
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MPP5-COX16 (FusionGDB2 ID:54745) |
Fusion Gene Summary for MPP5-COX16 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MPP5-COX16 | Fusion gene ID: 54745 | Hgene | Tgene | Gene symbol | MPP5 | COX16 | Gene ID | 64398 | 51241 |
| Gene name | membrane palmitoylated protein 5 | cytochrome c oxidase assembly factor COX16 | |
| Synonyms | PALS1 | C14orf112|HSPC203|hCOX16 | |
| Cytomap | 14q23.3 | 14q24.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | MAGUK p55 subfamily member 5membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5)protein associated with Lin-7 1protein associated with lin seven 1stardust | cytochrome c oxidase assembly protein COX16 homolog, mitochondrialCOX16, cytochrome c oxidase assembly homologcytochrome c oxidase assembly factor | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q8N3R9 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000261681, ENST00000555925, ENST00000554911, ENST00000556345, | ENST00000557612, ENST00000389912, | |
| Fusion gene scores | * DoF score | 8 X 10 X 12=960 | 2 X 1 X 2=4 |
| # samples | 23 | 2 | |
| ** MAII score | log2(23/960*10)=-2.06140054466414 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(2/4*10)=2.32192809488736 | |
| Context | PubMed: MPP5 [Title/Abstract] AND COX16 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MPP5(67790529)-COX16(70795953), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | COX16 | GO:0033617 | mitochondrial respiratory chain complex IV assembly | 29381136 |
 Fusion gene breakpoints across MPP5 (5'-gene) Fusion gene breakpoints across MPP5 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
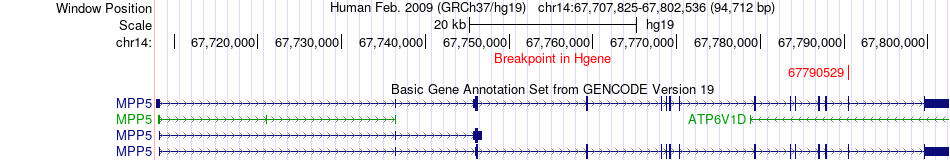 |
 Fusion gene breakpoints across COX16 (3'-gene) Fusion gene breakpoints across COX16 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
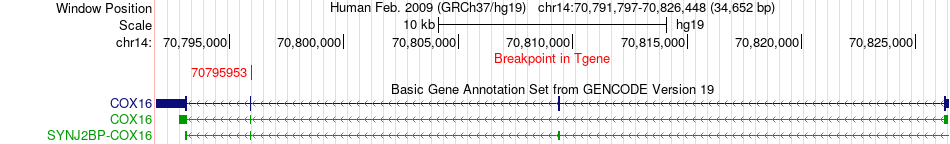 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | HNSC | TCGA-DQ-7594 | MPP5 | chr14 | 67790529 | + | COX16 | chr14 | 70795953 | - |
Top |
Fusion Gene ORF analysis for MPP5-COX16 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000261681 | ENST00000557612 | MPP5 | chr14 | 67790529 | + | COX16 | chr14 | 70795953 | - |
| 5CDS-5UTR | ENST00000555925 | ENST00000557612 | MPP5 | chr14 | 67790529 | + | COX16 | chr14 | 70795953 | - |
| In-frame | ENST00000261681 | ENST00000389912 | MPP5 | chr14 | 67790529 | + | COX16 | chr14 | 70795953 | - |
| In-frame | ENST00000555925 | ENST00000389912 | MPP5 | chr14 | 67790529 | + | COX16 | chr14 | 70795953 | - |
| intron-3CDS | ENST00000554911 | ENST00000389912 | MPP5 | chr14 | 67790529 | + | COX16 | chr14 | 70795953 | - |
| intron-3CDS | ENST00000556345 | ENST00000389912 | MPP5 | chr14 | 67790529 | + | COX16 | chr14 | 70795953 | - |
| intron-5UTR | ENST00000554911 | ENST00000557612 | MPP5 | chr14 | 67790529 | + | COX16 | chr14 | 70795953 | - |
