPH-SHARPIN | Fusion gene ID: 54339 | Hgene | Tgene | Gene symbol | MLPH | SHARPIN | Gene ID | 79083 | 81858 |
| Gene name | melanophilin | SHANK associated RH domain interactor | |
| Synonyms | SLAC2-A | SIPL1 | |
| Cytomap | 2q37.3 | 8q24.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | melanophilinexophilin-3slp homolog lacking C2 domains asynaptotagmin-like protein 2a | sharpinhSIPL1shank-associated RH domain-interacting proteinshank-interacting protein-like 1 | |
| Modification date | 20200320 | 20200313 | |
| UniProtAcc | Q9BV36 | SHARPIN | |
| Ensembl transtripts involved in fusion gene | ENST00000468178, ENST00000264605, ENST00000338530, ENST00000409373, ENST00000410032, ENST00000445024, | ENST00000398712, ENST00000533948, | |
| Fusion gene scores | * DoF score | 7 X 6 X 4=168 | 7 X 2 X 4=56 |
| # samples | 7 | 7 | |
| ** MAII score | log2(7/168*10)=-1.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/56*10)=0.321928094887362 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: MLPH [Title/Abstract] AND SHARPIN [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MLPH(238419744)-SHARPIN(145154972), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | SHARPIN | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway | 21455173 |
| Tgene | SHARPIN | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 21455173 |
| Tgene | SHARPIN | GO:0097039 | protein linear polyubiquitination | 21455173|21455180|21455181 |
| Tgene | SHARPIN | GO:2000348 | regulation of CD40 signaling pathway | 21455173 |
 Fusion gene breakpoints across MLPH (5'-gene) Fusion gene breakpoints across MLPH (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
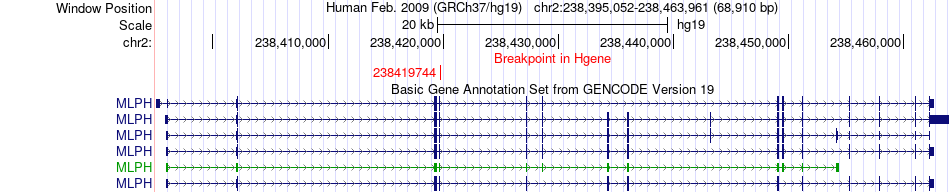 |
 Fusion gene breakpoints across SHARPIN (3'-gene) Fusion gene breakpoints across SHARPIN (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
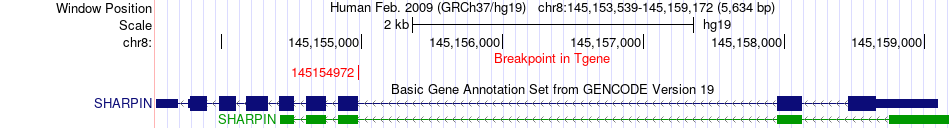 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-IN-7808-01A | MLPH | chr2 | 238419744 | + | SHARPIN | chr8 | 145154972 | - |
Top |
Fusion Gene ORF analysis for MLPH-SHARPIN |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000468178 | ENST00000398712 | MLPH | chr2 | 238419744 | + | SHARPIN | chr8 | 145154972 | - |
| 3UTR-5UTR | ENST00000468178 | ENST00000533948 | MLPH | chr2 | 238419744 | + | SHARPIN | chr8 | 145154972 | - |
