
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MPI-ESYT1 (FusionGDB2 ID:54715) |
Fusion Gene Summary for MPI-ESYT1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MPI-ESYT1 | Fusion gene ID: 54715 | Hgene | Tgene | Gene symbol | MPI | ESYT1 | Gene ID | 6590 | 23344 |
| Gene name | secretory leukocyte peptidase inhibitor | extended synaptotagmin 1 | |
| Synonyms | ALK1|ALP|BLPI|HUSI|HUSI-I|MPI|WAP4|WFDC4 | FAM62A|MBC2 | |
| Cytomap | 20q13.12 | 12q13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | antileukoproteinaseHUSI-1WAP four-disulfide core domain protein 4mucus proteinase inhibitorprotease inhibitor WAP4secretory leukocyte protease inhibitor (antileukoproteinase)seminal proteinase inhibitor | extended synaptotagmin-1extended synaptotagmin like protein 1extended synaptotagmin protein 1family with sequence similarity 62 (C2 domain containing), member Amembrane-bound C2 domain-containing protein | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P34949 | Q9BSJ8 | |
| Ensembl transtripts involved in fusion gene | ENST00000323744, ENST00000352410, ENST00000535694, ENST00000562606, ENST00000563422, ENST00000563786, ENST00000564003, ENST00000565576, ENST00000566377, | ENST00000550878, ENST00000267113, ENST00000394048, ENST00000541590, | |
| Fusion gene scores | * DoF score | 2 X 1 X 2=4 | 6 X 6 X 3=108 |
| # samples | 2 | 6 | |
| ** MAII score | log2(2/4*10)=2.32192809488736 | log2(6/108*10)=-0.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MPI [Title/Abstract] AND ESYT1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MPI(75185143)-ESYT1(56537606), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | MPI | GO:0019731 | antibacterial humoral response | 18714013 |
| Hgene | MPI | GO:0032091 | negative regulation of protein binding | 19333378 |
| Hgene | MPI | GO:0035821 | modification of morphology or physiology of other organism | 2467900 |
| Hgene | MPI | GO:0045071 | negative regulation of viral genome replication | 19333378 |
 Fusion gene breakpoints across MPI (5'-gene) Fusion gene breakpoints across MPI (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
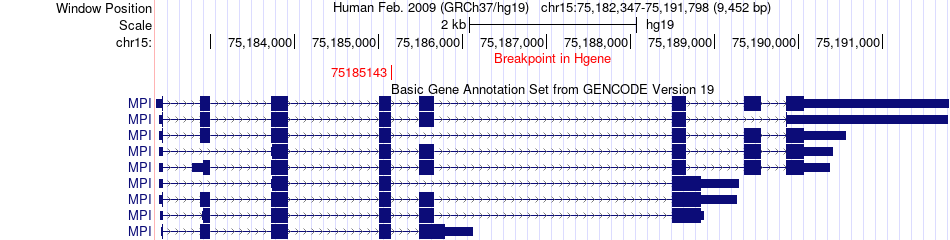 |
 Fusion gene breakpoints across ESYT1 (3'-gene) Fusion gene breakpoints across ESYT1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
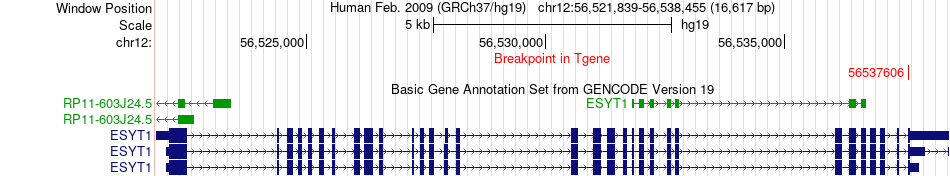 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-23-1023 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
Top |
Fusion Gene ORF analysis for MPI-ESYT1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000323744 | ENST00000550878 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| 5CDS-intron | ENST00000352410 | ENST00000550878 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| 5CDS-intron | ENST00000535694 | ENST00000550878 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| 5CDS-intron | ENST00000562606 | ENST00000550878 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| 5CDS-intron | ENST00000563422 | ENST00000550878 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| 5CDS-intron | ENST00000563786 | ENST00000550878 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| 5CDS-intron | ENST00000564003 | ENST00000550878 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| 5CDS-intron | ENST00000565576 | ENST00000550878 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| 5CDS-intron | ENST00000566377 | ENST00000550878 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000323744 | ENST00000267113 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000323744 | ENST00000394048 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000323744 | ENST00000541590 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000352410 | ENST00000267113 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000352410 | ENST00000394048 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000352410 | ENST00000541590 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000535694 | ENST00000267113 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000535694 | ENST00000394048 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000535694 | ENST00000541590 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000562606 | ENST00000267113 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000562606 | ENST00000394048 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000562606 | ENST00000541590 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000563422 | ENST00000267113 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000563422 | ENST00000394048 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000563422 | ENST00000541590 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000563786 | ENST00000267113 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000563786 | ENST00000394048 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000563786 | ENST00000541590 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000564003 | ENST00000267113 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000564003 | ENST00000394048 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000564003 | ENST00000541590 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000565576 | ENST00000267113 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000565576 | ENST00000394048 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000565576 | ENST00000541590 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000566377 | ENST00000267113 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000566377 | ENST00000394048 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
| In-frame | ENST00000566377 | ENST00000541590 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000352410 | MPI | chr15 | 75185143 | + | ENST00000394048 | ESYT1 | chr12 | 56537606 | + | 1403 | 554 | 49 | 564 | 171 |
| ENST00000352410 | MPI | chr15 | 75185143 | + | ENST00000267113 | ESYT1 | chr12 | 56537606 | + | 773 | 554 | 49 | 564 | 171 |
| ENST00000352410 | MPI | chr15 | 75185143 | + | ENST00000541590 | ESYT1 | chr12 | 56537606 | + | 923 | 554 | 49 | 564 | 171 |
| ENST00000566377 | MPI | chr15 | 75185143 | + | ENST00000394048 | ESYT1 | chr12 | 56537606 | + | 1371 | 522 | 17 | 532 | 171 |
| ENST00000566377 | MPI | chr15 | 75185143 | + | ENST00000267113 | ESYT1 | chr12 | 56537606 | + | 741 | 522 | 17 | 532 | 171 |
| ENST00000566377 | MPI | chr15 | 75185143 | + | ENST00000541590 | ESYT1 | chr12 | 56537606 | + | 891 | 522 | 17 | 532 | 171 |
| ENST00000563422 | MPI | chr15 | 75185143 | + | ENST00000394048 | ESYT1 | chr12 | 56537606 | + | 1368 | 519 | 14 | 529 | 171 |
| ENST00000563422 | MPI | chr15 | 75185143 | + | ENST00000267113 | ESYT1 | chr12 | 56537606 | + | 738 | 519 | 14 | 529 | 171 |
| ENST00000563422 | MPI | chr15 | 75185143 | + | ENST00000541590 | ESYT1 | chr12 | 56537606 | + | 888 | 519 | 14 | 529 | 171 |
| ENST00000564003 | MPI | chr15 | 75185143 | + | ENST00000394048 | ESYT1 | chr12 | 56537606 | + | 1240 | 391 | 48 | 401 | 117 |
| ENST00000564003 | MPI | chr15 | 75185143 | + | ENST00000267113 | ESYT1 | chr12 | 56537606 | + | 610 | 391 | 48 | 401 | 117 |
| ENST00000564003 | MPI | chr15 | 75185143 | + | ENST00000541590 | ESYT1 | chr12 | 56537606 | + | 760 | 391 | 48 | 401 | 117 |
| ENST00000563786 | MPI | chr15 | 75185143 | + | ENST00000394048 | ESYT1 | chr12 | 56537606 | + | 1457 | 608 | 115 | 618 | 167 |
| ENST00000563786 | MPI | chr15 | 75185143 | + | ENST00000267113 | ESYT1 | chr12 | 56537606 | + | 827 | 608 | 115 | 618 | 167 |
