
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MND1-ARFIP1 (FusionGDB2 ID:54469) |
Fusion Gene Summary for MND1-ARFIP1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MND1-ARFIP1 | Fusion gene ID: 54469 | Hgene | Tgene | Gene symbol | MND1 | ARFIP1 | Gene ID | 84057 | 27236 |
| Gene name | meiotic nuclear divisions 1 | ADP ribosylation factor interacting protein 1 | |
| Synonyms | GAJ | HSU52521 | |
| Cytomap | 4q31.3 | 4q31.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | meiotic nuclear division protein 1 homologhomolog of yeast MND1meiotic nuclear divisions 1 homolog | arfaptin-1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9BWT6 | P53367 | |
| Ensembl transtripts involved in fusion gene | ENST00000240488, ENST00000504860, ENST00000503967, | ENST00000511289, ENST00000353617, ENST00000356064, ENST00000405727, ENST00000429148, ENST00000451320, | |
| Fusion gene scores | * DoF score | 3 X 3 X 2=18 | 10 X 10 X 11=1100 |
| # samples | 3 | 19 | |
| ** MAII score | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(19/1100*10)=-2.53343220008107 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MND1 [Title/Abstract] AND ARFIP1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MND1(154279774)-ARFIP1(153784758), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | ARFIP1 | GO:0006886 | intracellular protein transport | 12606037 |
| Tgene | ARFIP1 | GO:0050708 | regulation of protein secretion | 12606037 |
 Fusion gene breakpoints across MND1 (5'-gene) Fusion gene breakpoints across MND1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
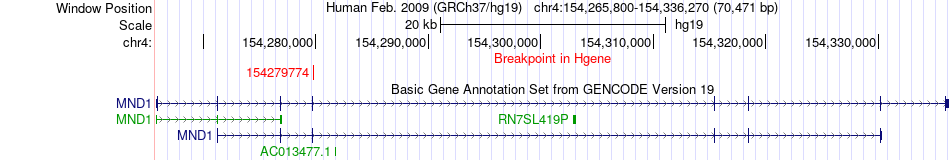 |
 Fusion gene breakpoints across ARFIP1 (3'-gene) Fusion gene breakpoints across ARFIP1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
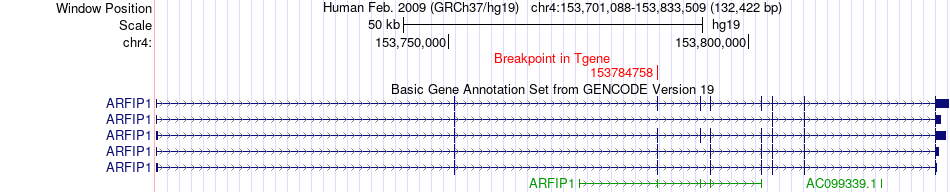 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-A7-A13D-01A | MND1 | chr4 | 154279774 | - | ARFIP1 | chr4 | 153784758 | + |
| ChimerDB4 | BRCA | TCGA-A7-A13D-01A | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + |
Top |
Fusion Gene ORF analysis for MND1-ARFIP1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000240488 | ENST00000511289 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + |
| 5CDS-3UTR | ENST00000504860 | ENST00000511289 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + |
| 5CDS-intron | ENST00000240488 | ENST00000353617 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + |
| 5CDS-intron | ENST00000240488 | ENST00000356064 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + |
| 5CDS-intron | ENST00000240488 | ENST00000405727 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + |
| 5CDS-intron | ENST00000240488 | ENST00000429148 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + |
| 5CDS-intron | ENST00000504860 | ENST00000353617 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + |
| 5CDS-intron | ENST00000504860 | ENST00000356064 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + |
| 5CDS-intron | ENST00000504860 | ENST00000405727 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + |
| 5CDS-intron | ENST00000504860 | ENST00000429148 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + |
| In-frame | ENST00000240488 | ENST00000451320 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + |
| In-frame | ENST00000504860 | ENST00000451320 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + |
| intron-3CDS | ENST00000503967 | ENST00000451320 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + |
| intron-3UTR | ENST00000503967 | ENST00000511289 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + |
| intron-intron | ENST00000503967 | ENST00000353617 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + |
| intron-intron | ENST00000503967 | ENST00000356064 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + |
| intron-intron | ENST00000503967 | ENST00000405727 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + |
| intron-intron | ENST00000503967 | ENST00000429148 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000240488 | MND1 | chr4 | 154279774 | + | ENST00000451320 | ARFIP1 | chr4 | 153784758 | + | 3532 | 365 | 89 | 1393 | 434 |
| ENST00000504860 | MND1 | chr4 | 154279774 | + | ENST00000451320 | ARFIP1 | chr4 | 153784758 | + | 3441 | 274 | 16 | 1302 | 428 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000240488 | ENST00000451320 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + | 0.000282229 | 0.9997178 |