| 5CDS-5UTR | ENST00000264605 | ENST00000533948 | MLPH | chr2 | 238419744 | + | SHARPIN | chr8 | 145154972 | - |
| 5CDS-5UTR | ENST00000338530 | ENST00000533948 | MLPH | chr2 | 238419744 | + | SHARPIN | chr8 | 145154972 | - |
| 5CDS-5UTR | ENST00000409373 | ENST00000533948 | MLPH | chr2 | 238419744 | + | SHARPIN | chr8 | 145154972 | - |
| 5CDS-5UTR | ENST00000410032 | ENST00000533948 | MLPH | chr2 | 238419744 | + | SHARPIN | chr8 | 145154972 | - |
| 5CDS-5UTR | ENST00000445024 | ENST00000533948 | MLPH | chr2 | 238419744 | + | SHARPIN | chr8 | 145154972 | - |
| In-frame | ENST00000264605 | ENST00000398712 | MLPH | chr2 | 238419744 | + | SHARPIN | chr8 | 145154972 | - |
| In-frame | ENST00000338530 | ENST00000398712 | MLPH | chr2 | 238419744 | + | SHARPIN | chr8 | 145154972 | - |
| In-frame | ENST00000409373 | ENST00000398712 | MLPH | chr2 | 238419744 | + | SHARPIN | chr8 | 145154972 | - |
| In-frame | ENST00000410032 | ENST00000398712 | MLPH | chr2 | 238419744 | + | SHARPIN | chr8 | 145154972 | - |
| In-frame | ENST00000445024 | ENST00000398712 | MLPH | chr2 | 238419744 | + | SHARPIN | chr8 | 145154972 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000410032 | MLPH | chr2 | 238419744 | + | ENST00000398712 | SHARPIN | chr8 | 145154972 | - | 1790 | 838 | 393 | 1625 | 410 |
| ENST00000264605 | MLPH | chr2 | 238419744 | + | ENST00000398712 | SHARPIN | chr8 | 145154972 | - | 1691 | 739 | 294 | 1526 | 410 |
| ENST00000445024 | MLPH | chr2 | 238419744 | + | ENST00000398712 | SHARPIN | chr8 | 145154972 | - | 1609 | 657 | 212 | 1444 | 410 |
| ENST00000338530 | MLPH | chr2 | 238419744 | + | ENST00000398712 | SHARPIN | chr8 | 145154972 | - | 1608 | 656 | 211 | 1443 | 410 |
| ENST00000409373 | MLPH | chr2 | 238419744 | + | ENST00000398712 | SHARPIN | chr8 | 145154972 | - | 1583 | 631 | 186 | 1418 | 410 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000410032 | ENST00000398712 | MLPH | chr2 | 238419744 | + | SHARPIN | chr8 | 145154972 | - | 0.19324152 | 0.8067585 |
| ENST00000264605 | ENST00000398712 | MLPH | chr2 | 238419744 | + | SHARPIN | chr8 | 145154972 | - | 0.21013698 | 0.78986305 |
| ENST00000445024 | ENST00000398712 | MLPH | chr2 | 238419744 | + | SHARPIN | chr8 | 145154972 | - | 0.21407509 | 0.7859249 |
| ENST00000338530 | ENST00000398712 | MLPH | chr2 | 238419744 | + | SHARPIN | chr8 | 145154972 | - | 0.21480581 | 0.7851942 |