| ENST00000563786 | MPI | chr15 | 75185143 | + | ENST00000541590 | ESYT1 | chr12 | 56537606 | + | 977 | 608 | 115 | 618 | 167 |
| ENST00000535694 | MPI | chr15 | 75185143 | + | ENST00000394048 | ESYT1 | chr12 | 56537606 | + | 1240 | 391 | 48 | 401 | 117 |
| ENST00000535694 | MPI | chr15 | 75185143 | + | ENST00000267113 | ESYT1 | chr12 | 56537606 | + | 610 | 391 | 48 | 401 | 117 |
| ENST00000535694 | MPI | chr15 | 75185143 | + | ENST00000541590 | ESYT1 | chr12 | 56537606 | + | 760 | 391 | 48 | 401 | 117 |
| ENST00000323744 | MPI | chr15 | 75185143 | + | ENST00000394048 | ESYT1 | chr12 | 56537606 | + | 1368 | 519 | 14 | 529 | 171 |
| ENST00000323744 | MPI | chr15 | 75185143 | + | ENST00000267113 | ESYT1 | chr12 | 56537606 | + | 738 | 519 | 14 | 529 | 171 |
| ENST00000323744 | MPI | chr15 | 75185143 | + | ENST00000541590 | ESYT1 | chr12 | 56537606 | + | 888 | 519 | 14 | 529 | 171 |
| ENST00000562606 | MPI | chr15 | 75185143 | + | ENST00000394048 | ESYT1 | chr12 | 56537606 | + | 1331 | 482 | 55 | 492 | 145 |
| ENST00000562606 | MPI | chr15 | 75185143 | + | ENST00000267113 | ESYT1 | chr12 | 56537606 | + | 701 | 482 | 55 | 492 | 145 |
| ENST00000562606 | MPI | chr15 | 75185143 | + | ENST00000541590 | ESYT1 | chr12 | 56537606 | + | 851 | 482 | 55 | 492 | 145 |
| ENST00000565576 | MPI | chr15 | 75185143 | + | ENST00000394048 | ESYT1 | chr12 | 56537606 | + | 1348 | 499 | 12 | 509 | 165 |
| ENST00000565576 | MPI | chr15 | 75185143 | + | ENST00000267113 | ESYT1 | chr12 | 56537606 | + | 718 | 499 | 12 | 509 | 165 |
| ENST00000565576 | MPI | chr15 | 75185143 | + | ENST00000541590 | ESYT1 | chr12 | 56537606 | + | 868 | 499 | 12 | 509 | 165 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000352410 | ENST00000394048 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.007561369 | 0.9924386 |
| ENST00000352410 | ENST00000267113 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.007626726 | 0.9923733 |
| ENST00000352410 | ENST00000541590 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.008637924 | 0.99136204 |
| ENST00000566377 | ENST00000394048 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.00886669 | 0.9911333 |
| ENST00000566377 | ENST00000267113 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.008683946 | 0.9913161 |
| ENST00000566377 | ENST00000541590 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.009918876 | 0.99008113 |
| ENST00000563422 | ENST00000394048 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.01036848 | 0.9896316 |
| ENST00000563422 | ENST00000267113 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.007935002 | 0.992065 |
| ENST00000563422 | ENST00000541590 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.008556526 | 0.99144346 |
| ENST00000564003 | ENST00000394048 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.03148293 | 0.96851707 |
| ENST00000564003 | ENST00000267113 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.030811511 | 0.96918845 |
| ENST00000564003 | ENST00000541590 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.05302476 | 0.9469753 |
| ENST00000563786 | ENST00000394048 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.021176491 | 0.97882354 |
| ENST00000563786 | ENST00000267113 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.02094996 | 0.97905 |
| ENST00000563786 | ENST00000541590 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.027646776 | 0.9723532 |
| ENST00000535694 | ENST00000394048 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.03148293 | 0.96851707 |
| ENST00000535694 | ENST00000267113 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.030811511 | 0.96918845 |
| ENST00000535694 | ENST00000541590 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.05302476 | 0.9469753 |
| ENST00000323744 | ENST00000394048 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.01036848 | 0.9896316 |
| ENST00000323744 | ENST00000267113 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.007935002 | 0.992065 |
| ENST00000323744 | ENST00000541590 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.008556526 | 0.99144346 |
| ENST00000562606 | ENST00000394048 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.013255601 | 0.9867444 |
| ENST00000562606 | ENST00000267113 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.010485845 | 0.9895141 |
| ENST00000562606 | ENST00000541590 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.015105047 | 0.98489493 |
| ENST00000565576 | ENST00000394048 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.009676292 | 0.99032366 |
| ENST00000565576 | ENST00000267113 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.008751325 | 0.9912486 |
| ENST00000565576 | ENST00000541590 | MPI | chr15 | 75185143 | + | ESYT1 | chr12 | 56537606 | + | 0.010830508 | 0.9891695 |
Top |
Fusion Genomic Features for MPI-ESYT1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
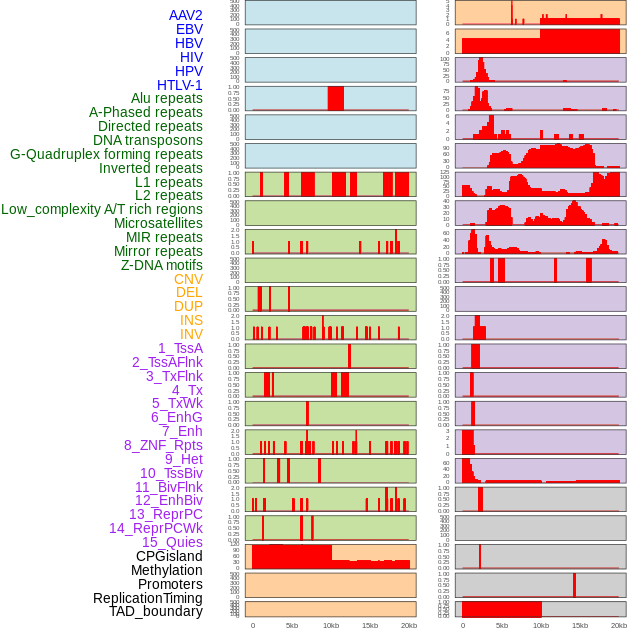 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
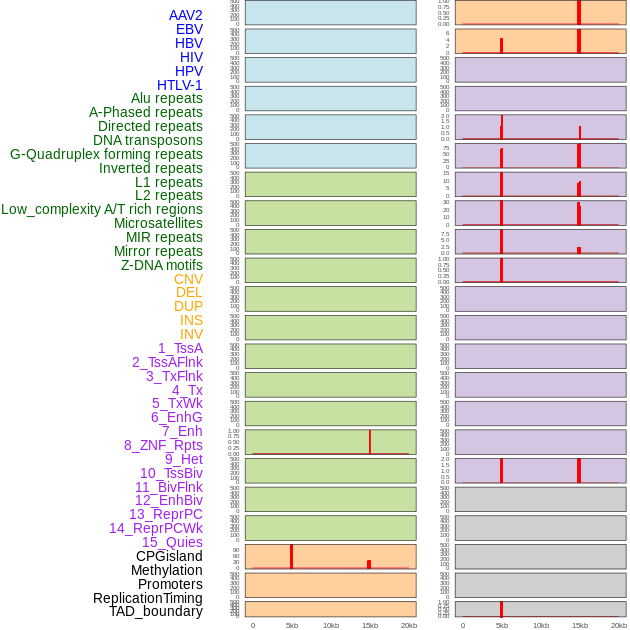 |
Top |
Fusion Protein Features for MPI-ESYT1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr15:75185143/chr12:56537606) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MPI | ESYT1 |
| FUNCTION: Involved in the synthesis of the GDP-mannose and dolichol-phosphate-mannose required for a number of critical mannosyl transfer reactions. | FUNCTION: Binds glycerophospholipids in a barrel-like domain and may play a role in cellular lipid transport (By similarity). Binds calcium (via the C2 domains) and translocates to sites of contact between the endoplasmic reticulum and the cell membrane in response to increased cytosolic calcium levels. Helps tether the endoplasmic reticulum to the cell membrane and promotes the formation of appositions between the endoplasmic reticulum and the cell membrane. {ECO:0000250, ECO:0000269|PubMed:23791178, ECO:0000269|PubMed:24183667}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000267113 | 29 | 31 | 91_116 | 1101 | 1115.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000394048 | 29 | 31 | 91_116 | 1091 | 1105.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000541590 | 29 | 32 | 91_116 | 1101 | 975.3333333333334 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000267113 | 29 | 31 | 1012_1015 | 1101 | 1115.0 | Compositional bias | Note=Poly-Leu | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000267113 | 29 | 31 | 924_935 | 1101 | 1115.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000394048 | 29 | 31 | 1012_1015 | 1091 | 1105.0 | Compositional bias | Note=Poly-Leu | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000394048 | 29 | 31 | 924_935 | 1091 | 1105.