| ENST00000504860 | ENST00000451320 | MND1 | chr4 | 154279774 | + | ARFIP1 | chr4 | 153784758 | + | 0.000162916 | 0.99983704 |
Top |
Fusion Genomic Features for MND1-ARFIP1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
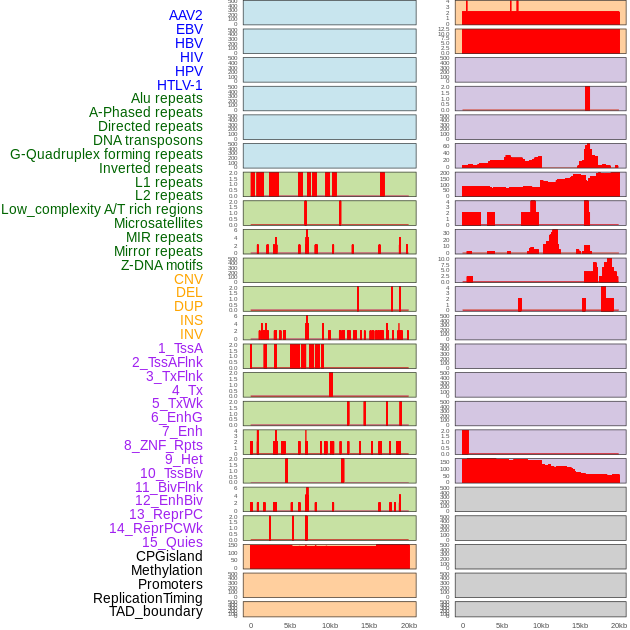 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
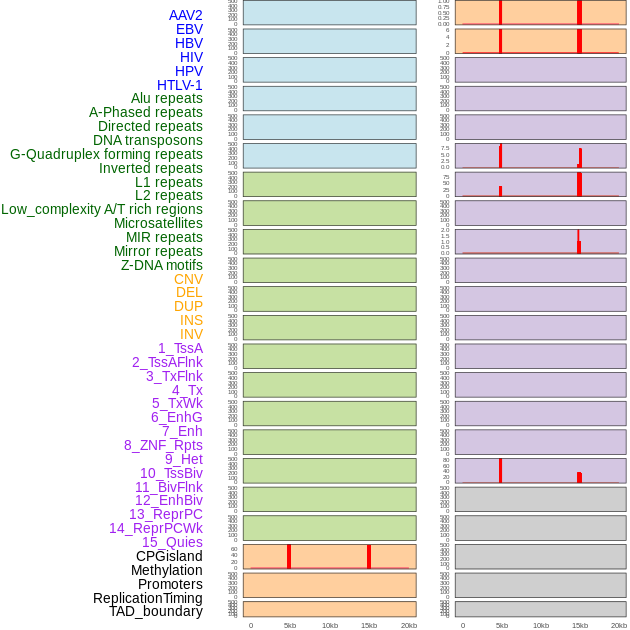 |
Top |
Fusion Protein Features for MND1-ARFIP1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr4:154279774/chr4:153784758) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MND1 | ARFIP1 |
| FUNCTION: Required for proper homologous chromosome pairing and efficient cross-over and intragenic recombination during meiosis (By similarity). Stimulates both DMC1- and RAD51-mediated homologous strand assimilation, which is required for the resolution of meiotic double-strand breaks. {ECO:0000250|UniProtKB:Q8K396, ECO:0000269|PubMed:16407260}. | FUNCTION: Plays a role in controlling biogenesis of secretory granules at the trans-Golgi network (PubMed:22981988). Mechanisitically, binds ARF-GTP at the neck of a growing secretory granule precursor and forms a protective scaffold (PubMed:9038142, PubMed:22981988). Once the granule precursor has been completely loaded, active PRKD1 phosphorylates ARFIP1 and releases it from ARFs (PubMed:22981988). In turn, ARFs induce fission (PubMed:22981988). Through this mechanism, ensures proper secretory granule formation at the Golgi of pancreatic beta cells (PubMed:22981988). {ECO:0000269|PubMed:22981988, ECO:0000269|PubMed:9038142}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ARFIP1 | chr4:154279774 | chr4:153784758 | ENST00000353617 | 1 | 9 | 153_353 | 31 | 374.0 | Domain | AH | |
| Tgene | ARFIP1 | chr4:154279774 | chr4:153784758 | ENST00000356064 | 1 | 8 | 153_353 | 31 | 342.0 | Domain | AH | |
| Tgene | ARFIP1 | chr4:154279774 | chr4:153784758 | ENST00000405727 | 1 | 8 | 153_353 | 31 | 342.0 | Domain | AH | |
| Tgene | ARFIP1 | chr4:154279774 | chr4:153784758 | ENST00000451320 | 1 | 9 | 153_353 | 31 | 374.0 | Domain | AH |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MND1 | chr4:154279774 | chr4:153784758 | ENST00000240488 | + | 4 | 8 | 84_173 | 92 | 206.0 | Coiled coil | Ontology_term=ECO:0000255 |