| intron-5UTR | ENST00000556345 | ENST00000557612 | MPP5 | chr14 | 67790529 | + | COX16 | chr14 | 70795953 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000261681 | MPP5 | chr14 | 67790529 | + | ENST00000389912 | COX16 | chr14 | 70795953 | - | 3944 | 2512 | 661 | 2691 | 676 |
| ENST00000555925 | MPP5 | chr14 | 67790529 | + | ENST00000389912 | COX16 | chr14 | 70795953 | - | 3398 | 1966 | 217 | 2145 | 642 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000261681 | ENST00000389912 | MPP5 | chr14 | 67790529 | + | COX16 | chr14 | 70795953 | - | 0.001082823 | 0.9989172 |
| ENST00000555925 | ENST00000389912 | MPP5 | chr14 | 67790529 | + | COX16 | chr14 | 70795953 | - | 0.000722207 | 0.9992778 |
Top |
Fusion Genomic Features for MPP5-COX16 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
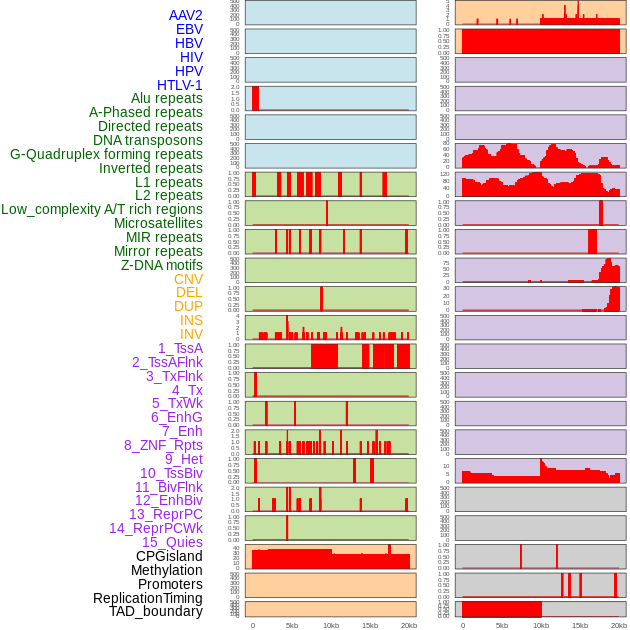 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for MPP5-COX16 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr14:67790529/chr14:70795953) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MPP5 | . |
| FUNCTION: Plays a role in tight junction biogenesis and in the establishment of cell polarity in epithelial cells (PubMed:16678097, PubMed:25385611). Also involved in adherens junction biogenesis by ensuring correct localization of the exocyst complex protein EXOC4/SEC8 which allows trafficking of adherens junction structural component CDH1 to the cell surface (By similarity). Plays a role through its interaction with CDH5 in vascular lumen formation and endothelial membrane polarity (PubMed:27466317). Required during embryonic and postnatal retinal development (By similarity). Required for the maintenance of cerebellar progenitor cells in an undifferentiated proliferative state, preventing premature differentiation, and is required for cerebellar histogenesis, fissure formation, cerebellar layer organization and cortical development (By similarity). Plays a role in neuronal progenitor cell survival, potentially via promotion of mTOR signaling (By similarity). Plays a role in the radial and longitudinal extension of the myelin sheath in Schwann cells (By similarity). May modulate SC6A1/GAT1-mediated GABA uptake by stabilizing the transporter (By similarity). Plays a role in the T-cell receptor-mediated activation of NF-kappa-B (PubMed:21479189). Required for localization of EZR to the apical membrane of parietal cells and may play a role in the dynamic remodeling of the apical cytoskeleton (By similarity). Required for the normal polarized localization of the vesicular marker STX4 (By similarity). Required for the correct trafficking of the myelin proteins PMP22 and MAG (By similarity). Involved in promoting phosphorylation and cytoplasmic retention of transcriptional coactivators YAP1 and WWTR1/TAZ which leads to suppression of TGFB1-dependent transcription of target genes such as