| ENST00000409373 | ENST00000398712 | MLPH | chr2 | 238419744 | + | SHARPIN | chr8 | 145154972 | - | 0.21757989 | 0.78242016 |
Top |
Fusion Genomic Features for MLPH-SHARPIN |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for MLPH-SHARPIN |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:238419744/chr8:145154972) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MLPH | SHARPIN |
| FUNCTION: Rab effector protein involved in melanosome transport. Serves as link between melanosome-bound RAB27A and the motor protein MYO5A. {ECO:0000269|PubMed:12062444}. | 387 |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MLPH | chr2:238419744 | chr8:145154972 | ENST00000264605 | + | 4 | 16 | 4_124 | 148 | 601.0 | Domain | RabBD |
| Hgene | MLPH | chr2:238419744 | chr8:145154972 | ENST00000338530 | + | 4 | 15 | 4_124 | 148 | 573.0 | Domain | RabBD |
| Hgene | MLPH | chr2:238419744 | chr8:145154972 | ENST00000409373 | + | 4 | 13 | 4_124 | 148 | 481.0 | Domain | RabBD |
| Hgene | MLPH | chr2:238419744 | chr8:145154972 | ENST00000264605 | + | 4 | 16 | 64_107 | 148 | 601.0 | Zinc finger | Note=FYVE-type |
| Hgene | MLPH | chr2:238419744 | chr8:145154972 | ENST00000338530 | + | 4 | 15 | 64_107 | 148 | 573.0 | Zinc finger | Note=FYVE-type |
| Hgene | MLPH | chr2:238419744 | chr8:145154972 | ENST00000409373 | + | 4 | 13 | 64_107 | 148 | 481.0 | Zinc finger | Note=FYVE-type |
| Tgene | SHARPIN | chr2:238419744 | chr8:145154972 | ENST00000398712 | 1 | 9 | 219_288 | 125 | 210.66666666666666 | Domain | Note=Ubiquitin-like | |
| Tgene | SHARPIN | chr2:238419744 | chr8:145154972 | ENST00000398712 | 1 | 9 | 348_377 | 125 | 210.66666666666666 | Zinc finger | RanBP2-type |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MLPH | chr2:238419744 | chr8:145154972 | ENST00000264605 | + | 4 | 16 | 373_496 | 148 | 601.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MLPH | chr2:238419744 | chr8:145154972 | ENST00000338530 | + | 4 | 15 | 373_496 | 148 | 573.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MLPH | chr2:238419744 | chr8:145154972 | ENST00000409373 | + | 4 | 13 | 373_496 | 148 | 481.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Tgene | SHARPIN | chr2:238419744 | chr8:145154972 | ENST00000398712 | 1 | 9 | 1_180 | 125 | 210.66666666666666 | Region | Self-association |
Top |