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000541590 | 29 | 32 | 1012_1015 | 1101 | 975.3333333333334 | Compositional bias | Note=Poly-Leu | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000541590 | 29 | 32 | 924_935 | 1101 | 975.3333333333334 | Compositional bias | Note=Poly-Ser | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000267113 | 29 | 31 | 135_313 | 1101 | 1115.0 | Domain | SMP-LTD | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000267113 | 29 | 31 | 312_433 | 1101 | 1115.0 | Domain | C2 1 | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000267113 | 29 | 31 | 460_580 | 1101 | 1115.0 | Domain | C2 2 | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000267113 | 29 | 31 | 627_751 | 1101 | 1115.0 | Domain | C2 3 | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000267113 | 29 | 31 | 777_899 | 1101 | 1115.0 | Domain | C2 4 | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000267113 | 29 | 31 | 971_1093 | 1101 | 1115.0 | Domain | C2 5 | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000394048 | 29 | 31 | 135_313 | 1091 | 1105.0 | Domain | SMP-LTD | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000394048 | 29 | 31 | 312_433 | 1091 | 1105.0 | Domain | C2 1 | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000394048 | 29 | 31 | 460_580 | 1091 | 1105.0 | Domain | C2 2 | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000394048 | 29 | 31 | 627_751 | 1091 | 1105.0 | Domain | C2 3 | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000394048 | 29 | 31 | 777_899 | 1091 | 1105.0 | Domain | C2 4 | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000394048 | 29 | 31 | 971_1093 | 1091 | 1105.0 | Domain | C2 5 | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000541590 | 29 | 32 | 135_313 | 1101 | 975.3333333333334 | Domain | SMP-LTD | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000541590 | 29 | 32 | 312_433 | 1101 | 975.3333333333334 | Domain | C2 1 | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000541590 | 29 | 32 | 460_580 | 1101 | 975.3333333333334 | Domain | C2 2 | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000541590 | 29 | 32 | 627_751 | 1101 | 975.3333333333334 | Domain | C2 3 | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000541590 | 29 | 32 | 777_899 | 1101 | 975.3333333333334 | Domain | C2 4 | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000541590 | 29 | 32 | 971_1093 | 1101 | 975.3333333333334 | Domain | C2 5 | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000267113 | 29 | 31 | 1018_1025 | 1101 | 1115.0 | Region | Required for phosphatidylinositol 4%2C5-bisphosphate-dependent location at the cell membrane | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000394048 | 29 | 31 | 1018_1025 | 1091 | 1105.0 | Region | Required for phosphatidylinositol 4%2C5-bisphosphate-dependent location at the cell membrane | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000541590 | 29 | 32 | 1018_1025 | 1101 | 975.3333333333334 | Region | Required for phosphatidylinositol 4%2C5-bisphosphate-dependent location at the cell membrane | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000267113 | 29 | 31 | 1_38 | 1101 | 1115.0 | Topological domain | Cytoplasmic | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000267113 | 29 | 31 | 60_62 | 1101 | 1115.0 | Topological domain | Lumenal | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000267113 | 29 | 31 | 84_1104 | 1101 | 1115.0 | Topological domain | Cytoplasmic | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000394048 | 29 | 31 | 1_38 | 1091 | 1105.0 | Topological domain | Cytoplasmic | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000394048 | 29 | 31 | 60_62 | 1091 | 1105.0 | Topological domain | Lumenal | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000394048 | 29 | 31 | 84_1104 | 1091 | 1105.0 | Topological domain | Cytoplasmic | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000541590 | 29 | 32 | 1_38 | 1101 | 975.3333333333334 | Topological domain | Cytoplasmic | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000541590 | 29 | 32 | 60_62 | 1101 | 975.3333333333334 | Topological domain | Lumenal | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000541590 | 29 | 32 | 84_1104 | 1101 | 975.3333333333334 | Topological domain | Cytoplasmic | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000267113 | 29 | 31 | 39_59 | 1101 | 1115.0 | Transmembrane | Helical | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000267113 | 29 | 31 | 63_83 | 1101 | 1115.0 | Transmembrane | Helical | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000394048 | 29 | 31 | 39_59 | 1091 | 1105.0 | Transmembrane | Helical | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000394048 | 29 | 31 | 63_83 | 1091 | 1105.0 | Transmembrane | Helical | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000541590 | 29 | 32 | 39_59 | 1101 | 975.3333333333334 | Transmembrane | Helical | |
| Tgene | ESYT1 | chr15:75185143 | chr12:56537606 | ENST00000541590 | 29 | 32 | 63_83 | 1101 | 975.3333333333334 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for MPI-ESYT1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >54715_54715_1_MPI-ESYT1_MPI_chr15_75185143_ENST00000323744_ESYT1_chr12_56537606_ENST00000267113_length(transcript)=738nt_BP=519nt ATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTATTCCCACTTTCCTGTGCGGTGCAGCAGTATGCCTGGGGGA AGATGGGTTCCAACAGCGAAGTGGCGCGGCTGTTGGCCAGCAGTGATCCACTGGCCCAGATCGCAGAGGACAAGCCTTATGCAGAGTTGT GGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTTGACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGG ACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGCAACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCC AGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCACCTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCA TTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGGCCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAA GGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCCTGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGA TGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCCCTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCAAAG >54715_54715_1_MPI-ESYT1_MPI_chr15_75185143_ENST00000323744_ESYT1_chr12_56537606_ENST00000267113_length(amino acids)=171AA_BP= MVQGASMAAPRVFPLSCAVQQYAWGKMGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGS -------------------------------------------------------------- >54715_54715_2_MPI-ESYT1_MPI_chr15_75185143_ENST00000323744_ESYT1_chr12_56537606_ENST00000394048_length(transcript)=1368nt_BP=519nt ATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTATTCCCACTTTCCTGTGCGGTGCAGCAGTATGCCTGGGGGA AGATGGGTTCCAACAGCGAAGTGGCGCGGCTGTTGGCCAGCAGTGATCCACTGGCCCAGATCGCAGAGGACAAGCCTTATGCAGAGTTGT GGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTTGACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGG ACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGCAACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCC AGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCACCTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCA TTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGGCCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAA GGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCCTGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGA TGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCCCTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCAAAG AGAAGAACCGTATGTTCCCTTTACTGCACGGCCTTTATCCTTCTGGGCCCCTGGGGCGGGGACCTGAGCTGGCTGTTTCCTGCTTTGCCT GCACATTGTTCTCCCTTCCTCCCAACTCCTCAGGGCCTTCTGTATCTGTGCCTGGCCAGTGGCAGCACTAGCAGTGGTATTAGCTTATGC CAAATACAGCTTTGGAAGGATCTTTTTTTCTTTAACTAGATGGTCACCTTCTTCCCTACCACACATGGGTGGGAAGGTGGACAGGCTAAC CTCTCCAGCTGTGAGCCTCTTAGACTACTGCATGTAGCAAATGTTCAGCAGCTCAGGCCCCCATGTCCAGTTCTGTCCCCACTGTCCTCA ACCCTGTCCTGAAAATTCTACTGCTTTGATGGCTGGGGCCAGTCTCTTGTCACTTTGGAAACTGAGGACGCGTGGATTCTACTCAAGCCT CCAAGTAGTGGCATATCAGTCTTGGAGCTCCTAGCTGGTGATACGGAGAGGGCTTTGGAGGACTTGGGACAGCAGGGCCAATTTTTTTGC CCAAGTGCCTAGGCTGCTAACTCACTGACTAGAACTTAATCTGGTACTTTACAGTTTTGCACCAACTCTGCCAAGCCACTGGATCTTACA >54715_54715_2_MPI-ESYT1_MPI_chr15_75185143_ENST00000323744_ESYT1_chr12_56537606_ENST00000394048_length(amino acids)=171AA_BP= MVQGASMAAPRVFPLSCAVQQYAWGKMGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGS -------------------------------------------------------------- >54715_54715_3_MPI-ESYT1_MPI_chr15_75185143_ENST00000323744_ESYT1_chr12_56537606_ENST00000541590_length(transcript)=888nt_BP=519nt ATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTATTCCCACTTTCCTGTGCGGTGCAGCAGTATGCCTGGGGGA AGATGGGTTCCAACAGCGAAGTGGCGCGGCTGTTGGCCAGCAGTGATCCACTGGCCCAGATCGCAGAGGACAAGCCTTATGCAGAGTTGT GGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTTGACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGG ACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGCAACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCC AGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCACCTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCA TTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGGCCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAA GGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCCTGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGA TGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCCCTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCAAAG AGAAGAACCGTATGTTCCCTTTACTGCACGGCCTTTATCCTTCTGGGCCCCTGGGGCGGGGACCTGAGCTGGCTGTTTCCTGCTTTGCCT >54715_54715_3_MPI-ESYT1_MPI_chr15_75185143_ENST00000323744_ESYT1_chr12_56537606_ENST00000541590_length(amino acids)=171AA_BP= MVQGASMAAPRVFPLSCAVQQYAWGKMGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGS -------------------------------------------------------------- >54715_54715_4_MPI-ESYT1_MPI_chr15_75185143_ENST00000352410_ESYT1_chr12_56537606_ENST00000267113_length(transcript)=773nt_BP=554nt GGGCGCAGAGGGCCGCGGGGGCTGCCGGGAAAGGCATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTATTCCC ACTTTCCTGTGCGGTGCAGCAGTATGCCTGGGGGAAGATGGGTTCCAACAGCGAAGTGGCGCGGCTGTTGGCCAGCAGTGATCCACTGGC CCAGATCGCAGAGGACAAGCCTTATGCAGAGTTGTGGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTTGACAACCGCATCTCACA GAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGGACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGCAACCTGCCCTTCCTCTT CAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCCAGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCACCTCCAGGCTCCGCAGCA CTACCCCGATGCCAACCACAAGCCAGAGATGGCCATTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGGCCAGTTGAGGAGATTGT AACCTTTCTAAAGAGTATGACCTGATGGACAACAAGGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCCTGACTGCTCTGTCTTCC TGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGATGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCCCTTCAGCCCTTTCACCT >54715_54715_4_MPI-ESYT1_MPI_chr15_75185143_ENST00000352410_ESYT1_chr12_56537606_ENST00000267113_length(amino acids)=171AA_BP= MVQGASMAAPRVFPLSCAVQQYAWGKMGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGS -------------------------------------------------------------- >54715_54715_5_MPI-ESYT1_MPI_chr15_75185143_ENST00000352410_ESYT1_chr12_56537606_ENST00000394048_length(transcript)=1403nt_BP=554nt GGGCGCAGAGGGCCGCGGGGGCTGCCGGGAAAGGCATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTATTCCC ACTTTCCTGTGCGGTGCAGCAGTATGCCTGGGGGAAGATGGGTTCCAACAGCGAAGTGGCGCGGCTGTTGGCCAGCAGTGATCCACTGGC CCAGATCGCAGAGGACAAGCCTTATGCAGAGTTGTGGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTTGACAACCGCATCTCACA GAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGGACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGCAACCTGCCCTTCCTCTT CAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCCAGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCACCTCCAGGCTCCGCAGCA CTACCCCGATGCCAACCACAAGCCAGAGATGGCCATTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGGCCAGTTGAGGAGATTGT AACCTTTCTAAAGAGTATGACCTGATGGACAACAAGGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCCTGACTGCTCTGTCTTCC TGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGATGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCCCTTCAGCCCTTTCACCT AACAGGCCCATATTCGGGCCTTTGCCTGACCAAAGAGAAGAACCGTATGTTCCCTTTACTGCACGGCCTTTATCCTTCTGGGCCCCTGGG GCGGGGACCTGAGCTGGCTGTTTCCTGCTTTGCCTGCACATTGTTCTCCCTTCCTCCCAACTCCTCAGGGCCTTCTGTATCTGTGCCTGG CCAGTGGCAGCACTAGCAGTGGTATTAGCTTATGCCAAATACAGCTTTGGAAGGATCTTTTTTTCTTTAACTAGATGGTCACCTTCTTCC CTACCACACATGGGTGGGAAGGTGGACAGGCTAACCTCTCCAGCTGTGAGCCTCTTAGACTACTGCATGTAGCAAATGTTCAGCAGCTCA GGCCCCCATGTCCAGTTCTGTCCCCACTGTCCTCAACCCTGTCCTGAAAATTCTACTGCTTTGATGGCTGGGGCCAGTCTCTTGTCACTT TGGAAACTGAGGACGCGTGGATTCTACTCAAGCCTCCAAGTAGTGGCATATCAGTCTTGGAGCTCCTAGCTGGTGATACGGAGAGGGCTT TGGAGGACTTGGGACAGCAGGGCCAATTTTTTTGCCCAAGTGCCTAGGCTGCTAACTCACTGACTAGAACTTAATCTGGTACTTTACAGT >54715_54715_5_MPI-ESYT1_MPI_chr15_75185143_ENST00000352410_ESYT1_chr12_56537606_ENST00000394048_length(amino acids)=171AA_BP= MVQGASMAAPRVFPLSCAVQQYAWGKMGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGS -------------------------------------------------------------- >54715_54715_6_MPI-ESYT1_MPI_chr15_75185143_ENST00000352410_ESYT1_chr12_56537606_ENST00000541590_length(transcript)=923nt_BP=554nt GGGCGCAGAGGGCCGCGGGGGCTGCCGGGAAAGGCATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTATTCCC ACTTTCCTGTGCGGTGCAGCAGTATGCCTGGGGGAAGATGGGTTCCAACAGCGAAGTGGCGCGGCTGTTGGCCAGCAGTGATCCACTGGC CCAGATCGCAGAGGACAAGCCTTATGCAGAGTTGTGGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTTGACAACCGCATCTCACA GAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGGACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGCAACCTGCCCTTCCTCTT CAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCCAGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCACCTCCAGGCTCCGCAGCA CTACCCCGATGCCAACCACAAGCCAGAGATGGCCATTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGGCCAGTTGAGGAGATTGT AACCTTTCTAAAGAGTATGACCTGATGGACAACAAGGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCCTGACTGCTCTGTCTTCC TGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGATGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCCCTTCAGCCCTTTCACCT AACAGGCCCATATTCGGGCCTTTGCCTGACCAAAGAGAAGAACCGTATGTTCCCTTTACTGCACGGCCTTTATCCTTCTGGGCCCCTGGG GCGGGGACCTGAGCTGGCTGTTTCCTGCTTTGCCTGCACATTGTTCTCCCTTCCTCCCAACTCCTCAGGGCCTTCTGTATCTGTGCCTGG >54715_54715_6_MPI-ESYT1_MPI_chr15_75185143_ENST00000352410_ESYT1_chr12_56537606_ENST00000541590_length(amino acids)=171AA_BP= MVQGASMAAPRVFPLSCAVQQYAWGKMGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGS -------------------------------------------------------------- >54715_54715_7_MPI-ESYT1_MPI_chr15_75185143_ENST00000535694_ESYT1_chr12_56537606_ENST00000267113_length(transcript)=610nt_BP=391nt ATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTTGTGGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTT GACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGGACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGC AACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCCAGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCAC CTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCATTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGG CCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAAGGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCC TGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGATGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCC >54715_54715_7_MPI-ESYT1_MPI_chr15_75185143_ENST00000535694_ESYT1_chr12_56537606_ENST00000267113_length(amino acids)=117AA_BP= MWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGSKVKDTFNGNLPFLFKVLSVETPLSIQAHPNKELAEKLHLQAPQHYPDANHKPEM -------------------------------------------------------------- >54715_54715_8_MPI-ESYT1_MPI_chr15_75185143_ENST00000535694_ESYT1_chr12_56537606_ENST00000394048_length(transcript)=1240nt_BP=391nt ATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTTGTGGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTT GACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGGACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGC AACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCCAGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCAC CTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCATTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGG CCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAAGGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCC TGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGATGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCC CTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCAAAGAGAAGAACCGTATGTTCCCTTTACTGCACGGCCTTTAT CCTTCTGGGCCCCTGGGGCGGGGACCTGAGCTGGCTGTTTCCTGCTTTGCCTGCACATTGTTCTCCCTTCCTCCCAACTCCTCAGGGCCT TCTGTATCTGTGCCTGGCCAGTGGCAGCACTAGCAGTGGTATTAGCTTATGCCAAATACAGCTTTGGAAGGATCTTTTTTTCTTTAACTA GATGGTCACCTTCTTCCCTACCACACATGGGTGGGAAGGTGGACAGGCTAACCTCTCCAGCTGTGAGCCTCTTAGACTACTGCATGTAGC AAATGTTCAGCAGCTCAGGCCCCCATGTCCAGTTCTGTCCCCACTGTCCTCAACCCTGTCCTGAAAATTCTACTGCTTTGATGGCTGGGG CCAGTCTCTTGTCACTTTGGAAACTGAGGACGCGTGGATTCTACTCAAGCCTCCAAGTAGTGGCATATCAGTCTTGGAGCTCCTAGCTGG TGATACGGAGAGGGCTTTGGAGGACTTGGGACAGCAGGGCCAATTTTTTTGCCCAAGTGCCTAGGCTGCTAACTCACTGACTAGAACTTA >54715_54715_8_MPI-ESYT1_MPI_chr15_75185143_ENST00000535694_ESYT1_chr12_56537606_ENST00000394048_length(amino acids)=117AA_BP= MWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGSKVKDTFNGNLPFLFKVLSVETPLSIQAHPNKELAEKLHLQAPQHYPDANHKPEM -------------------------------------------------------------- >54715_54715_9_MPI-ESYT1_MPI_chr15_75185143_ENST00000535694_ESYT1_chr12_56537606_ENST00000541590_length(transcript)=760nt_BP=391nt ATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTTGTGGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTT GACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGGACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGC AACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCCAGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCAC CTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCATTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGG CCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAAGGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCC TGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGATGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCC CTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCAAAGAGAAGAACCGTATGTTCCCTTTACTGCACGGCCTTTAT CCTTCTGGGCCCCTGGGGCGGGGACCTGAGCTGGCTGTTTCCTGCTTTGCCTGCACATTGTTCTCCCTTCCTCCCAACTCCTCAGGGCCT >54715_54715_9_MPI-ESYT1_MPI_chr15_75185143_ENST00000535694_ESYT1_chr12_56537606_ENST00000541590_length(amino acids)=117AA_BP= MWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGSKVKDTFNGNLPFLFKVLSVETPLSIQAHPNKELAEKLHLQAPQHYPDANHKPEM -------------------------------------------------------------- >54715_54715_10_MPI-ESYT1_MPI_chr15_75185143_ENST00000562606_ESYT1_chr12_56537606_ENST00000267113_length(transcript)=701nt_BP=482nt ATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGCAGTATGCCTGGGGGAAGATGGGTTCCAACAGCGAAGTGGCGCGGCTGTTGGC CAGCAGTGATCCACTGGCCCAGATCGCAGAGGACAAGCCTTATGCAGAGTTGTGGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCT TGACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGGACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGG CAACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCCAGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCA CCTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCATTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCG GCCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAAGGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGC CTGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGATGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGC >54715_54715_10_MPI-ESYT1_MPI_chr15_75185143_ENST00000562606_ESYT1_chr12_56537606_ENST00000267113_length(amino acids)=145AA_BP= MGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGSKVKDTFNGNLPFLFKVLSVETPLSIQ -------------------------------------------------------------- >54715_54715_11_MPI-ESYT1_MPI_chr15_75185143_ENST00000562606_ESYT1_chr12_56537606_ENST00000394048_length(transcript)=1331nt_BP=482nt ATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGCAGTATGCCTGGGGGAAGATGGGTTCCAACAGCGAAGTGGCGCGGCTGTTGGC CAGCAGTGATCCACTGGCCCAGATCGCAGAGGACAAGCCTTATGCAGAGTTGTGGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCT TGACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGGACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGG CAACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCCAGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCA CCTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCATTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCG GCCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAAGGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGC CTGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGATGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGC CCTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCAAAGAGAAGAACCGTATGTTCCCTTTACTGCACGGCCTTTA TCCTTCTGGGCCCCTGGGGCGGGGACCTGAGCTGGCTGTTTCCTGCTTTGCCTGCACATTGTTCTCCCTTCCTCCCAACTCCTCAGGGCC TTCTGTATCTGTGCCTGGCCAGTGGCAGCACTAGCAGTGGTATTAGCTTATGCCAAATACAGCTTTGGAAGGATCTTTTTTTCTTTAACT AGATGGTCACCTTCTTCCCTACCACACATGGGTGGGAAGGTGGACAGGCTAACCTCTCCAGCTGTGAGCCTCTTAGACTACTGCATGTAG CAAATGTTCAGCAGCTCAGGCCCCCATGTCCAGTTCTGTCCCCACTGTCCTCAACCCTGTCCTGAAAATTCTACTGCTTTGATGGCTGGG GCCAGTCTCTTGTCACTTTGGAAACTGAGGACGCGTGGATTCTACTCAAGCCTCCAAGTAGTGGCATATCAGTCTTGGAGCTCCTAGCTG GTGATACGGAGAGGGCTTTGGAGGACTTGGGACAGCAGGGCCAATTTTTTTGCCCAAGTGCCTAGGCTGCTAACTCACTGACTAGAACTT >54715_54715_11_MPI-ESYT1_MPI_chr15_75185143_ENST00000562606_ESYT1_chr12_56537606_ENST00000394048_length(amino acids)=145AA_BP= MGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGSKVKDTFNGNLPFLFKVLSVETPLSIQ -------------------------------------------------------------- >54715_54715_12_MPI-ESYT1_MPI_chr15_75185143_ENST00000562606_ESYT1_chr12_56537606_ENST00000541590_length(transcript)=851nt_BP=482nt ATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGCAGTATGCCTGGGGGAAGATGGGTTCCAACAGCGAAGTGGCGCGGCTGTTGGC CAGCAGTGATCCACTGGCCCAGATCGCAGAGGACAAGCCTTATGCAGAGTTGTGGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCT TGACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGGACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGG CAACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCCAGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCA CCTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCATTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCG GCCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAAGGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGC CTGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGATGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGC CCTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCAAAGAGAAGAACCGTATGTTCCCTTTACTGCACGGCCTTTA TCCTTCTGGGCCCCTGGGGCGGGGACCTGAGCTGGCTGTTTCCTGCTTTGCCTGCACATTGTTCTCCCTTCCTCCCAACTCCTCAGGGCC >54715_54715_12_MPI-ESYT1_MPI_chr15_75185143_ENST00000562606_ESYT1_chr12_56537606_ENST00000541590_length(amino acids)=145AA_BP= MGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGSKVKDTFNGNLPFLFKVLSVETPLSIQ -------------------------------------------------------------- >54715_54715_13_MPI-ESYT1_MPI_chr15_75185143_ENST00000563422_ESYT1_chr12_56537606_ENST00000267113_length(transcript)=738nt_BP=519nt ATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTATTCCCACTTTCCTGTGCGGTGCAGCAGTATGCCTGGGGGA AGATGGGTTCCAACAGCGAAGTGGCGCGGCTGTTGGCCAGCAGTGATCCACTGGCCCAGATCGCAGAGGACAAGCCTTATGCAGAGTTGT GGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTTGACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGG ACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGCAACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCC AGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCACCTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCA TTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGGCCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAA GGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCCTGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGA TGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCCCTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCAAAG >54715_54715_13_MPI-ESYT1_MPI_chr15_75185143_ENST00000563422_ESYT1_chr12_56537606_ENST00000267113_length(amino acids)=171AA_BP= MVQGASMAAPRVFPLSCAVQQYAWGKMGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGS -------------------------------------------------------------- >54715_54715_14_MPI-ESYT1_MPI_chr15_75185143_ENST00000563422_ESYT1_chr12_56537606_ENST00000394048_length(transcript)=1368nt_BP=519nt ATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTATTCCCACTTTCCTGTGCGGTGCAGCAGTATGCCTGGGGGA AGATGGGTTCCAACAGCGAAGTGGCGCGGCTGTTGGCCAGCAGTGATCCACTGGCCCAGATCGCAGAGGACAAGCCTTATGCAGAGTTGT GGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTTGACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGG ACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGCAACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCC AGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCACCTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCA TTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGGCCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAA GGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCCTGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGA TGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCCCTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCAAAG AGAAGAACCGTATGTTCCCTTTACTGCACGGCCTTTATCCTTCTGGGCCCCTGGGGCGGGGACCTGAGCTGGCTGTTTCCTGCTTTGCCT GCACATTGTTCTCCCTTCCTCCCAACTCCTCAGGGCCTTCTGTATCTGTGCCTGGCCAGTGGCAGCACTAGCAGTGGTATTAGCTTATGC CAAATACAGCTTTGGAAGGATCTTTTTTTCTTTAACTAGATGGTCACCTTCTTCCCTACCACACATGGGTGGGAAGGTGGACAGGCTAAC CTCTCCAGCTGTGAGCCTCTTAGACTACTGCATGTAGCAAATGTTCAGCAGCTCAGGCCCCCATGTCCAGTTCTGTCCCCACTGTCCTCA ACCCTGTCCTGAAAATTCTACTGCTTTGATGGCTGGGGCCAGTCTCTTGTCACTTTGGAAACTGAGGACGCGTGGATTCTACTCAAGCCT CCAAGTAGTGGCATATCAGTCTTGGAGCTCCTAGCTGGTGATACGGAGAGGGCTTTGGAGGACTTGGGACAGCAGGGCCAATTTTTTTGC CCAAGTGCCTAGGCTGCTAACTCACTGACTAGAACTTAATCTGGTACTTTACAGTTTTGCACCAACTCTGCCAAGCCACTGGATCTTACA >54715_54715_14_MPI-ESYT1_MPI_chr15_75185143_ENST00000563422_ESYT1_chr12_56537606_ENST00000394048_length(amino acids)=171AA_BP= MVQGASMAAPRVFPLSCAVQQYAWGKMGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGS -------------------------------------------------------------- >54715_54715_15_MPI-ESYT1_MPI_chr15_75185143_ENST00000563422_ESYT1_chr12_56537606_ENST00000541590_length(transcript)=888nt_BP=519nt ATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTATTCCCACTTTCCTGTGCGGTGCAGCAGTATGCCTGGGGGA AGATGGGTTCCAACAGCGAAGTGGCGCGGCTGTTGGCCAGCAGTGATCCACTGGCCCAGATCGCAGAGGACAAGCCTTATGCAGAGTTGT GGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTTGACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGG ACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGCAACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCC AGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCACCTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCA TTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGGCCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAA GGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCCTGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGA TGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCCCTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCAAAG AGAAGAACCGTATGTTCCCTTTACTGCACGGCCTTTATCCTTCTGGGCCCCTGGGGCGGGGACCTGAGCTGGCTGTTTCCTGCTTTGCCT >54715_54715_15_MPI-ESYT1_MPI_chr15_75185143_ENST00000563422_ESYT1_chr12_56537606_ENST00000541590_length(amino acids)=171AA_BP= MVQGASMAAPRVFPLSCAVQQYAWGKMGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGS -------------------------------------------------------------- >54715_54715_16_MPI-ESYT1_MPI_chr15_75185143_ENST00000563786_ESYT1_chr12_56537606_ENST00000267113_length(transcript)=827nt_BP=608nt ATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTGAGCACCCCCAGCTCTTGCCTGGTTTCCCGGGACACTAGGG TGGGGCCTGAGGAGTGGAGTGGCAGCTGACCCTGTCTGTGCCCCTAGTATTCCCACTTTCCTGTGCGGTGCAGCAGTATGCCTGGGGGAA GATGGGTTCCAACAGCGAAGTGGCGCGGCTGTTGGCCAGCAGTGATCCACTGGCCCAGATCGCAGAGGACAAGCCTTATGCAGAGTTGTG GATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTTGACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGGA CAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGCAACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCCA GGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCACCTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCAT TGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGGCCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAAG GACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCCTGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGAT GAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCCCTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCAAAGA >54715_54715_16_MPI-ESYT1_MPI_chr15_75185143_ENST00000563786_ESYT1_chr12_56537606_ENST00000267113_length(amino acids)=167AA_BP= MTLSVPLVFPLSCAVQQYAWGKMGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGSKVKD -------------------------------------------------------------- >54715_54715_17_MPI-ESYT1_MPI_chr15_75185143_ENST00000563786_ESYT1_chr12_56537606_ENST00000394048_length(transcript)=1457nt_BP=608nt ATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTGAGCACCCCCAGCTCTTGCCTGGTTTCCCGGGACACTAGGG TGGGGCCTGAGGAGTGGAGTGGCAGCTGACCCTGTCTGTGCCCCTAGTATTCCCACTTTCCTGTGCGGTGCAGCAGTATGCCTGGGGGAA GATGGGTTCCAACAGCGAAGTGGCGCGGCTGTTGGCCAGCAGTGATCCACTGGCCCAGATCGCAGAGGACAAGCCTTATGCAGAGTTGTG GATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTTGACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGGA CAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGCAACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCCA GGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCACCTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCAT TGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGGCCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAAG GACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCCTGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGAT GAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCCCTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCAAAGA GAAGAACCGTATGTTCCCTTTACTGCACGGCCTTTATCCTTCTGGGCCCCTGGGGCGGGGACCTGAGCTGGCTGTTTCCTGCTTTGCCTG CACATTGTTCTCCCTTCCTCCCAACTCCTCAGGGCCTTCTGTATCTGTGCCTGGCCAGTGGCAGCACTAGCAGTGGTATTAGCTTATGCC AAATACAGCTTTGGAAGGATCTTTTTTTCTTTAACTAGATGGTCACCTTCTTCCCTACCACACATGGGTGGGAAGGTGGACAGGCTAACC TCTCCAGCTGTGAGCCTCTTAGACTACTGCATGTAGCAAATGTTCAGCAGCTCAGGCCCCCATGTCCAGTTCTGTCCCCACTGTCCTCAA