
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MORF4L1-PSMA4 (FusionGDB2 ID:54605) |
Fusion Gene Summary for MORF4L1-PSMA4 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MORF4L1-PSMA4 | Fusion gene ID: 54605 | Hgene | Tgene | Gene symbol | MORF4L1 | PSMA4 | Gene ID | 10933 | 5685 |
| Gene name | mortality factor 4 like 1 | proteasome 20S subunit alpha 4 | |
| Synonyms | Eaf3|FWP006|HsT17725|MEAF3|MORFRG15|MRG15|S863-6 | HC9|HsT17706|PSC9 | |
| Cytomap | 15q25.1 | 15q25.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | mortality factor 4-like protein 1Esa1p-associated factor 3 homologMORF-related gene 15 proteinMORF-related gene on chromosome 15protein MSL3-1transcription factor-like protein MRG15 | proteasome subunit alpha type-4macropain subunit C9multicatalytic endopeptidase complex subunit C9proteasome (prosome, macropain) subunit, alpha type, 4proteasome component C9proteasome subunit HC9proteasome subunit Lproteasome subunit alpha 4prot | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | Q9UBU8 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000331268, ENST00000379535, ENST00000426013, ENST00000558502, ENST00000558746, ENST00000559345, ENST00000561171, | ENST00000557929, ENST00000558094, ENST00000558281, ENST00000558341, ENST00000044462, ENST00000560217, ENST00000413382, ENST00000559082, | |
| Fusion gene scores | * DoF score | 9 X 8 X 4=288 | 11 X 11 X 5=605 |
| # samples | 11 | 11 | |
| ** MAII score | log2(11/288*10)=-1.38856528791765 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(11/605*10)=-2.4594316186373 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MORF4L1 [Title/Abstract] AND PSMA4 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MORF4L1(79186572)-PSMA4(78841131), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | MORF4L1-PSMA4 seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. MORF4L1-PSMA4 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. MORF4L1-PSMA4 seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. MORF4L1-PSMA4 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | MORF4L1 | GO:0000724 | double-strand break repair via homologous recombination | 20332121 |
| Hgene | MORF4L1 | GO:0016575 | histone deacetylation | 14966270 |
| Hgene | MORF4L1 | GO:0043967 | histone H4 acetylation | 14966270 |
| Hgene | MORF4L1 | GO:0043968 | histone H2A acetylation | 14966270 |
 Fusion gene breakpoints across MORF4L1 (5'-gene) Fusion gene breakpoints across MORF4L1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
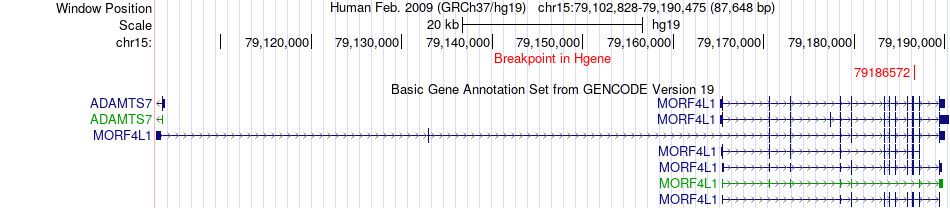 |
 Fusion gene breakpoints across PSMA4 (3'-gene) Fusion gene breakpoints across PSMA4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
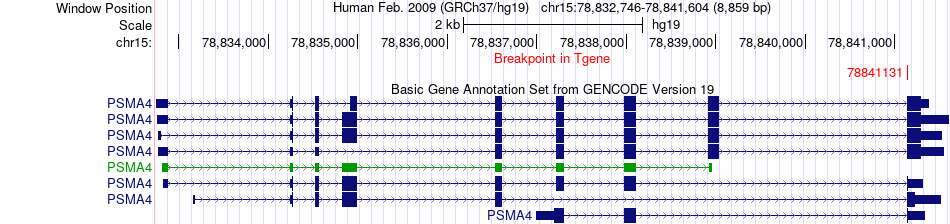 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-24-1562 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
Top |
Fusion Gene ORF analysis for MORF4L1-PSMA4 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000331268 | ENST00000557929 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000331268 | ENST00000558094 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000331268 | ENST00000558281 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000331268 | ENST00000558341 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000379535 | ENST00000557929 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000379535 | ENST00000558094 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000379535 | ENST00000558281 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000379535 | ENST00000558341 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000426013 | ENST00000557929 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000426013 | ENST00000558094 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000426013 | ENST00000558281 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000426013 | ENST00000558341 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000558502 | ENST00000557929 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000558502 | ENST00000558094 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000558502 | ENST00000558281 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000558502 | ENST00000558341 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000558746 | ENST00000557929 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000558746 | ENST00000558094 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000558746 | ENST00000558281 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000558746 | ENST00000558341 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000559345 | ENST00000557929 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000559345 | ENST00000558094 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000559345 | ENST00000558281 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| 5CDS-intron | ENST00000559345 | ENST00000558341 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| Frame-shift | ENST00000331268 | ENST00000044462 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| Frame-shift | ENST00000331268 | ENST00000560217 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| Frame-shift | ENST00000379535 | ENST00000044462 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| Frame-shift | ENST00000379535 | ENST00000560217 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| Frame-shift | ENST00000426013 | ENST00000044462 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| Frame-shift | ENST00000426013 | ENST00000560217 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| Frame-shift | ENST00000558502 | ENST00000044462 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| Frame-shift | ENST00000558502 | ENST00000560217 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| Frame-shift | ENST00000558746 | ENST00000044462 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| Frame-shift | ENST00000558746 | ENST00000560217 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| Frame-shift | ENST00000559345 | ENST00000044462 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| Frame-shift | ENST00000559345 | ENST00000560217 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| In-frame | ENST00000331268 | ENST00000413382 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| In-frame | ENST00000331268 | ENST00000559082 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| In-frame | ENST00000379535 | ENST00000413382 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| In-frame | ENST00000379535 | ENST00000559082 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| In-frame | ENST00000426013 | ENST00000413382 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| In-frame | ENST00000426013 | ENST00000559082 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| In-frame | ENST00000558502 | ENST00000413382 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| In-frame | ENST00000558502 | ENST00000559082 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| In-frame | ENST00000558746 | ENST00000413382 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| In-frame | ENST00000558746 | ENST00000559082 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| In-frame | ENST00000559345 | ENST00000413382 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| In-frame | ENST00000559345 | ENST00000559082 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| intron-3CDS | ENST00000561171 | ENST00000044462 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| intron-3CDS | ENST00000561171 | ENST00000413382 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| intron-3CDS | ENST00000561171 | ENST00000559082 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| intron-3CDS | ENST00000561171 | ENST00000560217 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| intron-intron | ENST00000561171 | ENST00000557929 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| intron-intron | ENST00000561171 | ENST00000558094 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| intron-intron | ENST00000561171 | ENST00000558281 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
| intron-intron | ENST00000561171 | ENST00000558341 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000379535 | MORF4L1 | chr15 | 79186572 | + | ENST00000559082 | PSMA4 | chr15 | 78841131 | + | 1838 | 1441 | 411 | 1595 | 394 |
| ENST00000379535 | MORF4L1 | chr15 | 79186572 | + | ENST00000413382 | PSMA4 | chr15 | 78841131 | + | 1856 | 1441 | 411 | 1595 | 394 |
| ENST00000426013 | MORF4L1 | chr15 | 79186572 | + | ENST00000559082 | PSMA4 | chr15 | 78841131 | + | 1441 | 1044 | 242 | 1198 | 318 |
| ENST00000426013 | MORF4L1 | chr15 | 79186572 | + | ENST00000413382 | PSMA4 | chr15 | 78841131 | + | 1459 | 1044 | 242 | 1198 | 318 |
| ENST00000331268 | MORF4L1 | chr15 | 79186572 | + | ENST00000559082 | PSMA4 | chr15 | 78841131 | + | 1520 | 1123 | 204 | 1277 | 357 |
| ENST00000331268 | MORF4L1 | chr15 | 79186572 | + | ENST00000413382 | PSMA4 | chr15 | 78841131 | + | 1538 | 1123 | 204 | 1277 | 357 |
| ENST00000558746 | MORF4L1 | chr15 | 79186572 | + | ENST00000559082 | PSMA4 | chr15 | 78841131 | + | 1182 | 785 | 64 | 939 | 291 |
| ENST00000558746 | MORF4L1 | chr15 | 79186572 | + | ENST00000413382 | PSMA4 | chr15 | 78841131 | + | 1200 | 785 | 64 | 939 | 291 |
| ENST00000559345 | MORF4L1 | chr15 | 79186572 | + | ENST00000559082 | PSMA4 | chr15 | 78841131 | + | 1373 | 976 | 225 | 1130 | 301 |
| ENST00000559345 | MORF4L1 | chr15 | 79186572 | + | ENST00000413382 | PSMA4 | chr15 | 78841131 | + | 1391 | 976 | 225 | 1130 | 301 |
| ENST00000558502 | MORF4L1 | chr15 | 79186572 | + | ENST00000559082 | PSMA4 | chr15 | 78841131 | + | 1072 | 675 | 80 | 829 | 249 |
| ENST00000558502 | MORF4L1 | chr15 | 79186572 | + | ENST00000413382 | PSMA4 | chr15 | 78841131 | + | 1090 | 675 | 80 | 829 | 249 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000379535 | ENST00000559082 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + | 0.002340482 | 0.99765956 |