CCN2/CTGF, SERPINE1/PAI1, SNAI1/SNAIL1 and SMAD7 (By similarity). {ECO:0000250|UniProtKB:B4F7E7, ECO:0000250|UniProtKB:Q9JLB2, ECO:0000269|PubMed:16678097, ECO:0000269|PubMed:21479189, ECO:0000269|PubMed:25385611, ECO:0000269|PubMed:27466317}.; FUNCTION: (Microbial infection) Acts as an interaction partner for human coronaviruses SARS-CoV and, probably, SARS-CoV-2 envelope protein E which results in delayed formation of tight junctions and disregulation of cell polarity. {ECO:0000269|PubMed:20861307, ECO:0000303|PubMed:32891874}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MPP5 | chr14:67790529 | chr14:70795953 | ENST00000261681 | + | 14 | 15 | 120_177 | 617 | 676.0 | Domain | L27 1 |
| Hgene | MPP5 | chr14:67790529 | chr14:70795953 | ENST00000261681 | + | 14 | 15 | 179_235 | 617 | 676.0 | Domain | L27 2 |
| Hgene | MPP5 | chr14:67790529 | chr14:70795953 | ENST00000261681 | + | 14 | 15 | 256_336 | 617 | 676.0 | Domain | PDZ |
| Hgene | MPP5 | chr14:67790529 | chr14:70795953 | ENST00000261681 | + | 14 | 15 | 345_417 | 617 | 676.0 | Domain | SH3 |
| Hgene | MPP5 | chr14:67790529 | chr14:70795953 | ENST00000555925 | + | 14 | 15 | 120_177 | 583 | 642.0 | Domain | L27 1 |
| Hgene | MPP5 | chr14:67790529 | chr14:70795953 | ENST00000555925 | + | 14 | 15 | 179_235 | 583 | 642.0 | Domain | L27 2 |
| Hgene | MPP5 | chr14:67790529 | chr14:70795953 | ENST00000555925 | + | 14 | 15 | 256_336 | 583 | 642.0 | Domain | PDZ |
| Hgene | MPP5 | chr14:67790529 | chr14:70795953 | ENST00000555925 | + | 14 | 15 | 345_417 | 583 | 642.0 | Domain | SH3 |
| Hgene | MPP5 | chr14:67790529 | chr14:70795953 | ENST00000261681 | + | 14 | 15 | 486_493 | 617 | 676.0 | Nucleotide binding | ATP |
| Hgene | MPP5 | chr14:67790529 | chr14:70795953 | ENST00000555925 | + | 14 | 15 | 486_493 | 583 | 642.0 | Nucleotide binding | ATP |
| Hgene | MPP5 | chr14:67790529 | chr14:70795953 | ENST00000261681 | + | 14 | 15 | 1_345 | 617 | 676.0 | Region | Required for the correct localization of MPP5 and PATJ at cell-cell contacts and the normal formation of tight junctions and adherens junctions |
| Hgene | MPP5 | chr14:67790529 | chr14:70795953 | ENST00000555925 | + | 14 | 15 | 1_345 | 583 | 642.0 | Region | Required for the correct localization of MPP5 and PATJ at cell-cell contacts and the normal formation of tight junctions and adherens junctions |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MPP5 | chr14:67790529 | chr14:70795953 | ENST00000261681 | + | 14 | 15 | 479_660 | 617 | 676.0 | Domain | Guanylate kinase-like |
| Hgene | MPP5 | chr14:67790529 | chr14:70795953 | ENST00000555925 | + | 14 | 15 | 479_660 | 583 | 642.0 | Domain | Guanylate kinase-like |
| Tgene | COX16 | chr14:67790529 | chr14:70795953 | ENST00000389912 | 1 | 4 | 1_14 | 47 | 107.0 | Topological domain | Mitochondrial matrix | |
| Tgene | COX16 | chr14:67790529 | chr14:70795953 | ENST00000389912 | 1 | 4 | 38_106 | 47 | 107.0 | Topological domain | Mitochondrial intermembrane | |