Top |
Fusion Gene Sequence for MND1-ARFIP1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >54469_54469_1_MND1-ARFIP1_MND1_chr4_154279774_ENST00000240488_ARFIP1_chr4_153784758_ENST00000451320_length(transcript)=3532nt_BP=365nt AGTCGGCGCCAAAATCAAACGCGTCCTGGCCTGTCCCGCCCCTCTCCCCAAGCGCGGGCCCGGCCAGCGGAAGCCCCTGCGCCCGCGCCA TGTCAAAGAAAAAAGGACTGAGTGCAGAAGAAAAGAGAACTCGCATGATGGAAATATTTTCTGAAACAAAAGATGTATTTCAATTAAAAG ACTTGGAGAAGATTGCTCCCAAAGAGAAAGGCATTACTGCTATGTCAGTAAAAGAAGTCCTTCAAAGCTTAGTTGATGATGGTATGGTTG ACTGTGAGAGGATCGGAACTTCTAATTATTATTGGGCTTTTCCAAGTAAAGCTCTTCATGCAAGGAAACATAAGTTGGAGGTTCTGGAAT CTCAGGATTTGAAGCATTCATTACCATCTGGACTTGGTCTCTCAGAAACCCAAATTACATCTCATGGCTTTGACAATACCAAAGAGGGTG TTATTGAAGCAGGAGCATTTCAAGGTTCCCCAGCACCGCCACTGCCATCTGTTATGTCTCCTAGCAGGGTTGCAGCTAGTCGACTGGCTC AGCAAGGAAGTGATTTAATTGTTCCTGCAGGTGGCCAGAGAACACAGACAAAAAGTGGACCAGTTATTCTAGCAGATGAAATTAAAAATC CTGCAATGGAAAAGTTAGAACTTGTTAGAAAATGGAGTCTAAACACCTATAAGTGTACTCGACAGATTATCTCTGAGAAGCTAGGCCGTG GCTCAAGAACTGTGGACCTTGAACTTGAAGCTCAGATTGATATATTAAGGGATAACAAGAAAAAATATGAAAATATTTTAAAACTGGCTC AAACATTGTCGACCCAGCTTTTCCAGATGGTACATACCCAAAGGCAACTTGGAGATGCATTTGCTGACCTGAGTTTGAAGTCACTAGAAC TTCATGAAGAATTTGGCTATAATGCCGATACCCAGAAACTGCTGGCTAAAAATGGAGAGACTCTTCTTGGGGCCATTAATTTTTTCATTG CTAGTGTGAACACTTTGGTGAATAAAACCATTGAAGATACATTAATGACTGTGAAACAGTATGAAAGTGCCAGGATTGAATATGATGCAT ATCGCACTGATTTGGAAGAACTGAATCTTGGACCACGTGACGCAAACACTCTGCCAAAGATTGAGCAGTCACAGCATCTCTTCCAAGCAC ATAAGGAAAAATATGATAAAATGCGCAATGATGTTTCTGTCAAATTGAAATTTCTAGAAGAAAATAAGGTTAAAGTATTGCACAATCAGC TGGTCCTTTTCCACAATGCCATTGCCGCTTACTTTGCTGGGAATCAGAAGCAGCTTGAACAGACACTTAAACAGTTCCATATCAAATTGA AAACCCCTGGAGTGGATGCCCCATCTTGGCTTGAAGAACAGTAAAATCACAGCGGAAAATAAAAAGAAAGTCGCGTTGTTATATTTCTAA ACCAACCTAACAAGAATTAAGCAGAGTTGGGGGAAGTGGGAGGGGTGACAAGCATTATAGTGATTCTTGCACAAACAGCTTTAATTTTTC CCTTTTTCATACTTTAACAATTGAACTGTTAAGGGTGGTTTTAATGTAAACATAGTTTCTATAATCTGAACACAATAATTTTCCTTTTTG AGAAATCCTTTTGTATTGTGAGGGTTGATGGCTATTTCTGTAGTTTGAAAACATCTAATAGAGATTTGCACTGACAAGATAAAAATTCAA GGTTTTTGGTCCTGGAAATGAGGGCCGATTGTAACTTTTCCAAAGGGAGGTTTACATGATTTGTATCAACATCAGATATTTTATATGTGA