| ENST00000379535 | ENST00000413382 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + | 0.002403392 | 0.99759656 |
| ENST00000426013 | ENST00000559082 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + | 0.000295169 | 0.9997048 |
| ENST00000426013 | ENST00000413382 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + | 0.000299596 | 0.9997004 |
| ENST00000331268 | ENST00000559082 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + | 0.000635608 | 0.99936444 |
| ENST00000331268 | ENST00000413382 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + | 0.000644455 | 0.9993555 |
| ENST00000558746 | ENST00000559082 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + | 0.001212594 | 0.99878746 |
| ENST00000558746 | ENST00000413382 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + | 0.00128777 | 0.9987122 |
| ENST00000559345 | ENST00000559082 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + | 0.001173123 | 0.9988269 |
| ENST00000559345 | ENST00000413382 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + | 0.001220037 | 0.99877995 |
| ENST00000558502 | ENST00000559082 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + | 0.001614159 | 0.9983859 |
| ENST00000558502 | ENST00000413382 | MORF4L1 | chr15 | 79186572 | + | PSMA4 | chr15 | 78841131 | + | 0.001743921 | 0.998256 |
Top |
Fusion Genomic Features for MORF4L1-PSMA4 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
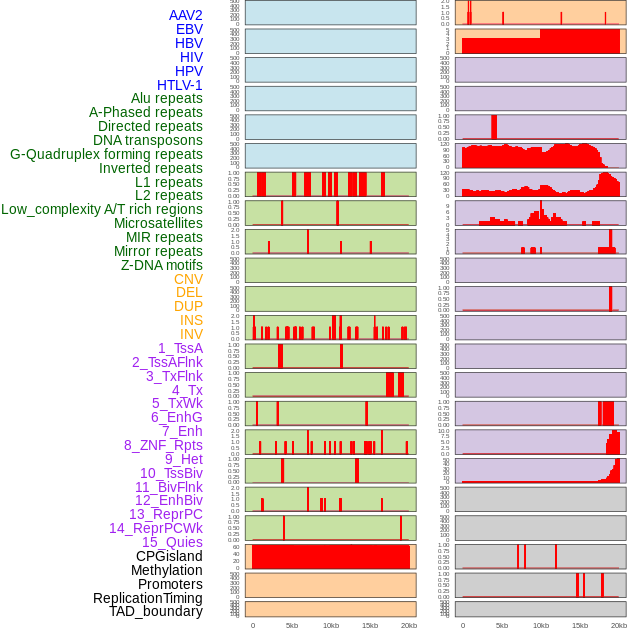 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
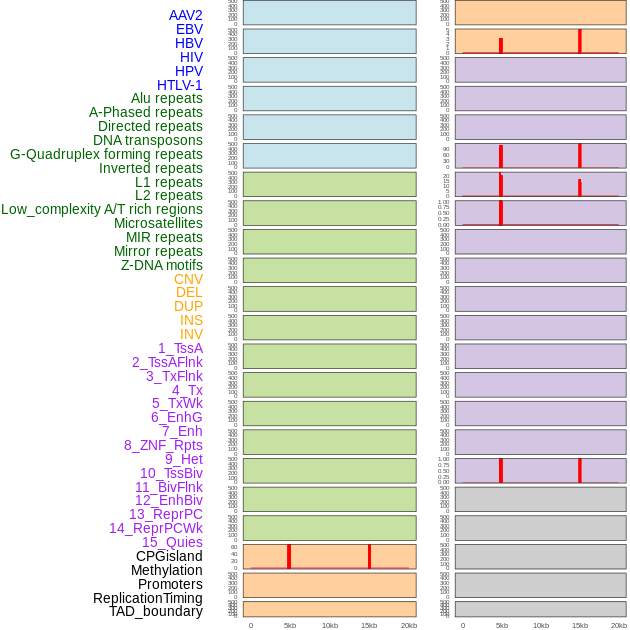 |
Top |
Fusion Protein Features for MORF4L1-PSMA4 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr15:79186572/chr15:78841131) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MORF4L1 | . |
| FUNCTION: Component of the NuA4 histone acetyltransferase (HAT) complex which is involved in transcriptional activation of select genes principally by acetylation of nucleosomal histones H4 and H2A. This modification may both alter nucleosome - DNA interactions and promote interaction of the modified histones with other proteins which positively regulate transcription. This complex may be required for the activation of transcriptional programs associated with oncogene and proto-oncogene mediated growth induction, tumor suppressor mediated growth arrest and replicative senescence, apoptosis, and DNA repair. The NuA4 complex ATPase and helicase activities seem to be, at least in part, contributed by the association of RUVBL1 and RUVBL2 with EP400. NuA4 may also play a direct role in DNA repair when directly recruited to sites of DNA damage. Also component of the mSin3A complex which acts to repress transcription by deacetylation of nucleosomal histones. Required for homologous recombination repair (HRR) and resistance to mitomycin C (MMC). Involved in the localization of PALB2, BRCA2 and RAD51, but not BRCA1, to DNA-damage foci. {ECO:0000269|PubMed:14966270, ECO:0000269|PubMed:20332121}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000331268 | + | 11 | 13 | 135_146 | 306 | 363.0 | Motif | Nuclear localization signal |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000426013 | + | 10 | 12 | 135_146 | 267 | 324.0 | Motif | Nuclear localization signal |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000558502 | + | 8 | 10 | 135_146 | 179 | 236.0 | Motif | Nuclear localization signal |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000559345 | + | 10 | 12 | 135_146 | 179 | 236.0 | Motif | Nuclear localization signal |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000331268 | + | 11 | 13 | 191_362 | 306 | 363.0 | Domain | MRG |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000426013 | + | 10 | 12 | 191_362 | 267 | 324.0 | Domain | MRG |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000558502 | + | 8 | 10 | 191_362 | 179 | 236.0 | Domain | MRG |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000559345 | + | 10 | 12 | 191_362 | 179 | 236.0 | Domain | MRG |
Top |
Fusion Gene Sequence for MORF4L1-PSMA4 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >54605_54605_1_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000331268_PSMA4_chr15_78841131_ENST00000413382_length(transcript)=1538nt_BP=1123nt GCGTGCGCGGCGGGCTGTCGTTGGCTGGAGCAGCGGCTGCGCGGGTCGCGGTGCTGTGAGGTCTGCGGGCGCTGGCAAATCCGGCCCAGG ATGTAGAGCTGGCAGTGCCTGACGGCGCGTCTGACGCGGAGTTGGGTGGGGTAGAGAGTAGGGGGCGGTAGTCGGGGGTGGTGGGAGAAG GAGGAGGCGGCGAATCACTTATAAATGGCGCCGAAGCAGGACCCGAAGCCTAAATTCCAGGAGGGTGAGCGAGTGCTGTGCTTTCATGGG CCTCTTCTTTATGAAGCAAAGTGTGTAAAGGTTGCCATAAAGGACAAACAAGTGAAATACTTCATACATTACAGTGGTTGGAATAAAAAA AGTGCTGTGAGGCCCAGGCGCTCTGAAAAATCTTTGAAGACACATGAGGATATTGTAGCCCTTTTTCCTGTTCCTGAAGGAGCTCCCTCA GTACACCACCCCCTCCTGACCTCTAGTTGGGATGAATGGGTTCCGGAGAGCAGAGTACTCAAATACGTGGACACCAATTTGCAGAAACAG CGAGAACTTCAAAAAGCCAATCAGGAGCAGTATGCAGAGGGGAAGATGAGAGGGGCTGCCCCAGGAAAGAAGACATCTGGTCTGCAACAG AAAAATGTTGAAGTGAAAACGAAAAAGAACAAACAGAAAACACCTGGAAATGGAGATGGTGGCAGTACCAGTGAGACCCCTCAGCCTCCT CGGAAGAAAAGGGCCCGGGTAGATCCTACTGTTGAAAATGAGGAAACATTCATGAACAGAGTTGAAGTTAAAGTAAAGATTCCTGAAGAG