CCCTGTCCTGAAAATTCTACTGCTTTGATGGCTGGGGCCAGTCTCTTGTCACTTTGGAAACTGAGGACGCGTGGATTCTACTCAAGCCTC CAAGTAGTGGCATATCAGTCTTGGAGCTCCTAGCTGGTGATACGGAGAGGGCTTTGGAGGACTTGGGACAGCAGGGCCAATTTTTTTGCC CAAGTGCCTAGGCTGCTAACTCACTGACTAGAACTTAATCTGGTACTTTACAGTTTTGCACCAACTCTGCCAAGCCACTGGATCTTACAT >54715_54715_17_MPI-ESYT1_MPI_chr15_75185143_ENST00000563786_ESYT1_chr12_56537606_ENST00000394048_length(amino acids)=167AA_BP= MTLSVPLVFPLSCAVQQYAWGKMGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGSKVKD -------------------------------------------------------------- >54715_54715_18_MPI-ESYT1_MPI_chr15_75185143_ENST00000563786_ESYT1_chr12_56537606_ENST00000541590_length(transcript)=977nt_BP=608nt ATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTGAGCACCCCCAGCTCTTGCCTGGTTTCCCGGGACACTAGGG TGGGGCCTGAGGAGTGGAGTGGCAGCTGACCCTGTCTGTGCCCCTAGTATTCCCACTTTCCTGTGCGGTGCAGCAGTATGCCTGGGGGAA GATGGGTTCCAACAGCGAAGTGGCGCGGCTGTTGGCCAGCAGTGATCCACTGGCCCAGATCGCAGAGGACAAGCCTTATGCAGAGTTGTG GATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTTGACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGGA CAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGCAACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCCA GGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCACCTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCAT TGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGGCCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAAG GACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCCTGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGAT GAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCCCTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCAAAGA GAAGAACCGTATGTTCCCTTTACTGCACGGCCTTTATCCTTCTGGGCCCCTGGGGCGGGGACCTGAGCTGGCTGTTTCCTGCTTTGCCTG >54715_54715_18_MPI-ESYT1_MPI_chr15_75185143_ENST00000563786_ESYT1_chr12_56537606_ENST00000541590_length(amino acids)=167AA_BP= MTLSVPLVFPLSCAVQQYAWGKMGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGSKVKD -------------------------------------------------------------- >54715_54715_19_MPI-ESYT1_MPI_chr15_75185143_ENST00000564003_ESYT1_chr12_56537606_ENST00000267113_length(transcript)=610nt_BP=391nt ATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTTGTGGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTT GACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGGACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGC AACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCCAGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCAC CTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCATTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGG CCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAAGGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCC TGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGATGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCC >54715_54715_19_MPI-ESYT1_MPI_chr15_75185143_ENST00000564003_ESYT1_chr12_56537606_ENST00000267113_length(amino acids)=117AA_BP= MWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGSKVKDTFNGNLPFLFKVLSVETPLSIQAHPNKELAEKLHLQAPQHYPDANHKPEM -------------------------------------------------------------- >54715_54715_20_MPI-ESYT1_MPI_chr15_75185143_ENST00000564003_ESYT1_chr12_56537606_ENST00000394048_length(transcript)=1240nt_BP=391nt ATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTTGTGGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTT GACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGGACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGC AACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCCAGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCAC CTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCATTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGG CCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAAGGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCC TGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGATGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCC CTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCAAAGAGAAGAACCGTATGTTCCCTTTACTGCACGGCCTTTAT CCTTCTGGGCCCCTGGGGCGGGGACCTGAGCTGGCTGTTTCCTGCTTTGCCTGCACATTGTTCTCCCTTCCTCCCAACTCCTCAGGGCCT TCTGTATCTGTGCCTGGCCAGTGGCAGCACTAGCAGTGGTATTAGCTTATGCCAAATACAGCTTTGGAAGGATCTTTTTTTCTTTAACTA GATGGTCACCTTCTTCCCTACCACACATGGGTGGGAAGGTGGACAGGCTAACCTCTCCAGCTGTGAGCCTCTTAGACTACTGCATGTAGC AAATGTTCAGCAGCTCAGGCCCCCATGTCCAGTTCTGTCCCCACTGTCCTCAACCCTGTCCTGAAAATTCTACTGCTTTGATGGCTGGGG CCAGTCTCTTGTCACTTTGGAAACTGAGGACGCGTGGATTCTACTCAAGCCTCCAAGTAGTGGCATATCAGTCTTGGAGCTCCTAGCTGG TGATACGGAGAGGGCTTTGGAGGACTTGGGACAGCAGGGCCAATTTTTTTGCCCAAGTGCCTAGGCTGCTAACTCACTGACTAGAACTTA >54715_54715_20_MPI-ESYT1_MPI_chr15_75185143_ENST00000564003_ESYT1_chr12_56537606_ENST00000394048_length(amino acids)=117AA_BP= MWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGSKVKDTFNGNLPFLFKVLSVETPLSIQAHPNKELAEKLHLQAPQHYPDANHKPEM -------------------------------------------------------------- >54715_54715_21_MPI-ESYT1_MPI_chr15_75185143_ENST00000564003_ESYT1_chr12_56537606_ENST00000541590_length(transcript)=760nt_BP=391nt ATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTTGTGGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTT GACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGGACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGC AACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCCAGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCAC CTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCATTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGG CCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAAGGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCC TGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGATGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCC CTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCAAAGAGAAGAACCGTATGTTCCCTTTACTGCACGGCCTTTAT CCTTCTGGGCCCCTGGGGCGGGGACCTGAGCTGGCTGTTTCCTGCTTTGCCTGCACATTGTTCTCCCTTCCTCCCAACTCCTCAGGGCCT >54715_54715_21_MPI-ESYT1_MPI_chr15_75185143_ENST00000564003_ESYT1_chr12_56537606_ENST00000541590_length(amino acids)=117AA_BP= MWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGSKVKDTFNGNLPFLFKVLSVETPLSIQAHPNKELAEKLHLQAPQHYPDANHKPEM -------------------------------------------------------------- >54715_54715_22_MPI-ESYT1_MPI_chr15_75185143_ENST00000565576_ESYT1_chr12_56537606_ENST00000267113_length(transcript)=718nt_BP=499nt CAGGGGGCGAGCATGGCCGCTCCGCGAGTATTCCCACTTTCCTGTGCGGTGCAGCAGTATGCCTGGGGGAAGATGGGTTCCAACAGCGAA GTGGCGCGGCTGTTGGCCAGCAGTGATCCACTGGCCCAGATCGCAGAGGACAAGCCTTATGCAGAGTTGTGGATGGGGACTCACCCCCGA GGGGATGCCAAGATCCTTGACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGGACAGCTTGGGCTCAAAGGTC AAGGACACCTTTAATGGCAACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCCAGGCACACCCTAACAAGGAG CTGGCAGAGAAGCTGCACCTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCATTGCCCTCACCCCCTTCCAG GGCTTGTGTGGCTTCCGGCCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAAGGACAAGGGCAGCTCCTAGG AGCTGGCGAGTCCCAGCCTGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGATGAGCCTAAAGCTAGGGTCC >54715_54715_22_MPI-ESYT1_MPI_chr15_75185143_ENST00000565576_ESYT1_chr12_56537606_ENST00000267113_length(amino acids)=165AA_BP= MAAPRVFPLSCAVQQYAWGKMGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGSKVKDTF -------------------------------------------------------------- >54715_54715_23_MPI-ESYT1_MPI_chr15_75185143_ENST00000565576_ESYT1_chr12_56537606_ENST00000394048_length(transcript)=1348nt_BP=499nt CAGGGGGCGAGCATGGCCGCTCCGCGAGTATTCCCACTTTCCTGTGCGGTGCAGCAGTATGCCTGGGGGAAGATGGGTTCCAACAGCGAA