ATATATTAAACATCTTTAAGAGATTCATTTTCATGTATTAAAGAAAAAAGAAACTATTTTGCAGAGTAAAGTTATATGGTGTTCCTGCAG CGTCCACACAGAGGCAGCAGCTCTAATGAATGATTAGCTTGTGGAGTTGTGGCAAATACCTTTTTAAAAATGAATCTGTACCACTGTAAT TTTATTTAGACTTTTTTTTAAAGACACAGAAATCACACTGTCATTTCTGTGAGGCTTTTAAGCTTATGAATTTGCTTCACCCGAAGTCAT TGGACAGTTACATTTGCAATTTCTTTTGGTTCATAACAAAACCTGCTTAAGAGAATTTTTTTTTTTGACAGGTGGACGTGGGAATGGAAA TTCTTAATGGTTTCTGGAGTAGAAATCTGTACTTAGAAAACTAGTTTTTGGGTCAGTTCCATTTTGATAGAAGTGAATACATAGACAGCT TTTATTGACTTCCATATGTAACAATACCGTTTAAGCCTTAAATCACAGATATGTGCCTTCTCAGATACAGTAATTCTTGTTCAACCAATG TACAAAATCCATAAAGAAACCCCTGTTGGATAATGTTTGATGTGTCTATTCCTTCCATGGAATGGAGCACTATGTATGAATGTTGGGTTT CTTTGTACTTGTTAAGCCACATTTGAGGTTTATGGTAAAAATCATCTTTTGAGTTTGCTCTTTGGTTTTTCTTCATTCCTTTTGAGGATT GGGAAAACAGAAAGATTCTTTGATTTGGGTAATGAAGAGGTAATTTGGGACAGTGTGGTGGTACCAGGAAGAAAGAGGATTGGAAAGGCC AGTACTGTTTTAGTTGCTCGGCACTGTTGGTTTTGTTTTAATGTGGTTGCCCTGTCCACTACATGGTTCTATCAGTAGTGTAATCCATTT TCAATGTAAAGCTCTTTTAGTTTTTGTCATAGACATAAATTAATATTTTGAGAGGCATCCCTCACCTGTTCATTTCTTCTGTGTTGAAAT GAAGTACTTAAAATTACCGTTATACATGAACTTTGTGGACTGTAAGATTTGTTATATATGTTCAAATGCCTTTTAGCTGGCTTTTTAATT AATATGCCTGTTTTGAGTGCTTAATACAATGTAATGTGATTGTAAATCATACCTATTTTAAATCATTCCTTCCTGTATATTTGTACTCAG AGAGCCTTATTTTATTCTTCCAGCAGAATTACTACTTGTGATAGTTCATTTACATTCCCCAGAATGTGCCTCTAGTGTGAATTTTGTATT CTTATTAAACGTGTTTGCTGCAGTAAATCGTTTCTCTGGACCTTCATAGTTTAAGAAGTAACTATTTGGAAATTCTGAAGACACATAATT TGCGTTTTCTGAACCTTAAAAAAAAAGGTACTTCTGTCACTTATAATTGTTTTAGCTCAGTAAGCTATTTTTTTTCAATGTGAATCTCTT TTGAGAGCTGAACATGTCCATTCTTAACCACATTTCAATAACAGTTACTGTAGCTAAGGAGCTAAATAGTTTCATGGATTAATTTTTTTC AGAATAAATTTAAAATATCCATGATTCTGATAGTGGGCACATGACCAAAAGAAAAAAGTAAATCAATATATTTAGCCAATGGTACATTTA CTTTGTTAGGAATGTAATTTATGTTCATTATAGAAAAGAATTAGAAGAAAATTTCAAGTAATAGAAGATAAGTACCTTCCCAAGTGAATA >54469_54469_1_MND1-ARFIP1_MND1_chr4_154279774_ENST00000240488_ARFIP1_chr4_153784758_ENST00000451320_length(amino acids)=434AA_BP=92 MSKKKGLSAEEKRTRMMEIFSETKDVFQLKDLEKIAPKEKGITAMSVKEVLQSLVDDGMVDCERIGTSNYYWAFPSKALHARKHKLEVLE SQDLKHSLPSGLGLSETQITSHGFDNTKEGVIEAGAFQGSPAPPLPSVMSPSRVAASRLAQQGSDLIVPAGGQRTQTKSGPVILADEIKN PAMEKLELVRKWSLNTYKCTRQIISEKLGRGSRTVDLELEAQIDILRDNKKKYENILKLAQTLSTQLFQMVHTQRQLGDAFADLSLKSLE LHEEFGYNADTQKLLAKNGETLLGAINFFIASVNTLVNKTIEDTLMTVKQYESARIEYDAYRTDLEELNLGPRDANTLPKIEQSQHLFQA -------------------------------------------------------------- >54469_54469_2_MND1-ARFIP1_MND1_chr4_154279774_ENST00000504860_ARFIP1_chr4_153784758_ENST00000451320_length(transcript)=3441nt_BP=274nt GTCAAAGAAAAAAGGACTGAGTGCAGAAGAAAAGAGAACTCGCATGATGGAAATATTTTCTGAAACAAAAGATGTATTTCAATTAAAAGA CTTGGAGAAGATTGCTCCCAAAGAGAAAGGCATTACTGCTATGTCAGTAAAAGAAGTCCTTCAAAGCTTAGTTGATGATGGTATGGTTGA CTGTGAGAGGATCGGAACTTCTAATTATTATTGGGCTTTTCCAAGTAAAGCTCTTCATGCAAGGAAACATAAGTTGGAGGTTCTGGAATC TCAGGATTTGAAGCATTCATTACCATCTGGACTTGGTCTCTCAGAAACCCAAATTACATCTCATGGCTTTGACAATACCAAAGAGGGTGT TATTGAAGCAGGAGCATTTCAAGGTTCCCCAGCACCGCCACTGCCATCTGTTATGTCTCCTAGCAGGGTTGCAGCTAGTCGACTGGCTCA GCAAGGAAGTGATTTAATTGTTCCTGCAGGTGGCCAGAGAACACAGACAAAAAGTGGACCAGTTATTCTAGCAGATGAAATTAAAAATCC TGCAATGGAAAAGTTAGAACTTGTTAGAAAATGGAGTCTAAACACCTATAAGTGTACTCGACAGATTATCTCTGAGAAGCTAGGCCGTGG CTCAAGAACTGTGGACCTTGAACTTGAAGCTCAGATTGATATATTAAGGGATAACAAGAAAAAATATGAAAATATTTTAAAACTGGCTCA AACATTGTCGACCCAGCTTTTCCAGATGGTACATACCCAAAGGCAACTTGGAGATGCATTTGCTGACCTGAGTTTGAAGTCACTAGAACT TCATGAAGAATTTGGCTATAATGCCGATACCCAGAAACTGCTGGCTAAAAATGGAGAGACTCTTCTTGGGGCCATTAATTTTTTCATTGC TAGTGTGAACACTTTGGTGAATAAAACCATTGAAGATACATTAATGACTGTGAAACAGTATGAAAGTGCCAGGATTGAATATGATGCATA TCGCACTGATTTGGAAGAACTGAATCTTGGACCACGTGACGCAAACACTCTGCCAAAGATTGAGCAGTCACAGCATCTCTTCCAAGCACA TAAGGAAAAATATGATAAAATGCGCAATGATGTTTCTGTCAAATTGAAATTTCTAGAAGAAAATAAGGTTAAAGTATTGCACAATCAGCT GGTCCTTTTCCACAATGCCATTGCCGCTTACTTTGCTGGGAATCAGAAGCAGCTTGAACAGACACTTAAACAGTTCCATATCAAATTGAA AACCCCTGGAGTGGATGCCCCATCTTGGCTTGAAGAACAGTAAAATCACAGCGGAAAATAAAAAGAAAGTCGCGTTGTTATATTTCTAAA CCAACCTAACAAGAATTAAGCAGAGTTGGGGGAAGTGGGAGGGGTGACAAGCATTATAGTGATTCTTGCACAAACAGCTTTAATTTTTCC CTTTTTCATACTTTAACAATTGAACTGTTAAGGGTGGTTTTAATGTAAACATAGTTTCTATAATCTGAACACAATAATTTTCCTTTTTGA GAAATCCTTTTGTATTGTGAGGGTTGATGGCTATTTCTGTAGTTTGAAAACATCTAATAGAGATTTGCACTGACAAGATAAAAATTCAAG GTTTTTGGTCCTGGAAATGAGGGCCGATTGTAACTTTTCCAAAGGGAGGTTTACATGATTTGTATCAACATCAGATATTTTATATGTGAA TATATTAAACATCTTTAAGAGATTCATTTTCATGTATTAAAGAAAAAAGAAACTATTTTGCAGAGTAAAGTTATATGGTGTTCCTGCAGC GTCCACACAGAGGCAGCAGCTCTAATGAATGATTAGCTTGTGGAGTTGTGGCAAATACCTTTTTAAAAATGAATCTGTACCACTGTAATT TTATTTAGACTTTTTTTTAAAGACACAGAAATCACACTGTCATTTCTGTGAGGCTTTTAAGCTTATGAATTTGCTTCACCCGAAGTCATT