CTAAAACCGTGGCTTGTTGATGACTGGGACTTAATTACCAGGCAAAAACAGCTCTTTTATCTTCCTGCCAAGAAGAATGTGGATTCCATT CTTGAGGATTATGCAAATTACAAGAAATCTCGTGGAAACACAGATAATAAGGAGTATGCGGTTAATGAAGTTGTGGCAGGGATAAAAGAA TACTTCAACGTAATGTTGGGTACCCAGCTACTCTATAAATTTGAGAGACCACAGTATGCTGAAATTCTTGCAGATCATCCCGATGCACCC ATGTCCCAGGTGTATGGAGCGCCACATCTCCTGAGATTATTTGTGGAAATTGCAACACTAACAAGAGAGAATGGAAAGACAGTAATCAGA GTTCTCAAACAAAAAGAAGTGGAGCAGTTGATCAAAAAACATGAGGAAGAAGAAGCCAAAGCTGAGCGTGAGAAGAAAGAAAAAGAACAG AAAGAAAAGGATAAATAGAATCAGAGATTTTATTACTCATTTGGGGCACCATTTCAGTGTAAAAGCAGTCCTACTCTTCCACACTAGGAA GGCTTTACTTTTTTTAACTGGTGCAGTGGGAAAATAGGACATTACATACTGAATTGGGTCCTTGTCATTTCTGTCCAATTGAATACTTTA TTGTAACGATGATGGTTACCCTTCATGGACGTCTTAATCTTCCACACACATCCCCTTTTTTTGGAATAAAATTTGGAAAATGGAAATGAA >54605_54605_1_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000331268_PSMA4_chr15_78841131_ENST00000413382_length(amino acids)=357AA_BP=301 MAPKQDPKPKFQEGERVLCFHGPLLYEAKCVKVAIKDKQVKYFIHYSGWNKKSAVRPRRSEKSLKTHEDIVALFPVPEGAPSVHHPLLTS SWDEWVPESRVLKYVDTNLQKQRELQKANQEQYAEGKMRGAAPGKKTSGLQQKNVEVKTKKNKQKTPGNGDGGSTSETPQPPRKKRARVD PTVENEETFMNRVEVKVKIPEELKPWLVDDWDLITRQKQLFYLPAKKNVDSILEDYANYKKSRGNTDNKEYAVNEVVAGIKEYFNVMLGT -------------------------------------------------------------- >54605_54605_2_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000331268_PSMA4_chr15_78841131_ENST00000559082_length(transcript)=1520nt_BP=1123nt GCGTGCGCGGCGGGCTGTCGTTGGCTGGAGCAGCGGCTGCGCGGGTCGCGGTGCTGTGAGGTCTGCGGGCGCTGGCAAATCCGGCCCAGG ATGTAGAGCTGGCAGTGCCTGACGGCGCGTCTGACGCGGAGTTGGGTGGGGTAGAGAGTAGGGGGCGGTAGTCGGGGGTGGTGGGAGAAG GAGGAGGCGGCGAATCACTTATAAATGGCGCCGAAGCAGGACCCGAAGCCTAAATTCCAGGAGGGTGAGCGAGTGCTGTGCTTTCATGGG CCTCTTCTTTATGAAGCAAAGTGTGTAAAGGTTGCCATAAAGGACAAACAAGTGAAATACTTCATACATTACAGTGGTTGGAATAAAAAA AGTGCTGTGAGGCCCAGGCGCTCTGAAAAATCTTTGAAGACACATGAGGATATTGTAGCCCTTTTTCCTGTTCCTGAAGGAGCTCCCTCA GTACACCACCCCCTCCTGACCTCTAGTTGGGATGAATGGGTTCCGGAGAGCAGAGTACTCAAATACGTGGACACCAATTTGCAGAAACAG CGAGAACTTCAAAAAGCCAATCAGGAGCAGTATGCAGAGGGGAAGATGAGAGGGGCTGCCCCAGGAAAGAAGACATCTGGTCTGCAACAG AAAAATGTTGAAGTGAAAACGAAAAAGAACAAACAGAAAACACCTGGAAATGGAGATGGTGGCAGTACCAGTGAGACCCCTCAGCCTCCT CGGAAGAAAAGGGCCCGGGTAGATCCTACTGTTGAAAATGAGGAAACATTCATGAACAGAGTTGAAGTTAAAGTAAAGATTCCTGAAGAG CTAAAACCGTGGCTTGTTGATGACTGGGACTTAATTACCAGGCAAAAACAGCTCTTTTATCTTCCTGCCAAGAAGAATGTGGATTCCATT CTTGAGGATTATGCAAATTACAAGAAATCTCGTGGAAACACAGATAATAAGGAGTATGCGGTTAATGAAGTTGTGGCAGGGATAAAAGAA TACTTCAACGTAATGTTGGGTACCCAGCTACTCTATAAATTTGAGAGACCACAGTATGCTGAAATTCTTGCAGATCATCCCGATGCACCC ATGTCCCAGGTGTATGGAGCGCCACATCTCCTGAGATTATTTGTGGAAATTGCAACACTAACAAGAGAGAATGGAAAGACAGTAATCAGA GTTCTCAAACAAAAAGAAGTGGAGCAGTTGATCAAAAAACATGAGGAAGAAGAAGCCAAAGCTGAGCGTGAGAAGAAAGAAAAAGAACAG AAAGAAAAGGATAAATAGAATCAGAGATTTTATTACTCATTTGGGGCACCATTTCAGTGTAAAAGCAGTCCTACTCTTCCACACTAGGAA GGCTTTACTTTTTTTAACTGGTGCAGTGGGAAAATAGGACATTACATACTGAATTGGGTCCTTGTCATTTCTGTCCAATTGAATACTTTA >54605_54605_2_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000331268_PSMA4_chr15_78841131_ENST00000559082_length(amino acids)=357AA_BP=301 MAPKQDPKPKFQEGERVLCFHGPLLYEAKCVKVAIKDKQVKYFIHYSGWNKKSAVRPRRSEKSLKTHEDIVALFPVPEGAPSVHHPLLTS SWDEWVPESRVLKYVDTNLQKQRELQKANQEQYAEGKMRGAAPGKKTSGLQQKNVEVKTKKNKQKTPGNGDGGSTSETPQPPRKKRARVD PTVENEETFMNRVEVKVKIPEELKPWLVDDWDLITRQKQLFYLPAKKNVDSILEDYANYKKSRGNTDNKEYAVNEVVAGIKEYFNVMLGT -------------------------------------------------------------- >54605_54605_3_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000379535_PSMA4_chr15_78841131_ENST00000413382_length(transcript)=1856nt_BP=1441nt AACCCATGGCCGGGAAAAGAAGCTCCTGCTCCCCGCTCCACTCCCGGCGTCTTCACACTTGGGGAAGGGAGAGGCAGGAAGTGCAAGGAC CGCGCAAGACCCGGCCCCCCGAGCCGCCAGGGGCCGCGCCAGCAGCCCAGATGCGGTGGGCGGGAGCTGGGGGGGCGGCGGGGTCCCGCG AAGACCTGCGACCTGGCGCCCTCGCCGGCTGTGTCCCGGCCAGCCCGCTTTCCAGGCGAGCCTCGCTAAGGAGTGTGAGAAACCAAATCC TCAGTGGGTAAACCGCGCTCCGTCCAGCCCACTCGGACGAGAGCTCTCCCGGGAGCCAGGCGCGTTGCTGCACCCGAGGTGGGGAAACCG GAGCCCGAAGAGCGGAAGGGGCTTATTCCAGGCGACAGGCAGTGTCAGTAACTGAATCAGGGGCCGAGGAAGTTTCCCCAGAAGCGCGGC TTCGCTCCCGGCCCGGCCCGACCCCGACTACTGTCCCCTAATACCCAGGGAGCGGAAGACGCGACCAACTCCAGGCACAGCGGAGCCGGC GCGGGGTCCACAAAGCAAAAAAGAATGATCCCCTTCCCCCATCCTCAAGTGACTGATGCTCTTGGAAGATACTCCAACTATATCCTGAGT AGCACTGTGGAGAGCCCCAGTCTGAGAAGCATGAACTTTGAGAAGCAAGGTGAGCGAGTGCTGTGCTTTCATGGGCCTCTTCTTTATGAA GCAAAGTGTGTAAAGGTTGCCATAAAGGACAAACAAGTGAAATACTTCATACATTACAGTGGTTGGAATAAAAATTGGGATGAATGGGTT CCGGAGAGCAGAGTACTCAAATACGTGGACACCAATTTGCAGAAACAGCGAGAACTTCAAAAAGCCAATCAGGAGCAGTATGCAGAGGGG AAGATGAGAGGGGCTGCCCCAGGAAAGAAGACATCTGGTCTGCAACAGAAAAATGTTGAAGTGAAAACGAAAAAGAACAAACAGAAAACA CCTGGAAATGGAGATGGTGGCAGTACCAGTGAGACCCCTCAGCCTCCTCGGAAGAAAAGGGCCCGGGTAGATCCTACTGTTGAAAATGAG GAAACATTCATGAACAGAGTTGAAGTTAAAGTAAAGATTCCTGAAGAGCTAAAACCGTGGCTTGTTGATGACTGGGACTTAATTACCAGG CAAAAACAGCTCTTTTATCTTCCTGCCAAGAAGAATGTGGATTCCATTCTTGAGGATTATGCAAATTACAAGAAATCTCGTGGAAACACA GATAATAAGGAGTATGCGGTTAATGAAGTTGTGGCAGGGATAAAAGAATACTTCAACGTAATGTTGGGTACCCAGCTACTCTATAAATTT GAGAGACCACAGTATGCTGAAATTCTTGCAGATCATCCCGATGCACCCATGTCCCAGGTGTATGGAGCGCCACATCTCCTGAGATTATTT GTGGAAATTGCAACACTAACAAGAGAGAATGGAAAGACAGTAATCAGAGTTCTCAAACAAAAAGAAGTGGAGCAGTTGATCAAAAAACAT