GTGGCGCGGCTGTTGGCCAGCAGTGATCCACTGGCCCAGATCGCAGAGGACAAGCCTTATGCAGAGTTGTGGATGGGGACTCACCCCCGA GGGGATGCCAAGATCCTTGACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGGACAGCTTGGGCTCAAAGGTC AAGGACACCTTTAATGGCAACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCCAGGCACACCCTAACAAGGAG CTGGCAGAGAAGCTGCACCTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCATTGCCCTCACCCCCTTCCAG GGCTTGTGTGGCTTCCGGCCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAAGGACAAGGGCAGCTCCTAGG AGCTGGCGAGTCCCAGCCTGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGATGAGCCTAAAGCTAGGGTCC AAGGGCAGAGCCTGTGCCCTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCAAAGAGAAGAACCGTATGTTCCCT TTACTGCACGGCCTTTATCCTTCTGGGCCCCTGGGGCGGGGACCTGAGCTGGCTGTTTCCTGCTTTGCCTGCACATTGTTCTCCCTTCCT CCCAACTCCTCAGGGCCTTCTGTATCTGTGCCTGGCCAGTGGCAGCACTAGCAGTGGTATTAGCTTATGCCAAATACAGCTTTGGAAGGA TCTTTTTTTCTTTAACTAGATGGTCACCTTCTTCCCTACCACACATGGGTGGGAAGGTGGACAGGCTAACCTCTCCAGCTGTGAGCCTCT TAGACTACTGCATGTAGCAAATGTTCAGCAGCTCAGGCCCCCATGTCCAGTTCTGTCCCCACTGTCCTCAACCCTGTCCTGAAAATTCTA CTGCTTTGATGGCTGGGGCCAGTCTCTTGTCACTTTGGAAACTGAGGACGCGTGGATTCTACTCAAGCCTCCAAGTAGTGGCATATCAGT CTTGGAGCTCCTAGCTGGTGATACGGAGAGGGCTTTGGAGGACTTGGGACAGCAGGGCCAATTTTTTTGCCCAAGTGCCTAGGCTGCTAA >54715_54715_23_MPI-ESYT1_MPI_chr15_75185143_ENST00000565576_ESYT1_chr12_56537606_ENST00000394048_length(amino acids)=165AA_BP= MAAPRVFPLSCAVQQYAWGKMGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGSKVKDTF -------------------------------------------------------------- >54715_54715_24_MPI-ESYT1_MPI_chr15_75185143_ENST00000565576_ESYT1_chr12_56537606_ENST00000541590_length(transcript)=868nt_BP=499nt CAGGGGGCGAGCATGGCCGCTCCGCGAGTATTCCCACTTTCCTGTGCGGTGCAGCAGTATGCCTGGGGGAAGATGGGTTCCAACAGCGAA GTGGCGCGGCTGTTGGCCAGCAGTGATCCACTGGCCCAGATCGCAGAGGACAAGCCTTATGCAGAGTTGTGGATGGGGACTCACCCCCGA GGGGATGCCAAGATCCTTGACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACCAGGACAGCTTGGGCTCAAAGGTC AAGGACACCTTTAATGGCAACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCATCCAGGCACACCCTAACAAGGAG CTGGCAGAGAAGCTGCACCTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGGCCATTGCCCTCACCCCCTTCCAG GGCTTGTGTGGCTTCCGGCCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAACAAGGACAAGGGCAGCTCCTAGG AGCTGGCGAGTCCCAGCCTGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATGTGATGAGCCTAAAGCTAGGGTCC AAGGGCAGAGCCTGTGCCCTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCAAAGAGAAGAACCGTATGTTCCCT TTACTGCACGGCCTTTATCCTTCTGGGCCCCTGGGGCGGGGACCTGAGCTGGCTGTTTCCTGCTTTGCCTGCACATTGTTCTCCCTTCCT >54715_54715_24_MPI-ESYT1_MPI_chr15_75185143_ENST00000565576_ESYT1_chr12_56537606_ENST00000541590_length(amino acids)=165AA_BP= MAAPRVFPLSCAVQQYAWGKMGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGSKVKDTF -------------------------------------------------------------- >54715_54715_25_MPI-ESYT1_MPI_chr15_75185143_ENST00000566377_ESYT1_chr12_56537606_ENST00000267113_length(transcript)=741nt_BP=522nt GGCATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTATTCCCACTTTCCTGTGCGGTGCAGCAGTATGCCTGGG GGAAGATGGGTTCCAACAGCGAAGTGGCGCGGCTGTTGGCCAGCAGTGATCCACTGGCCCAGATCGCAGAGGACAAGCCTTATGCAGAGT TGTGGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTTGACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACC AGGACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGCAACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCA TCCAGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCACCTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGG CCATTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGGCCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAA CAAGGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCCTGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATG TGATGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCCCTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCA >54715_54715_25_MPI-ESYT1_MPI_chr15_75185143_ENST00000566377_ESYT1_chr12_56537606_ENST00000267113_length(amino acids)=171AA_BP= MVQGASMAAPRVFPLSCAVQQYAWGKMGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGS -------------------------------------------------------------- >54715_54715_26_MPI-ESYT1_MPI_chr15_75185143_ENST00000566377_ESYT1_chr12_56537606_ENST00000394048_length(transcript)=1371nt_BP=522nt GGCATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTATTCCCACTTTCCTGTGCGGTGCAGCAGTATGCCTGGG GGAAGATGGGTTCCAACAGCGAAGTGGCGCGGCTGTTGGCCAGCAGTGATCCACTGGCCCAGATCGCAGAGGACAAGCCTTATGCAGAGT TGTGGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTTGACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACC AGGACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGCAACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCA TCCAGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCACCTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGG CCATTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGGCCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAA CAAGGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCCTGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATG TGATGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCCCTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCA AAGAGAAGAACCGTATGTTCCCTTTACTGCACGGCCTTTATCCTTCTGGGCCCCTGGGGCGGGGACCTGAGCTGGCTGTTTCCTGCTTTG CCTGCACATTGTTCTCCCTTCCTCCCAACTCCTCAGGGCCTTCTGTATCTGTGCCTGGCCAGTGGCAGCACTAGCAGTGGTATTAGCTTA TGCCAAATACAGCTTTGGAAGGATCTTTTTTTCTTTAACTAGATGGTCACCTTCTTCCCTACCACACATGGGTGGGAAGGTGGACAGGCT AACCTCTCCAGCTGTGAGCCTCTTAGACTACTGCATGTAGCAAATGTTCAGCAGCTCAGGCCCCCATGTCCAGTTCTGTCCCCACTGTCC TCAACCCTGTCCTGAAAATTCTACTGCTTTGATGGCTGGGGCCAGTCTCTTGTCACTTTGGAAACTGAGGACGCGTGGATTCTACTCAAG CCTCCAAGTAGTGGCATATCAGTCTTGGAGCTCCTAGCTGGTGATACGGAGAGGGCTTTGGAGGACTTGGGACAGCAGGGCCAATTTTTT TGCCCAAGTGCCTAGGCTGCTAACTCACTGACTAGAACTTAATCTGGTACTTTACAGTTTTGCACCAACTCTGCCAAGCCACTGGATCTT >54715_54715_26_MPI-ESYT1_MPI_chr15_75185143_ENST00000566377_ESYT1_chr12_56537606_ENST00000394048_length(amino acids)=171AA_BP= MVQGASMAAPRVFPLSCAVQQYAWGKMGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGS -------------------------------------------------------------- >54715_54715_27_MPI-ESYT1_MPI_chr15_75185143_ENST00000566377_ESYT1_chr12_56537606_ENST00000541590_length(transcript)=891nt_BP=522nt GGCATACGTGCTTAATCCTGGTGCAGGGGGCGAGCATGGCCGCTCCGCGAGTATTCCCACTTTCCTGTGCGGTGCAGCAGTATGCCTGGG GGAAGATGGGTTCCAACAGCGAAGTGGCGCGGCTGTTGGCCAGCAGTGATCCACTGGCCCAGATCGCAGAGGACAAGCCTTATGCAGAGT TGTGGATGGGGACTCACCCCCGAGGGGATGCCAAGATCCTTGACAACCGCATCTCACAGAAGACCCTAAGCCAGTGGATTGCTGAGAACC AGGACAGCTTGGGCTCAAAGGTCAAGGACACCTTTAATGGCAACCTGCCCTTCCTCTTCAAAGTGCTCTCAGTTGAAACACCCCTGTCCA TCCAGGCACACCCTAACAAGGAGCTGGCAGAGAAGCTGCACCTCCAGGCTCCGCAGCACTACCCCGATGCCAACCACAAGCCAGAGATGG CCATTGCCCTCACCCCCTTCCAGGGCTTGTGTGGCTTCCGGCCAGTTGAGGAGATTGTAACCTTTCTAAAGAGTATGACCTGATGGACAA CAAGGACAAGGGCAGCTCCTAGGAGCTGGCGAGTCCCAGCCTGACTGCTCTGTCTTCCTGCCTTCGTCTCGCTCCATCACCGCCTCAATG TGATGAGCCTAAAGCTAGGGTCCAAGGGCAGAGCCTGTGCCCTTCAGCCCTTTCACCTAACAGGCCCATATTCGGGCCTTTGCCTGACCA AAGAGAAGAACCGTATGTTCCCTTTACTGCACGGCCTTTATCCTTCTGGGCCCCTGGGGCGGGGACCTGAGCTGGCTGTTTCCTGCTTTG >54715_54715_27_MPI-ESYT1_MPI_chr15_75185143_ENST00000566377_ESYT1_chr12_56537606_ENST00000541590_length(amino acids)=171AA_BP= MVQGASMAAPRVFPLSCAVQQYAWGKMGSNSEVARLLASSDPLAQIAEDKPYAELWMGTHPRGDAKILDNRISQKTLSQWIAENQDSLGS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MPI-ESYT1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MPI-ESYT1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MPI-ESYT1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