Fusion Gene Sequence for MLPH-SHARPIN |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >54339_54339_1_MLPH-SHARPIN_MLPH_chr2_238419744_ENST00000264605_SHARPIN_chr8_145154972_ENST00000398712_length(transcript)=1691nt_BP=739nt ATCCAGAGGTGTCCGCGCTCCCGGAGCTGGGCGGAGCGGGGCGCAGCCCACGTGGTTCGGGCGGGAGGCGCCGGGACGTGGCCAGTTGCC CGCCTGCCCCGGAGAGCCAGGCGCTAACCAGCCGCTCTGCGCCCCGCGCCCTGCTTGCCCCCATTATCCAGCCTTGCCCCGGCGCCCTGA CCTGACGCCCTGGCCTGACGCCCTGCTTCGTCGCCTCCTTTCTCTCCCAGGTGCTGGACCAGGGACTGAGCGTCCCCCGGAGAGGGTCCG GTGTGACCCCGACAAGAAGCAGAAATGGGGAAGAAACTGGATCTTTCCAAGCTCACTGATGAAGAGGCCCAGCATGTCTTGGAAGTTGTT CAACGAGATTTTGACCTCCGAAGGAAAGAAGAGGAACGGCTAGAGGCGTTGAAGGGCAAGATTAAGAAGGAAAGCTCCAAGAGGGAGCTG CTTTCCGACACTGCCCATCTGAACGAGACCCACTGCGCCCGCTGCCTGCAGCCCTACCAGCTGCTTGTGAATAGCAAAAGGCAGTGCCTG GAATGTGGCCTCTTCACCTGCAAAAGCTGTGGCCGCGTCCACCCGGAGGAGCAGGGCTGGATCTGTGACCCCTGCCATCTGGCCAGAGTC GTGAAGATCGGCTCACTGGAGTGGTACTATGAGCATGTGAAAGCCCGCTTCAAGAGGTTCGGAAGTGCCAAGGTCATCCGGTCCCTCCAC GGGCGGCTGCAGGGTGGAGGCAGCAAGAGCAACTCACCACCAGCCTTGGGCCCAGAAGCATGCCCTGTCTCCCTGCCCAGTCCCCCGGAA GCCTCCACACTCAAGGGCCCTCCACCTGAGGCAGATCTTCCTAGGAGCCCTGGAAACTTGACGGAGAGAGAAGAGCTGGCAGGGAGCCTG GCCCGGGCTATTGCAGGTGGAGACGAGAAGGGGGCAGCCCAAGTGGCAGCCGTCCTGGCCCAGCATCGTGTGGCCCTGAGTGTTCAGCTT CAGGAGGCCTGCTTCCCACCTGGCCCCATCAGGCTGCAGGTCACACTTGAAGACGCTGCCTCTGCCGCATCCGCCGCGTCCTCTGCACAC GTTGCCCTGCAGGTCCACCCCCACTGCACTGTTGCAGCTCTCCAGGAGCAGGTGTTCTCAGAGCTCGGTTTCCCGCCAGCCGTGCAACGC TGGGTCATCGGACGGTGCCTGTGTGTGCCTGAGCGCAGCCTTGCCTCTTACGGGGTTCGGCAGGATGGGGACCCTGCTTTCCTCTACTTG CTGTCAGCTCCTCGAGAAGCCCCAGCCACAGGACCTAGCCCTCAGCACCCCCAGAAGATGGACGGGGAACTTGGACGCTTGTTTCCCCCA TCATTGGGGCTACCCCCAGGCCCCCAGCCAGCTGCCTCCAGCCTGCCCAGTCCACTCCAGCCCAGCTGGTCCTGTCCTTCCTGCACCTTC ATCAATGCCCCAGACCGCCCTGGCTGTGAGATGTGTAGCACCCAGAGGCCCTGCACTTGGGACCCCCTTGCTGCAGCTTCCACCTAGCAG CCACCAGAGGTTACAAGGGGAGAGTGGCCCTTCCCTCACAAGTCCGACATCTCCAGGCCCCCACTGAACTCCGGGGACCTCTACTGACTG >54339_54339_1_MLPH-SHARPIN_MLPH_chr2_238419744_ENST00000264605_SHARPIN_chr8_145154972_ENST00000398712_length(amino acids)=410AA_BP=148 MGKKLDLSKLTDEEAQHVLEVVQRDFDLRRKEEERLEALKGKIKKESSKRELLSDTAHLNETHCARCLQPYQLLVNSKRQCLECGLFTCK SCGRVHPEEQGWICDPCHLARVVKIGSLEWYYEHVKARFKRFGSAKVIRSLHGRLQGGGSKSNSPPALGPEACPVSLPSPPEASTLKGPP PEADLPRSPGNLTEREELAGSLARAIAGGDEKGAAQVAAVLAQHRVALSVQLQEACFPPGPIRLQVTLEDAASAASAASSAHVALQVHPH CTVAALQEQVFSELGFPPAVQRWVIGRCLCVPERSLASYGVRQDGDPAFLYLLSAPREAPATGPSPQHPQKMDGELGRLFPPSLGLPPGP -------------------------------------------------------------- >54339_54339_2_MLPH-SHARPIN_MLPH_chr2_238419744_ENST00000338530_SHARPIN_chr8_145154972_ENST00000398712_length(transcript)=1608nt_BP=656nt