GAGGAAGAAGAAGCCAAAGCTGAGCGTGAGAAGAAAGAAAAAGAACAGAAAGAAAAGGATAAATAGAATCAGAGATTTTATTACTCATTT GGGGCACCATTTCAGTGTAAAAGCAGTCCTACTCTTCCACACTAGGAAGGCTTTACTTTTTTTAACTGGTGCAGTGGGAAAATAGGACAT TACATACTGAATTGGGTCCTTGTCATTTCTGTCCAATTGAATACTTTATTGTAACGATGATGGTTACCCTTCATGGACGTCTTAATCTTC >54605_54605_3_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000379535_PSMA4_chr15_78841131_ENST00000413382_length(amino acids)=394AA_BP=338 MNQGPRKFPQKRGFAPGPARPRLLSPNTQGAEDATNSRHSGAGAGSTKQKRMIPFPHPQVTDALGRYSNYILSSTVESPSLRSMNFEKQG ERVLCFHGPLLYEAKCVKVAIKDKQVKYFIHYSGWNKNWDEWVPESRVLKYVDTNLQKQRELQKANQEQYAEGKMRGAAPGKKTSGLQQK NVEVKTKKNKQKTPGNGDGGSTSETPQPPRKKRARVDPTVENEETFMNRVEVKVKIPEELKPWLVDDWDLITRQKQLFYLPAKKNVDSIL EDYANYKKSRGNTDNKEYAVNEVVAGIKEYFNVMLGTQLLYKFERPQYAEILADHPDAPMSQVYGAPHLLRLFVEIATLTRENGKTVIRV -------------------------------------------------------------- >54605_54605_4_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000379535_PSMA4_chr15_78841131_ENST00000559082_length(transcript)=1838nt_BP=1441nt AACCCATGGCCGGGAAAAGAAGCTCCTGCTCCCCGCTCCACTCCCGGCGTCTTCACACTTGGGGAAGGGAGAGGCAGGAAGTGCAAGGAC CGCGCAAGACCCGGCCCCCCGAGCCGCCAGGGGCCGCGCCAGCAGCCCAGATGCGGTGGGCGGGAGCTGGGGGGGCGGCGGGGTCCCGCG AAGACCTGCGACCTGGCGCCCTCGCCGGCTGTGTCCCGGCCAGCCCGCTTTCCAGGCGAGCCTCGCTAAGGAGTGTGAGAAACCAAATCC TCAGTGGGTAAACCGCGCTCCGTCCAGCCCACTCGGACGAGAGCTCTCCCGGGAGCCAGGCGCGTTGCTGCACCCGAGGTGGGGAAACCG GAGCCCGAAGAGCGGAAGGGGCTTATTCCAGGCGACAGGCAGTGTCAGTAACTGAATCAGGGGCCGAGGAAGTTTCCCCAGAAGCGCGGC TTCGCTCCCGGCCCGGCCCGACCCCGACTACTGTCCCCTAATACCCAGGGAGCGGAAGACGCGACCAACTCCAGGCACAGCGGAGCCGGC GCGGGGTCCACAAAGCAAAAAAGAATGATCCCCTTCCCCCATCCTCAAGTGACTGATGCTCTTGGAAGATACTCCAACTATATCCTGAGT AGCACTGTGGAGAGCCCCAGTCTGAGAAGCATGAACTTTGAGAAGCAAGGTGAGCGAGTGCTGTGCTTTCATGGGCCTCTTCTTTATGAA GCAAAGTGTGTAAAGGTTGCCATAAAGGACAAACAAGTGAAATACTTCATACATTACAGTGGTTGGAATAAAAATTGGGATGAATGGGTT CCGGAGAGCAGAGTACTCAAATACGTGGACACCAATTTGCAGAAACAGCGAGAACTTCAAAAAGCCAATCAGGAGCAGTATGCAGAGGGG AAGATGAGAGGGGCTGCCCCAGGAAAGAAGACATCTGGTCTGCAACAGAAAAATGTTGAAGTGAAAACGAAAAAGAACAAACAGAAAACA CCTGGAAATGGAGATGGTGGCAGTACCAGTGAGACCCCTCAGCCTCCTCGGAAGAAAAGGGCCCGGGTAGATCCTACTGTTGAAAATGAG GAAACATTCATGAACAGAGTTGAAGTTAAAGTAAAGATTCCTGAAGAGCTAAAACCGTGGCTTGTTGATGACTGGGACTTAATTACCAGG CAAAAACAGCTCTTTTATCTTCCTGCCAAGAAGAATGTGGATTCCATTCTTGAGGATTATGCAAATTACAAGAAATCTCGTGGAAACACA GATAATAAGGAGTATGCGGTTAATGAAGTTGTGGCAGGGATAAAAGAATACTTCAACGTAATGTTGGGTACCCAGCTACTCTATAAATTT GAGAGACCACAGTATGCTGAAATTCTTGCAGATCATCCCGATGCACCCATGTCCCAGGTGTATGGAGCGCCACATCTCCTGAGATTATTT GTGGAAATTGCAACACTAACAAGAGAGAATGGAAAGACAGTAATCAGAGTTCTCAAACAAAAAGAAGTGGAGCAGTTGATCAAAAAACAT GAGGAAGAAGAAGCCAAAGCTGAGCGTGAGAAGAAAGAAAAAGAACAGAAAGAAAAGGATAAATAGAATCAGAGATTTTATTACTCATTT GGGGCACCATTTCAGTGTAAAAGCAGTCCTACTCTTCCACACTAGGAAGGCTTTACTTTTTTTAACTGGTGCAGTGGGAAAATAGGACAT TACATACTGAATTGGGTCCTTGTCATTTCTGTCCAATTGAATACTTTATTGTAACGATGATGGTTACCCTTCATGGACGTCTTAATCTTC >54605_54605_4_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000379535_PSMA4_chr15_78841131_ENST00000559082_length(amino acids)=394AA_BP=338 MNQGPRKFPQKRGFAPGPARPRLLSPNTQGAEDATNSRHSGAGAGSTKQKRMIPFPHPQVTDALGRYSNYILSSTVESPSLRSMNFEKQG ERVLCFHGPLLYEAKCVKVAIKDKQVKYFIHYSGWNKNWDEWVPESRVLKYVDTNLQKQRELQKANQEQYAEGKMRGAAPGKKTSGLQQK NVEVKTKKNKQKTPGNGDGGSTSETPQPPRKKRARVDPTVENEETFMNRVEVKVKIPEELKPWLVDDWDLITRQKQLFYLPAKKNVDSIL EDYANYKKSRGNTDNKEYAVNEVVAGIKEYFNVMLGTQLLYKFERPQYAEILADHPDAPMSQVYGAPHLLRLFVEIATLTRENGKTVIRV -------------------------------------------------------------- >54605_54605_5_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000426013_PSMA4_chr15_78841131_ENST00000413382_length(transcript)=1459nt_BP=1044nt AGTCCCGGCGTGCCCTGGGGCGGCGCGGGCGCAGGGGCGCGTGCGCGGCGGGCTGTCGTTGGCTGGAGCAGCGGCTGCGCGGGTCGCGGT GCTGTGAGGTCTGCGGGCGCTGGCAAATCCGGCCCAGGATGTAGAGCTGGCAGTGCCTGACGGCGCGTCTGACGCGGAGTTGGGTGGGGT AGAGAGTAGGGGGCGGTAGTCGGGGGTGGTGGGAGAAGGAGGAGGCGGCGAATCACTTATAAATGGCGCCGAAGCAGGACCCGAAGCCTA AATTCCAGGAGGGTGAGCGAGTGCTGTGCTTTCATGGGCCTCTTCTTTATGAAGCAAAGTGTGTAAAGGTTGCCATAAAGGACAAACAAG TGAAATACTTCATACATTACAGTGGTTGGAATAAAAATTGGGATGAATGGGTTCCGGAGAGCAGAGTACTCAAATACGTGGACACCAATT TGCAGAAACAGCGAGAACTTCAAAAAGCCAATCAGGAGCAGTATGCAGAGGGGAAGATGAGAGGGGCTGCCCCAGGAAAGAAGACATCTG GTCTGCAACAGAAAAATGTTGAAGTGAAAACGAAAAAGAACAAACAGAAAACACCTGGAAATGGAGATGGTGGCAGTACCAGTGAGACCC CTCAGCCTCCTCGGAAGAAAAGGGCCCGGGTAGATCCTACTGTTGAAAATGAGGAAACATTCATGAACAGAGTTGAAGTTAAAGTAAAGA TTCCTGAAGAGCTAAAACCGTGGCTTGTTGATGACTGGGACTTAATTACCAGGCAAAAACAGCTCTTTTATCTTCCTGCCAAGAAGAATG TGGATTCCATTCTTGAGGATTATGCAAATTACAAGAAATCTCGTGGAAACACAGATAATAAGGAGTATGCGGTTAATGAAGTTGTGGCAG GGATAAAAGAATACTTCAACGTAATGTTGGGTACCCAGCTACTCTATAAATTTGAGAGACCACAGTATGCTGAAATTCTTGCAGATCATC CCGATGCACCCATGTCCCAGGTGTATGGAGCGCCACATCTCCTGAGATTATTTGTGGAAATTGCAACACTAACAAGAGAGAATGGAAAGA CAGTAATCAGAGTTCTCAAACAAAAAGAAGTGGAGCAGTTGATCAAAAAACATGAGGAAGAAGAAGCCAAAGCTGAGCGTGAGAAGAAAG AAAAAGAACAGAAAGAAAAGGATAAATAGAATCAGAGATTTTATTACTCATTTGGGGCACCATTTCAGTGTAAAAGCAGTCCTACTCTTC CACACTAGGAAGGCTTTACTTTTTTTAACTGGTGCAGTGGGAAAATAGGACATTACATACTGAATTGGGTCCTTGTCATTTCTGTCCAAT TGAATACTTTATTGTAACGATGATGGTTACCCTTCATGGACGTCTTAATCTTCCACACACATCCCCTTTTTTTGGAATAAAATTTGGAAA >54605_54605_5_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000426013_PSMA4_chr15_78841131_ENST00000413382_length(amino acids)=318AA_BP=262 MAPKQDPKPKFQEGERVLCFHGPLLYEAKCVKVAIKDKQVKYFIHYSGWNKNWDEWVPESRVLKYVDTNLQKQRELQKANQEQYAEGKMR GAAPGKKTSGLQQKNVEVKTKKNKQKTPGNGDGGSTSETPQPPRKKRARVDPTVENEETFMNRVEVKVKIPEELKPWLVDDWDLITRQKQ LFYLPAKKNVDSILEDYANYKKSRGNTDNKEYAVNEVVAGIKEYFNVMLGTQLLYKFERPQYAEILADHPDAPMSQVYGAPHLLRLFVEI -------------------------------------------------------------- >54605_54605_6_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000426013_PSMA4_chr15_78841131_ENST00000559082_length(transcript)=1441nt_BP=1044nt AGTCCCGGCGTGCCCTGGGGCGGCGCGGGCGCAGGGGCGCGTGCGCGGCGGGCTGTCGTTGGCTGGAGCAGCGGCTGCGCGGGTCGCGGT GCTGTGAGGTCTGCGGGCGCTGGCAAATCCGGCCCAGGATGTAGAGCTGGCAGTGCCTGACGGCGCGTCTGACGCGGAGTTGGGTGGGGT AGAGAGTAGGGGGCGGTAGTCGGGGGTGGTGGGAGAAGGAGGAGGCGGCGAATCACTTATAAATGGCGCCGAAGCAGGACCCGAAGCCTA AATTCCAGGAGGGTGAGCGAGTGCTGTGCTTTCATGGGCCTCTTCTTTATGAAGCAAAGTGTGTAAAGGTTGCCATAAAGGACAAACAAG TGAAATACTTCATACATTACAGTGGTTGGAATAAAAATTGGGATGAATGGGTTCCGGAGAGCAGAGTACTCAAATACGTGGACACCAATT TGCAGAAACAGCGAGAACTTCAAAAAGCCAATCAGGAGCAGTATGCAGAGGGGAAGATGAGAGGGGCTGCCCCAGGAAAGAAGACATCTG GTCTGCAACAGAAAAATGTTGAAGTGAAAACGAAAAAGAACAAACAGAAAACACCTGGAAATGGAGATGGTGGCAGTACCAGTGAGACCC CTCAGCCTCCTCGGAAGAAAAGGGCCCGGGTAGATCCTACTGTTGAAAATGAGGAAACATTCATGAACAGAGTTGAAGTTAAAGTAAAGA TTCCTGAAGAGCTAAAACCGTGGCTTGTTGATGACTGGGACTTAATTACCAGGCAAAAACAGCTCTTTTATCTTCCTGCCAAGAAGAATG TGGATTCCATTCTTGAGGATTATGCAAATTACAAGAAATCTCGTGGAAACACAGATAATAAGGAGTATGCGGTTAATGAAGTTGTGGCAG GGATAAAAGAATACTTCAACGTAATGTTGGGTACCCAGCTACTCTATAAATTTGAGAGACCACAGTATGCTGAAATTCTTGCAGATCATC CCGATGCACCCATGTCCCAGGTGTATGGAGCGCCACATCTCCTGAGATTATTTGTGGAAATTGCAACACTAACAAGAGAGAATGGAAAGA CAGTAATCAGAGTTCTCAAACAAAAAGAAGTGGAGCAGTTGATCAAAAAACATGAGGAAGAAGAAGCCAAAGCTGAGCGTGAGAAGAAAG AAAAAGAACAGAAAGAAAAGGATAAATAGAATCAGAGATTTTATTACTCATTTGGGGCACCATTTCAGTGTAAAAGCAGTCCTACTCTTC CACACTAGGAAGGCTTTACTTTTTTTAACTGGTGCAGTGGGAAAATAGGACATTACATACTGAATTGGGTCCTTGTCATTTCTGTCCAAT TGAATACTTTATTGTAACGATGATGGTTACCCTTCATGGACGTCTTAATCTTCCACACACATCCCCTTTTTTTGGAATAAAATTTGGAAA >54605_54605_6_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000426013_PSMA4_chr15_78841131_ENST00000559082_length(amino acids)=318AA_BP=262 MAPKQDPKPKFQEGERVLCFHGPLLYEAKCVKVAIKDKQVKYFIHYSGWNKNWDEWVPESRVLKYVDTNLQKQRELQKANQEQYAEGKMR GAAPGKKTSGLQQKNVEVKTKKNKQKTPGNGDGGSTSETPQPPRKKRARVDPTVENEETFMNRVEVKVKIPEELKPWLVDDWDLITRQKQ LFYLPAKKNVDSILEDYANYKKSRGNTDNKEYAVNEVVAGIKEYFNVMLGTQLLYKFERPQYAEILADHPDAPMSQVYGAPHLLRLFVEI -------------------------------------------------------------- >54605_54605_7_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000558502_PSMA4_chr15_78841131_ENST00000413382_length(transcript)=1090nt_BP=675nt CAGGACCCGAAGCCTAAATTCCAGGAGGTTGGGATGAATGGGTTCCGGAGAGCAGAGTACTCAAATACGTGGACACCAATTTGCAGAAAC AGCGAGAACTTCAAAAAGCCAATCAGGAGCAGTATGCAGAGGGGAAGATGAGAGGGGCTGCCCCAGGAAAGAAGACATCTGGTCTGCAAC AGAAAAATGTTGAAGTGAAAACGAAAAAGAACAAACAGAAAACACCTGGAAATGGAGATGGTGGCAGTACCAGTGAGACCCCTCAGCCTC CTCGGAAGAAAAGGGCCCGGGTAGATCCTACTGTTGAAAATGAGGAAACATTCATGAACAGAGTTGAAGTTAAAGTAAAGATTCCTGAAG AGCTAAAACCGTGGCTTGTTGATGACTGGGACTTAATTACCAGGCAAAAACAGCTCTTTTATCTTCCTGCCAAGAAGAATGTGGATTCCA TTCTTGAGGATTATGCAAATTACAAGAAATCTCGTGGAAACACAGATAATAAGGAGTATGCGGTTAATGAAGTTGTGGCAGGGATAAAAG AATACTTCAACGTAATGTTGGGTACCCAGCTACTCTATAAATTTGAGAGACCACAGTATGCTGAAATTCTTGCAGATCATCCCGATGCAC CCATGTCCCAGGTGTATGGAGCGCCACATCTCCTGAGATTATTTGTGGAAATTGCAACACTAACAAGAGAGAATGGAAAGACAGTAATCA GAGTTCTCAAACAAAAAGAAGTGGAGCAGTTGATCAAAAAACATGAGGAAGAAGAAGCCAAAGCTGAGCGTGAGAAGAAAGAAAAAGAAC AGAAAGAAAAGGATAAATAGAATCAGAGATTTTATTACTCATTTGGGGCACCATTTCAGTGTAAAAGCAGTCCTACTCTTCCACACTAGG AAGGCTTTACTTTTTTTAACTGGTGCAGTGGGAAAATAGGACATTACATACTGAATTGGGTCCTTGTCATTTCTGTCCAATTGAATACTT TATTGTAACGATGATGGTTACCCTTCATGGACGTCTTAATCTTCCACACACATCCCCTTTTTTTGGAATAAAATTTGGAAAATGGAAATG >54605_54605_7_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000558502_PSMA4_chr15_78841131_ENST00000413382_length(amino acids)=249AA_BP=193 MQKQRELQKANQEQYAEGKMRGAAPGKKTSGLQQKNVEVKTKKNKQKTPGNGDGGSTSETPQPPRKKRARVDPTVENEETFMNRVEVKVK IPEELKPWLVDDWDLITRQKQLFYLPAKKNVDSILEDYANYKKSRGNTDNKEYAVNEVVAGIKEYFNVMLGTQLLYKFERPQYAEILADH -------------------------------------------------------------- >54605_54605_8_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000558502_PSMA4_chr15_78841131_ENST00000559082_length(transcript)=1072nt_BP=675nt CAGGACCCGAAGCCTAAATTCCAGGAGGTTGGGATGAATGGGTTCCGGAGAGCAGAGTACTCAAATACGTGGACACCAATTTGCAGAAAC AGCGAGAACTTCAAAAAGCCAATCAGGAGCAGTATGCAGAGGGGAAGATGAGAGGGGCTGCCCCAGGAAAGAAGACATCTGGTCTGCAAC AGAAAAATGTTGAAGTGAAAACGAAAAAGAACAAACAGAAAACACCTGGAAATGGAGATGGTGGCAGTACCAGTGAGACCCCTCAGCCTC CTCGGAAGAAAAGGGCCCGGGTAGATCCTACTGTTGAAAATGAGGAAACATTCATGAACAGAGTTGAAGTTAAAGTAAAGATTCCTGAAG AGCTAAAACCGTGGCTTGTTGATGACTGGGACTTAATTACCAGGCAAAAACAGCTCTTTTATCTTCCTGCCAAGAAGAATGTGGATTCCA TTCTTGAGGATTATGCAAATTACAAGAAATCTCGTGGAAACACAGATAATAAGGAGTATGCGGTTAATGAAGTTGTGGCAGGGATAAAAG AATACTTCAACGTAATGTTGGGTACCCAGCTACTCTATAAATTTGAGAGACCACAGTATGCTGAAATTCTTGCAGATCATCCCGATGCAC