GGACAGTTACATTTGCAATTTCTTTTGGTTCATAACAAAACCTGCTTAAGAGAATTTTTTTTTTTGACAGGTGGACGTGGGAATGGAAAT TCTTAATGGTTTCTGGAGTAGAAATCTGTACTTAGAAAACTAGTTTTTGGGTCAGTTCCATTTTGATAGAAGTGAATACATAGACAGCTT TTATTGACTTCCATATGTAACAATACCGTTTAAGCCTTAAATCACAGATATGTGCCTTCTCAGATACAGTAATTCTTGTTCAACCAATGT ACAAAATCCATAAAGAAACCCCTGTTGGATAATGTTTGATGTGTCTATTCCTTCCATGGAATGGAGCACTATGTATGAATGTTGGGTTTC TTTGTACTTGTTAAGCCACATTTGAGGTTTATGGTAAAAATCATCTTTTGAGTTTGCTCTTTGGTTTTTCTTCATTCCTTTTGAGGATTG GGAAAACAGAAAGATTCTTTGATTTGGGTAATGAAGAGGTAATTTGGGACAGTGTGGTGGTACCAGGAAGAAAGAGGATTGGAAAGGCCA GTACTGTTTTAGTTGCTCGGCACTGTTGGTTTTGTTTTAATGTGGTTGCCCTGTCCACTACATGGTTCTATCAGTAGTGTAATCCATTTT CAATGTAAAGCTCTTTTAGTTTTTGTCATAGACATAAATTAATATTTTGAGAGGCATCCCTCACCTGTTCATTTCTTCTGTGTTGAAATG AAGTACTTAAAATTACCGTTATACATGAACTTTGTGGACTGTAAGATTTGTTATATATGTTCAAATGCCTTTTAGCTGGCTTTTTAATTA ATATGCCTGTTTTGAGTGCTTAATACAATGTAATGTGATTGTAAATCATACCTATTTTAAATCATTCCTTCCTGTATATTTGTACTCAGA GAGCCTTATTTTATTCTTCCAGCAGAATTACTACTTGTGATAGTTCATTTACATTCCCCAGAATGTGCCTCTAGTGTGAATTTTGTATTC TTATTAAACGTGTTTGCTGCAGTAAATCGTTTCTCTGGACCTTCATAGTTTAAGAAGTAACTATTTGGAAATTCTGAAGACACATAATTT GCGTTTTCTGAACCTTAAAAAAAAAGGTACTTCTGTCACTTATAATTGTTTTAGCTCAGTAAGCTATTTTTTTTCAATGTGAATCTCTTT TGAGAGCTGAACATGTCCATTCTTAACCACATTTCAATAACAGTTACTGTAGCTAAGGAGCTAAATAGTTTCATGGATTAATTTTTTTCA GAATAAATTTAAAATATCCATGATTCTGATAGTGGGCACATGACCAAAAGAAAAAAGTAAATCAATATATTTAGCCAATGGTACATTTAC TTTGTTAGGAATGTAATTTATGTTCATTATAGAAAAGAATTAGAAGAAAATTTCAAGTAATAGAAGATAAGTACCTTCCCAAGTGAATAA >54469_54469_2_MND1-ARFIP1_MND1_chr4_154279774_ENST00000504860_ARFIP1_chr4_153784758_ENST00000451320_length(amino acids)=428AA_BP=86 MSAEEKRTRMMEIFSETKDVFQLKDLEKIAPKEKGITAMSVKEVLQSLVDDGMVDCERIGTSNYYWAFPSKALHARKHKLEVLESQDLKH SLPSGLGLSETQITSHGFDNTKEGVIEAGAFQGSPAPPLPSVMSPSRVAASRLAQQGSDLIVPAGGQRTQTKSGPVILADEIKNPAMEKL ELVRKWSLNTYKCTRQIISEKLGRGSRTVDLELEAQIDILRDNKKKYENILKLAQTLSTQLFQMVHTQRQLGDAFADLSLKSLELHEEFG YNADTQKLLAKNGETLLGAINFFIASVNTLVNKTIEDTLMTVKQYESARIEYDAYRTDLEELNLGPRDANTLPKIEQSQHLFQAHKEKYD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MND1-ARFIP1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MND1-ARFIP1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MND1-ARFIP1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