AGTTGCCCGCCTGCCCCGGAGAGCCAGGCGCTAACCAGCCGCTCTGCGCCCCGCGCCCTGCTTGCCCCCATTATCCAGCCTTGCCCCGGC GCCCTGACCTGACGCCCTGGCCTGACGCCCTGCTTCGTCGCCTCCTTTCTCTCCCAGGTGCTGGACCAGGGACTGAGCGTCCCCCGGAGA GGGTCCGGTGTGACCCCGACAAGAAGCAGAAATGGGGAAGAAACTGGATCTTTCCAAGCTCACTGATGAAGAGGCCCAGCATGTCTTGGA AGTTGTTCAACGAGATTTTGACCTCCGAAGGAAAGAAGAGGAACGGCTAGAGGCGTTGAAGGGCAAGATTAAGAAGGAAAGCTCCAAGAG GGAGCTGCTTTCCGACACTGCCCATCTGAACGAGACCCACTGCGCCCGCTGCCTGCAGCCCTACCAGCTGCTTGTGAATAGCAAAAGGCA GTGCCTGGAATGTGGCCTCTTCACCTGCAAAAGCTGTGGCCGCGTCCACCCGGAGGAGCAGGGCTGGATCTGTGACCCCTGCCATCTGGC CAGAGTCGTGAAGATCGGCTCACTGGAGTGGTACTATGAGCATGTGAAAGCCCGCTTCAAGAGGTTCGGAAGTGCCAAGGTCATCCGGTC CCTCCACGGGCGGCTGCAGGGTGGAGGCAGCAAGAGCAACTCACCACCAGCCTTGGGCCCAGAAGCATGCCCTGTCTCCCTGCCCAGTCC CCCGGAAGCCTCCACACTCAAGGGCCCTCCACCTGAGGCAGATCTTCCTAGGAGCCCTGGAAACTTGACGGAGAGAGAAGAGCTGGCAGG GAGCCTGGCCCGGGCTATTGCAGGTGGAGACGAGAAGGGGGCAGCCCAAGTGGCAGCCGTCCTGGCCCAGCATCGTGTGGCCCTGAGTGT TCAGCTTCAGGAGGCCTGCTTCCCACCTGGCCCCATCAGGCTGCAGGTCACACTTGAAGACGCTGCCTCTGCCGCATCCGCCGCGTCCTC TGCACACGTTGCCCTGCAGGTCCACCCCCACTGCACTGTTGCAGCTCTCCAGGAGCAGGTGTTCTCAGAGCTCGGTTTCCCGCCAGCCGT GCAACGCTGGGTCATCGGACGGTGCCTGTGTGTGCCTGAGCGCAGCCTTGCCTCTTACGGGGTTCGGCAGGATGGGGACCCTGCTTTCCT CTACTTGCTGTCAGCTCCTCGAGAAGCCCCAGCCACAGGACCTAGCCCTCAGCACCCCCAGAAGATGGACGGGGAACTTGGACGCTTGTT TCCCCCATCATTGGGGCTACCCCCAGGCCCCCAGCCAGCTGCCTCCAGCCTGCCCAGTCCACTCCAGCCCAGCTGGTCCTGTCCTTCCTG CACCTTCATCAATGCCCCAGACCGCCCTGGCTGTGAGATGTGTAGCACCCAGAGGCCCTGCACTTGGGACCCCCTTGCTGCAGCTTCCAC CTAGCAGCCACCAGAGGTTACAAGGGGAGAGTGGCCCTTCCCTCACAAGTCCGACATCTCCAGGCCCCCACTGAACTCCGGGGACCTCTA >54339_54339_2_MLPH-SHARPIN_MLPH_chr2_238419744_ENST00000338530_SHARPIN_chr8_145154972_ENST00000398712_length(amino acids)=410AA_BP=148 MGKKLDLSKLTDEEAQHVLEVVQRDFDLRRKEEERLEALKGKIKKESSKRELLSDTAHLNETHCARCLQPYQLLVNSKRQCLECGLFTCK SCGRVHPEEQGWICDPCHLARVVKIGSLEWYYEHVKARFKRFGSAKVIRSLHGRLQGGGSKSNSPPALGPEACPVSLPSPPEASTLKGPP PEADLPRSPGNLTEREELAGSLARAIAGGDEKGAAQVAAVLAQHRVALSVQLQEACFPPGPIRLQVTLEDAASAASAASSAHVALQVHPH CTVAALQEQVFSELGFPPAVQRWVIGRCLCVPERSLASYGVRQDGDPAFLYLLSAPREAPATGPSPQHPQKMDGELGRLFPPSLGLPPGP -------------------------------------------------------------- >54339_54339_3_MLPH-SHARPIN_MLPH_chr2_238419744_ENST00000409373_SHARPIN_chr8_145154972_ENST00000398712_length(transcript)=1583nt_BP=631nt