CCATGTCCCAGGTGTATGGAGCGCCACATCTCCTGAGATTATTTGTGGAAATTGCAACACTAACAAGAGAGAATGGAAAGACAGTAATCA GAGTTCTCAAACAAAAAGAAGTGGAGCAGTTGATCAAAAAACATGAGGAAGAAGAAGCCAAAGCTGAGCGTGAGAAGAAAGAAAAAGAAC AGAAAGAAAAGGATAAATAGAATCAGAGATTTTATTACTCATTTGGGGCACCATTTCAGTGTAAAAGCAGTCCTACTCTTCCACACTAGG AAGGCTTTACTTTTTTTAACTGGTGCAGTGGGAAAATAGGACATTACATACTGAATTGGGTCCTTGTCATTTCTGTCCAATTGAATACTT >54605_54605_8_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000558502_PSMA4_chr15_78841131_ENST00000559082_length(amino acids)=249AA_BP=193 MQKQRELQKANQEQYAEGKMRGAAPGKKTSGLQQKNVEVKTKKNKQKTPGNGDGGSTSETPQPPRKKRARVDPTVENEETFMNRVEVKVK IPEELKPWLVDDWDLITRQKQLFYLPAKKNVDSILEDYANYKKSRGNTDNKEYAVNEVVAGIKEYFNVMLGTQLLYKFERPQYAEILADH -------------------------------------------------------------- >54605_54605_9_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000558746_PSMA4_chr15_78841131_ENST00000413382_length(transcript)=1200nt_BP=785nt GTAGAGAGTAGGGGGCGGTAGTCGGGGGTGGTGGGAGAAGGAGGAGGCGGCGAATCACTTATAAATGGCGCCGAAGCAGGACCCGAAGCC TAAATTCCAGGAGGGTGAGCGAGTGCTGTGCTTTCATGGGCCTCTTCTTTATGAAGCAAAGTGTGTAAAGGTTGCCATAAAGGACAAACA AGTGAAATACTTCATACATTACAGTGGTTGGAATAAAAATTGGGATGAATGGGTTCCGGAGAGCAGAGTACTCAAATACGTGGACACCAA TTTGCAGAAACAGCGAGAACTTCAAAAAGCCAATCAGAAAACGAAAAAGAACAAACAGAAAACACCTGGAAATGGAGATGGTGGCAGTAC CAGTGAGACCCCTCAGCCTCCTCGGAAGAAAAGGGCCCGGGTAGATCCTACTGTTGAAAATGAGGAAACATTCATGAACAGAGTTGAAGT TAAAGTAAAGATTCCTGAAGAGCTAAAACCGTGGCTTGTTGATGACTGGGACTTAATTACCAGGCAAAAACAGCTCTTTTATCTTCCTGC CAAGAAGAATGTGGATTCCATTCTTGAGGATTATGCAAATTACAAGAAATCTCGTGGAAACACAGATAATAAGGAGTATGCGGTTAATGA AGTTGTGGCAGGGATAAAAGAATACTTCAACGTAATGTTGGGTACCCAGCTACTCTATAAATTTGAGAGACCACAGTATGCTGAAATTCT TGCAGATCATCCCGATGCACCCATGTCCCAGGTGTATGGAGCGCCACATCTCCTGAGATTATTTGTGGAAATTGCAACACTAACAAGAGA GAATGGAAAGACAGTAATCAGAGTTCTCAAACAAAAAGAAGTGGAGCAGTTGATCAAAAAACATGAGGAAGAAGAAGCCAAAGCTGAGCG TGAGAAGAAAGAAAAAGAACAGAAAGAAAAGGATAAATAGAATCAGAGATTTTATTACTCATTTGGGGCACCATTTCAGTGTAAAAGCAG TCCTACTCTTCCACACTAGGAAGGCTTTACTTTTTTTAACTGGTGCAGTGGGAAAATAGGACATTACATACTGAATTGGGTCCTTGTCAT TTCTGTCCAATTGAATACTTTATTGTAACGATGATGGTTACCCTTCATGGACGTCTTAATCTTCCACACACATCCCCTTTTTTTGGAATA >54605_54605_9_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000558746_PSMA4_chr15_78841131_ENST00000413382_length(amino acids)=291AA_BP=235 MAPKQDPKPKFQEGERVLCFHGPLLYEAKCVKVAIKDKQVKYFIHYSGWNKNWDEWVPESRVLKYVDTNLQKQRELQKANQKTKKNKQKT PGNGDGGSTSETPQPPRKKRARVDPTVENEETFMNRVEVKVKIPEELKPWLVDDWDLITRQKQLFYLPAKKNVDSILEDYANYKKSRGNT DNKEYAVNEVVAGIKEYFNVMLGTQLLYKFERPQYAEILADHPDAPMSQVYGAPHLLRLFVEIATLTRENGKTVIRVLKQKEVEQLIKKH -------------------------------------------------------------- >54605_54605_10_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000558746_PSMA4_chr15_78841131_ENST00000559082_length(transcript)=1182nt_BP=785nt GTAGAGAGTAGGGGGCGGTAGTCGGGGGTGGTGGGAGAAGGAGGAGGCGGCGAATCACTTATAAATGGCGCCGAAGCAGGACCCGAAGCC TAAATTCCAGGAGGGTGAGCGAGTGCTGTGCTTTCATGGGCCTCTTCTTTATGAAGCAAAGTGTGTAAAGGTTGCCATAAAGGACAAACA AGTGAAATACTTCATACATTACAGTGGTTGGAATAAAAATTGGGATGAATGGGTTCCGGAGAGCAGAGTACTCAAATACGTGGACACCAA TTTGCAGAAACAGCGAGAACTTCAAAAAGCCAATCAGAAAACGAAAAAGAACAAACAGAAAACACCTGGAAATGGAGATGGTGGCAGTAC CAGTGAGACCCCTCAGCCTCCTCGGAAGAAAAGGGCCCGGGTAGATCCTACTGTTGAAAATGAGGAAACATTCATGAACAGAGTTGAAGT TAAAGTAAAGATTCCTGAAGAGCTAAAACCGTGGCTTGTTGATGACTGGGACTTAATTACCAGGCAAAAACAGCTCTTTTATCTTCCTGC CAAGAAGAATGTGGATTCCATTCTTGAGGATTATGCAAATTACAAGAAATCTCGTGGAAACACAGATAATAAGGAGTATGCGGTTAATGA AGTTGTGGCAGGGATAAAAGAATACTTCAACGTAATGTTGGGTACCCAGCTACTCTATAAATTTGAGAGACCACAGTATGCTGAAATTCT TGCAGATCATCCCGATGCACCCATGTCCCAGGTGTATGGAGCGCCACATCTCCTGAGATTATTTGTGGAAATTGCAACACTAACAAGAGA GAATGGAAAGACAGTAATCAGAGTTCTCAAACAAAAAGAAGTGGAGCAGTTGATCAAAAAACATGAGGAAGAAGAAGCCAAAGCTGAGCG TGAGAAGAAAGAAAAAGAACAGAAAGAAAAGGATAAATAGAATCAGAGATTTTATTACTCATTTGGGGCACCATTTCAGTGTAAAAGCAG TCCTACTCTTCCACACTAGGAAGGCTTTACTTTTTTTAACTGGTGCAGTGGGAAAATAGGACATTACATACTGAATTGGGTCCTTGTCAT TTCTGTCCAATTGAATACTTTATTGTAACGATGATGGTTACCCTTCATGGACGTCTTAATCTTCCACACACATCCCCTTTTTTTGGAATA >54605_54605_10_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000558746_PSMA4_chr15_78841131_ENST00000559082_length(amino acids)=291AA_BP=235 MAPKQDPKPKFQEGERVLCFHGPLLYEAKCVKVAIKDKQVKYFIHYSGWNKNWDEWVPESRVLKYVDTNLQKQRELQKANQKTKKNKQKT PGNGDGGSTSETPQPPRKKRARVDPTVENEETFMNRVEVKVKIPEELKPWLVDDWDLITRQKQLFYLPAKKNVDSILEDYANYKKSRGNT DNKEYAVNEVVAGIKEYFNVMLGTQLLYKFERPQYAEILADHPDAPMSQVYGAPHLLRLFVEIATLTRENGKTVIRVLKQKEVEQLIKKH -------------------------------------------------------------- >54605_54605_11_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000559345_PSMA4_chr15_78841131_ENST00000413382_length(transcript)=1391nt_BP=976nt GAATCACTTATAAATGGCGCCGAAGCAGGACCCGAAGCCTAAATTCCAGGAGGGTGAGTGTGCGCCTTTGGGAAAAAGGCACCTAACGGC GCAGGAGATAGAGGCGGGCTCGAGGTGATTGAGGCTTGAGGGCCGGGGGCGGCGCGGGCTGCGCCCTGAGAAGGCGGCGGTCAGTGCTTT GTGCGGGGAAAGCGCATTGCGGATGGCAATTCCGGTGAGCGAGTGCTGTGCTTTCATGGGCCTCTTCTTTATGAAGCAAAGTGTGTAAAG GTTGCCATAAAGGACAAACAAGTGAAATACTTCATACATTACAGTGGTTGGAATAAAAATTGGGATGAATGGGTTCCGGAGAGCAGAGTA CTCAAATACGTGGACACCAATTTGCAGAAACAGCGAGAACTTCAAAAAGCCAATCAGGAGCAGTATGCAGAGGGGAAGATGAGAGGGGCT GCCCCAGGAAAGAAGACATCTGGTCTGCAACAGAAAAATGTTGAAGTGAAAACGAAAAAGAACAAACAGAAAACACCTGGAAATGGAGAT