AGGCGCTAACCAGCCGCTCTGCGCCCCGCGCCCTGCTTGCCCCCATTATCCAGCCTTGCCCCGGCGCCCTGACCTGACGCCCTGGCCTGA CGCCCTGCTTCGTCGCCTCCTTTCTCTCCCAGGTGCTGGACCAGGGACTGAGCGTCCCCCGGAGAGGGTCCGGTGTGACCCCGACAAGAA GCAGAAATGGGGAAGAAACTGGATCTTTCCAAGCTCACTGATGAAGAGGCCCAGCATGTCTTGGAAGTTGTTCAACGAGATTTTGACCTC CGAAGGAAAGAAGAGGAACGGCTAGAGGCGTTGAAGGGCAAGATTAAGAAGGAAAGCTCCAAGAGGGAGCTGCTTTCCGACACTGCCCAT CTGAACGAGACCCACTGCGCCCGCTGCCTGCAGCCCTACCAGCTGCTTGTGAATAGCAAAAGGCAGTGCCTGGAATGTGGCCTCTTCACC TGCAAAAGCTGTGGCCGCGTCCACCCGGAGGAGCAGGGCTGGATCTGTGACCCCTGCCATCTGGCCAGAGTCGTGAAGATCGGCTCACTG GAGTGGTACTATGAGCATGTGAAAGCCCGCTTCAAGAGGTTCGGAAGTGCCAAGGTCATCCGGTCCCTCCACGGGCGGCTGCAGGGTGGA GGCAGCAAGAGCAACTCACCACCAGCCTTGGGCCCAGAAGCATGCCCTGTCTCCCTGCCCAGTCCCCCGGAAGCCTCCACACTCAAGGGC CCTCCACCTGAGGCAGATCTTCCTAGGAGCCCTGGAAACTTGACGGAGAGAGAAGAGCTGGCAGGGAGCCTGGCCCGGGCTATTGCAGGT GGAGACGAGAAGGGGGCAGCCCAAGTGGCAGCCGTCCTGGCCCAGCATCGTGTGGCCCTGAGTGTTCAGCTTCAGGAGGCCTGCTTCCCA CCTGGCCCCATCAGGCTGCAGGTCACACTTGAAGACGCTGCCTCTGCCGCATCCGCCGCGTCCTCTGCACACGTTGCCCTGCAGGTCCAC CCCCACTGCACTGTTGCAGCTCTCCAGGAGCAGGTGTTCTCAGAGCTCGGTTTCCCGCCAGCCGTGCAACGCTGGGTCATCGGACGGTGC CTGTGTGTGCCTGAGCGCAGCCTTGCCTCTTACGGGGTTCGGCAGGATGGGGACCCTGCTTTCCTCTACTTGCTGTCAGCTCCTCGAGAA GCCCCAGCCACAGGACCTAGCCCTCAGCACCCCCAGAAGATGGACGGGGAACTTGGACGCTTGTTTCCCCCATCATTGGGGCTACCCCCA GGCCCCCAGCCAGCTGCCTCCAGCCTGCCCAGTCCACTCCAGCCCAGCTGGTCCTGTCCTTCCTGCACCTTCATCAATGCCCCAGACCGC CCTGGCTGTGAGATGTGTAGCACCCAGAGGCCCTGCACTTGGGACCCCCTTGCTGCAGCTTCCACCTAGCAGCCACCAGAGGTTACAAGG GGAGAGTGGCCCTTCCCTCACAAGTCCGACATCTCCAGGCCCCCACTGAACTCCGGGGACCTCTACTGACTGCTTGCTGGGACAGTCACC >54339_54339_3_MLPH-SHARPIN_MLPH_chr2_238419744_ENST00000409373_SHARPIN_chr8_145154972_ENST00000398712_length(amino acids)=410AA_BP=148 MGKKLDLSKLTDEEAQHVLEVVQRDFDLRRKEEERLEALKGKIKKESSKRELLSDTAHLNETHCARCLQPYQLLVNSKRQCLECGLFTCK SCGRVHPEEQGWICDPCHLARVVKIGSLEWYYEHVKARFKRFGSAKVIRSLHGRLQGGGSKSNSPPALGPEACPVSLPSPPEASTLKGPP PEADLPRSPGNLTEREELAGSLARAIAGGDEKGAAQVAAVLAQHRVALSVQLQEACFPPGPIRLQVTLEDAASAASAASSAHVALQVHPH CTVAALQEQVFSELGFPPAVQRWVIGRCLCVPERSLASYGVRQDGDPAFLYLLSAPREAPATGPSPQHPQKMDGELGRLFPPSLGLPPGP -------------------------------------------------------------- >54339_54339_4_MLPH-SHARPIN_MLPH_chr2_238419744_ENST00000410032_SHARPIN_chr8_145154972_ENST00000398712_length(transcript)=1790nt_BP=838nt ATTGGAATTCGGAAGGACAGGCAGGCCGGGCGGACTGGAGACGGCGGCTCGCCTTGGGACCCTCTGAGCTTGCGGGCCCCGCTGAGCACC CAAAGGCCGGCCCTAGAGTCCAGGAGAAGAGCGCAGCGGCGCGGAGCTCCCAGGCGTTCCCCGCAGCGCGTCCTCGGTCCTGGAACCACC GCGCCGCGCGTCCTGGCTTCCACATCTGCCCCATTTGCCCGCGGATCTTGACTTTTTCTTGGCGGGCAAGGCCTCGGTCTCCTCCCCGCA GCAGGGCTGCCCGCGGCTGCGGGACCCTCCTGGTCCTGATGCCTGGCAGAATCGAAGGGGTGCTGGACCAGGGACTGAGCGTCCCCCGGA GAGGGTCCGGTGTGACCCCGACAAGAAGCAGAAATGGGGAAGAAACTGGATCTTTCCAAGCTCACTGATGAAGAGGCCCAGCATGTCTTG GAAGTTGTTCAACGAGATTTTGACCTCCGAAGGAAAGAAGAGGAACGGCTAGAGGCGTTGAAGGGCAAGATTAAGAAGGAAAGCTCCAAG AGGGAGCTGCTTTCCGACACTGCCCATCTGAACGAGACCCACTGCGCCCGCTGCCTGCAGCCCTACCAGCTGCTTGTGAATAGCAAAAGG CAGTGCCTGGAATGTGGCCTCTTCACCTGCAAAAGCTGTGGCCGCGTCCACCCGGAGGAGCAGGGCTGGATCTGTGACCCCTGCCATCTG GCCAGAGTCGTGAAGATCGGCTCACTGGAGTGGTACTATGAGCATGTGAAAGCCCGCTTCAAGAGGTTCGGAAGTGCCAAGGTCATCCGG TCCCTCCACGGGCGGCTGCAGGGTGGAGGCAGCAAGAGCAACTCACCACCAGCCTTGGGCCCAGAAGCATGCCCTGTCTCCCTGCCCAGT CCCCCGGAAGCCTCCACACTCAAGGGCCCTCCACCTGAGGCAGATCTTCCTAGGAGCCCTGGAAACTTGACGGAGAGAGAAGAGCTGGCA GGGAGCCTGGCCCGGGCTATTGCAGGTGGAGACGAGAAGGGGGCAGCCCAAGTGGCAGCCGTCCTGGCCCAGCATCGTGTGGCCCTGAGT GTTCAGCTTCAGGAGGCCTGCTTCCCACCTGGCCCCATCAGGCTGCAGGTCACACTTGAAGACGCTGCCTCTGCCGCATCCGCCGCGTCC TCTGCACACGTTGCCCTGCAGGTCCACCCCCACTGCACTGTTGCAGCTCTCCAGGAGCAGGTGTTCTCAGAGCTCGGTTTCCCGCCAGCC GTGCAACGCTGGGTCATCGGACGGTGCCTGTGTGTGCCTGAGCGCAGCCTTGCCTCTTACGGGGTTCGGCAGGATGGGGACCCTGCTTTC CTCTACTTGCTGTCAGCTCCTCGAGAAGCCCCAGCCACAGGACCTAGCCCTCAGCACCCCCAGAAGATGGACGGGGAACTTGGACGCTTG TTTCCCCCATCATTGGGGCTACCCCCAGGCCCCCAGCCAGCTGCCTCCAGCCTGCCCAGTCCACTCCAGCCCAGCTGGTCCTGTCCTTCC TGCACCTTCATCAATGCCCCAGACCGCCCTGGCTGTGAGATGTGTAGCACCCAGAGGCCCTGCACTTGGGACCCCCTTGCTGCAGCTTCC ACCTAGCAGCCACCAGAGGTTACAAGGGGAGAGTGGCCCTTCCCTCACAAGTCCGACATCTCCAGGCCCCCACTGAACTCCGGGGACCTC >54339_54339_4_MLPH-SHARPIN_MLPH_chr2_238419744_ENST00000410032_SHARPIN_chr8_145154972_ENST00000398712_length(amino acids)=410AA_BP=148 MGKKLDLSKLTDEEAQHVLEVVQRDFDLRRKEEERLEALKGKIKKESSKRELLSDTAHLNETHCARCLQPYQLLVNSKRQCLECGLFTCK SCGRVHPEEQGWICDPCHLARVVKIGSLEWYYEHVKARFKRFGSAKVIRSLHGRLQGGGSKSNSPPALGPEACPVSLPSPPEASTLKGPP PEADLPRSPGNLTEREELAGSLARAIAGGDEKGAAQVAAVLAQHRVALSVQLQEACFPPGPIRLQVTLEDAASAASAASSAHVALQVHPH CTVAALQEQVFSELGFPPAVQRWVIGRCLCVPERSLASYGVRQDGDPAFLYLLSAPREAPATGPSPQHPQKMDGELGRLFPPSLGLPPGP -------------------------------------------------------------- >54339_54339_5_MLPH-SHARPIN_MLPH_chr2_238419744_ENST00000445024_SHARPIN_chr8_145154972_ENST00000398712_length(transcript)=1609nt_BP=657nt CAGTTGCCCGCCTGCCCCGGAGAGCCAGGCGCTAACCAGCCGCTCTGCGCCCCGCGCCCTGCTTGCCCCCATTATCCAGCCTTGCCCCGG