GGTGGCAGTACCAGTGAGACCCCTCAGCCTCCTCGGAAGAAAAGGGCCCGGGTAGATCCTACTGTTGAAAATGAGGAAACATTCATGAAC AGAGTTGAAGTTAAAGTAAAGATTCCTGAAGAGCTAAAACCGTGGCTTGTTGATGACTGGGACTTAATTACCAGGCAAAAACAGCTCTTT TATCTTCCTGCCAAGAAGAATGTGGATTCCATTCTTGAGGATTATGCAAATTACAAGAAATCTCGTGGAAACACAGATAATAAGGAGTAT GCGGTTAATGAAGTTGTGGCAGGGATAAAAGAATACTTCAACGTAATGTTGGGTACCCAGCTACTCTATAAATTTGAGAGACCACAGTAT GCTGAAATTCTTGCAGATCATCCCGATGCACCCATGTCCCAGGTGTATGGAGCGCCACATCTCCTGAGATTATTTGTGGAAATTGCAACA CTAACAAGAGAGAATGGAAAGACAGTAATCAGAGTTCTCAAACAAAAAGAAGTGGAGCAGTTGATCAAAAAACATGAGGAAGAAGAAGCC AAAGCTGAGCGTGAGAAGAAAGAAAAAGAACAGAAAGAAAAGGATAAATAGAATCAGAGATTTTATTACTCATTTGGGGCACCATTTCAG TGTAAAAGCAGTCCTACTCTTCCACACTAGGAAGGCTTTACTTTTTTTAACTGGTGCAGTGGGAAAATAGGACATTACATACTGAATTGG GTCCTTGTCATTTCTGTCCAATTGAATACTTTATTGTAACGATGATGGTTACCCTTCATGGACGTCTTAATCTTCCACACACATCCCCTT >54605_54605_11_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000559345_PSMA4_chr15_78841131_ENST00000413382_length(amino acids)=301AA_BP=245 MCFHGPLLYEAKCVKVAIKDKQVKYFIHYSGWNKNWDEWVPESRVLKYVDTNLQKQRELQKANQEQYAEGKMRGAAPGKKTSGLQQKNVE VKTKKNKQKTPGNGDGGSTSETPQPPRKKRARVDPTVENEETFMNRVEVKVKIPEELKPWLVDDWDLITRQKQLFYLPAKKNVDSILEDY ANYKKSRGNTDNKEYAVNEVVAGIKEYFNVMLGTQLLYKFERPQYAEILADHPDAPMSQVYGAPHLLRLFVEIATLTRENGKTVIRVLKQ -------------------------------------------------------------- >54605_54605_12_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000559345_PSMA4_chr15_78841131_ENST00000559082_length(transcript)=1373nt_BP=976nt GAATCACTTATAAATGGCGCCGAAGCAGGACCCGAAGCCTAAATTCCAGGAGGGTGAGTGTGCGCCTTTGGGAAAAAGGCACCTAACGGC GCAGGAGATAGAGGCGGGCTCGAGGTGATTGAGGCTTGAGGGCCGGGGGCGGCGCGGGCTGCGCCCTGAGAAGGCGGCGGTCAGTGCTTT GTGCGGGGAAAGCGCATTGCGGATGGCAATTCCGGTGAGCGAGTGCTGTGCTTTCATGGGCCTCTTCTTTATGAAGCAAAGTGTGTAAAG GTTGCCATAAAGGACAAACAAGTGAAATACTTCATACATTACAGTGGTTGGAATAAAAATTGGGATGAATGGGTTCCGGAGAGCAGAGTA CTCAAATACGTGGACACCAATTTGCAGAAACAGCGAGAACTTCAAAAAGCCAATCAGGAGCAGTATGCAGAGGGGAAGATGAGAGGGGCT GCCCCAGGAAAGAAGACATCTGGTCTGCAACAGAAAAATGTTGAAGTGAAAACGAAAAAGAACAAACAGAAAACACCTGGAAATGGAGAT GGTGGCAGTACCAGTGAGACCCCTCAGCCTCCTCGGAAGAAAAGGGCCCGGGTAGATCCTACTGTTGAAAATGAGGAAACATTCATGAAC AGAGTTGAAGTTAAAGTAAAGATTCCTGAAGAGCTAAAACCGTGGCTTGTTGATGACTGGGACTTAATTACCAGGCAAAAACAGCTCTTT TATCTTCCTGCCAAGAAGAATGTGGATTCCATTCTTGAGGATTATGCAAATTACAAGAAATCTCGTGGAAACACAGATAATAAGGAGTAT GCGGTTAATGAAGTTGTGGCAGGGATAAAAGAATACTTCAACGTAATGTTGGGTACCCAGCTACTCTATAAATTTGAGAGACCACAGTAT GCTGAAATTCTTGCAGATCATCCCGATGCACCCATGTCCCAGGTGTATGGAGCGCCACATCTCCTGAGATTATTTGTGGAAATTGCAACA CTAACAAGAGAGAATGGAAAGACAGTAATCAGAGTTCTCAAACAAAAAGAAGTGGAGCAGTTGATCAAAAAACATGAGGAAGAAGAAGCC AAAGCTGAGCGTGAGAAGAAAGAAAAAGAACAGAAAGAAAAGGATAAATAGAATCAGAGATTTTATTACTCATTTGGGGCACCATTTCAG TGTAAAAGCAGTCCTACTCTTCCACACTAGGAAGGCTTTACTTTTTTTAACTGGTGCAGTGGGAAAATAGGACATTACATACTGAATTGG GTCCTTGTCATTTCTGTCCAATTGAATACTTTATTGTAACGATGATGGTTACCCTTCATGGACGTCTTAATCTTCCACACACATCCCCTT >54605_54605_12_MORF4L1-PSMA4_MORF4L1_chr15_79186572_ENST00000559345_PSMA4_chr15_78841131_ENST00000559082_length(amino acids)=301AA_BP=245 MCFHGPLLYEAKCVKVAIKDKQVKYFIHYSGWNKNWDEWVPESRVLKYVDTNLQKQRELQKANQEQYAEGKMRGAAPGKKTSGLQQKNVE VKTKKNKQKTPGNGDGGSTSETPQPPRKKRARVDPTVENEETFMNRVEVKVKIPEELKPWLVDDWDLITRQKQLFYLPAKKNVDSILEDY ANYKKSRGNTDNKEYAVNEVVAGIKEYFNVMLGTQLLYKFERPQYAEILADHPDAPMSQVYGAPHLLRLFVEIATLTRENGKTVIRVLKQ -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MORF4L1-PSMA4 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000331268 | + | 11 | 13 | 26_62 | 306.3333333333333 | 363.0 | KAT8 |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000426013 | + | 10 | 12 | 26_62 | 267.3333333333333 | 324.0 | KAT8 |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000558502 | + | 8 | 10 | 26_62 | 179.33333333333334 | 236.0 | KAT8 |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000559345 | + | 10 | 12 | 26_62 | 179.33333333333334 | 236.0 | KAT8 |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000331268 | + | 11 | 13 | 164_230 | 306.3333333333333 | 363.0 | RB1-1 |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000426013 | + | 10 | 12 | 164_230 | 267.3333333333333 | 324.0 | RB1-1 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000558502 | + | 8 | 10 | 164_230 | 179.33333333333334 | 236.0 | RB1-1 |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000559345 | + | 10 | 12 | 164_230 | 179.33333333333334 | 236.0 | RB1-1 |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000331268 | + | 11 | 13 | 323_344 | 306.3333333333333 | 363.0 | RB1-2 |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000426013 | + | 10 | 12 | 323_344 | 267.3333333333333 | 324.0 | RB1-2 |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000558502 | + | 8 | 10 | 323_344 | 179.33333333333334 | 236.0 | RB1-2 |
| Hgene | MORF4L1 | chr15:79186572 | chr15:78841131 | ENST00000559345 | + | 10 | 12 | 323_344 | 179.33333333333334 | 236.0 | RB1-2 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MORF4L1-PSMA4 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MORF4L1-PSMA4 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