CGCCCTGACCTGACGCCCTGGCCTGACGCCCTGCTTCGTCGCCTCCTTTCTCTCCCAGGTGCTGGACCAGGGACTGAGCGTCCCCCGGAG AGGGTCCGGTGTGACCCCGACAAGAAGCAGAAATGGGGAAGAAACTGGATCTTTCCAAGCTCACTGATGAAGAGGCCCAGCATGTCTTGG AAGTTGTTCAACGAGATTTTGACCTCCGAAGGAAAGAAGAGGAACGGCTAGAGGCGTTGAAGGGCAAGATTAAGAAGGAAAGCTCCAAGA GGGAGCTGCTTTCCGACACTGCCCATCTGAACGAGACCCACTGCGCCCGCTGCCTGCAGCCCTACCAGCTGCTTGTGAATAGCAAAAGGC AGTGCCTGGAATGTGGCCTCTTCACCTGCAAAAGCTGTGGCCGCGTCCACCCGGAGGAGCAGGGCTGGATCTGTGACCCCTGCCATCTGG CCAGAGTCGTGAAGATCGGCTCACTGGAGTGGTACTATGAGCATGTGAAAGCCCGCTTCAAGAGGTTCGGAAGTGCCAAGGTCATCCGGT CCCTCCACGGGCGGCTGCAGGGTGGAGGCAGCAAGAGCAACTCACCACCAGCCTTGGGCCCAGAAGCATGCCCTGTCTCCCTGCCCAGTC CCCCGGAAGCCTCCACACTCAAGGGCCCTCCACCTGAGGCAGATCTTCCTAGGAGCCCTGGAAACTTGACGGAGAGAGAAGAGCTGGCAG GGAGCCTGGCCCGGGCTATTGCAGGTGGAGACGAGAAGGGGGCAGCCCAAGTGGCAGCCGTCCTGGCCCAGCATCGTGTGGCCCTGAGTG TTCAGCTTCAGGAGGCCTGCTTCCCACCTGGCCCCATCAGGCTGCAGGTCACACTTGAAGACGCTGCCTCTGCCGCATCCGCCGCGTCCT CTGCACACGTTGCCCTGCAGGTCCACCCCCACTGCACTGTTGCAGCTCTCCAGGAGCAGGTGTTCTCAGAGCTCGGTTTCCCGCCAGCCG TGCAACGCTGGGTCATCGGACGGTGCCTGTGTGTGCCTGAGCGCAGCCTTGCCTCTTACGGGGTTCGGCAGGATGGGGACCCTGCTTTCC TCTACTTGCTGTCAGCTCCTCGAGAAGCCCCAGCCACAGGACCTAGCCCTCAGCACCCCCAGAAGATGGACGGGGAACTTGGACGCTTGT TTCCCCCATCATTGGGGCTACCCCCAGGCCCCCAGCCAGCTGCCTCCAGCCTGCCCAGTCCACTCCAGCCCAGCTGGTCCTGTCCTTCCT GCACCTTCATCAATGCCCCAGACCGCCCTGGCTGTGAGATGTGTAGCACCCAGAGGCCCTGCACTTGGGACCCCCTTGCTGCAGCTTCCA CCTAGCAGCCACCAGAGGTTACAAGGGGAGAGTGGCCCTTCCCTCACAAGTCCGACATCTCCAGGCCCCCACTGAACTCCGGGGACCTCT >54339_54339_5_MLPH-SHARPIN_MLPH_chr2_238419744_ENST00000445024_SHARPIN_chr8_145154972_ENST00000398712_length(amino acids)=410AA_BP=148 MGKKLDLSKLTDEEAQHVLEVVQRDFDLRRKEEERLEALKGKIKKESSKRELLSDTAHLNETHCARCLQPYQLLVNSKRQCLECGLFTCK SCGRVHPEEQGWICDPCHLARVVKIGSLEWYYEHVKARFKRFGSAKVIRSLHGRLQGGGSKSNSPPALGPEACPVSLPSPPEASTLKGPP PEADLPRSPGNLTEREELAGSLARAIAGGDEKGAAQVAAVLAQHRVALSVQLQEACFPPGPIRLQVTLEDAASAASAASSAHVALQVHPH CTVAALQEQVFSELGFPPAVQRWVIGRCLCVPERSLASYGVRQDGDPAFLYLLSAPREAPATGPSPQHPQKMDGELGRLFPPSLGLPPGP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MLPH-SHARPIN |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | SHARPIN | chr2:238419744 | chr8:145154972 | ENST00000398712 | 1 | 9 | 175_310 | 125.33333333333333 | 210.66666666666666 | SHANK1 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MLPH-SHARPIN |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MLPH-SHARPIN |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