| Tgene | COX16 | chr14:67790529 | chr14:70795953 | ENST00000389912 | 1 | 4 | 15_37 | 47 | 107.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for MPP5-COX16 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >54745_54745_1_MPP5-COX16_MPP5_chr14_67790529_ENST00000261681_COX16_chr14_70795953_ENST00000389912_length(transcript)=3944nt_BP=2512nt GTTTTAGACTTAGAGGCGCGCGACTGCGCGCACCCGGCTGGAGCTGCGGAGAGGACCAATGAGCGGGCGCTCTCTCCGCTCCGGCTCGGG ATTGGGCAAGCCGCGCCAGCCTCTCACGGGCGGGGTGGGACTCGGATCCGCGCGCCCACTGCGCCAGAGTCCCGGGGGCGCGTGCGCGGT CTCGCGGCGGCCGCAGGGGCCGGTCTCGCGCGGTCTAGAAGTGGAGTTGCTGGCGGCTGCGGCGGTGACGGCGGCGACGGAGGAGGCAGG CGGTGGGGCGGGGGCGGGGACTAAGGATTCTGAGGTGGGGAGTCGGGAGTTTCTGGATTCTTTATCCGGAATTTCAAGGGCCGCCGGAGG GCTGTCGCTTCTGCAGTGCGTAGGAGCGGCCGGGGCGGGAGGCTCCGCGGAGCCGAGGCGTGGAGAATAAGAAAGCCTTGAGTTTTGTGA AAAACCGGAGAAGAGAAACTAAAAGGACAGTAGAAAAGGCTTTTCCAGTTTGCATAATATTTCCTTCATGGATACTTTTTCATAGCATTA TTATGTGATGTGAGAAGTTTTTTTTTTTGAAGTAACATGGATTTTATACTACAGAATCAAGAGAATTGGCTTATAGGAAAAATTGATTTA TAAAAAGTGGTACAGGTTTTCATAGATAACCATGACAACATCCCATATGAATGGGCATGTTACAGAGGAATCAGACAGCGAAGTAAAAAA TGTTGATCTTGCATCACCAGAGGAACATCAGAAGCACCGAGAGATGGCTGTTGACTGCCCTGGAGATTTGGGCACCAGGATGATGCCAAT ACGTCGAAGTGCACAGTTGGAGCGTATTCGGCAACAACAGGAGGACATGAGGCGTAGGAGAGAGGAAGAAGGGAAAAAGCAAGAACTTGA CCTTAATTCTTCCATGAGACTTAAGAAACTAGCCCAAATTCCTCCAAAGACCGGAATAGATAACCCTATGTTTGATACAGAGGAAGGAAT TGTCTTAGAAAGTCCTCATTATGCTGTGAAAATATTAGAAATAGAAGACTTGTTTTCTTCACTTAAACATATCCAACATACTTTGGTAGA TTCTCAGAGCCAGGAGGATATTTCACTGCTTTTACAACTTGTTCAAAATAAGGATTTCCAGAATGCATTTAAGATACACAATGCCATCAC AGTACACATGAACAAGGCCAGTCCTCCATTTCCTCTTATCTCCAACGCACAAGATCTTGCTCAAGAGGTACAAACTGTTTTGAAGCCAGT TCATCATAAGGAAGGACAAGAACTAACTGCTTTGCTGAATACTCCACATATTCAGGCACTTTTACTGGCCCACGATAAGGTTGCTGAGCA GGAAATGCAGCTAGAGCCCATTACAGATGAGAGAGTTTATGAAAGTATTGGCCAGTATGGAGGAGAAACTGTAAAAATAGTTCGTATAGA AAAGGCTCGTGATATTCCGTTGGGTGCTACAGTTCGTAATGAAATGGACTCTGTCATCATTAGCCGGATAGTAAAAGGGGGTGCTGCAGA GAAAAGTGGTCTGTTGCATGAAGGAGATGAAGTTCTAGAGATTAATGGCATTGAAATTCGGGGGAAAGATGTCAATGAGGTTTTTGACTT GTTGTCTGATATGCATGGTACTTTGACTTTTGTCCTGATTCCCAGTCAACAGATCAAGCCGCCTCCTGCCAAGGAAACAGTAATCCATGT AAAAGCTCATTTTGACTATGACCCCTCAGATGACCCTTATGTTCCATGTCGAGAGTTAGGTCTGTCTTTTCAAAAAGGTGATATACTTCA TGTGATCAGTCAAGAAGATCCAAACTGGTGGCAGGCCTACAGGGAAGGGGACGAAGATAATCAACCTCTAGCCGGGCTTGTTCCAGGGAA AAGCTTTCAGCAGCAAAGGGAAGCCATGAAACAAACCATAGAAGAAGATAAGGAGCCAGAAAAATCAGGAAAACTGTGGTGTGCAAAGAA GAATAAAAAGAAGAGGAAAAAGGTTTTATATAATGCCAATAAAAATGATGATTATGACAACGAGGAGATCTTAACCTATGAGGAAATGTC ACTTTATCATCAGCCAGCAAATAGGAAGAGACCTATCATCTTGATTGGTCCACAGAACTGTGGCCAGAATGAATTGCGTCAGAGGCTCAT GAACAAAGAAAAGGACCGCTTTGCATCTGCAGTTCCTCATACAACCCGGAGTAGGCGAGACCAAGAAGTAGCCGGTAGAGATTACCACTT TGTTTCGCGGCAAGCATTCGAGGCAGACATAGCAGCTGGAAAGTTCATTGAGCATGGTGAATTTGAGAAGAATTTGTATGGAACTAGCAT AGATTCTGTACGGCAAGTGATCAACTCTGGCAAAATATGTCTTTTAAGTCTTCGTACACAGTCATTGAAGACTCTCCGGAATTCAGATTT GAAACCATATATTATCTTCATTGCACCCCCTTCACAAGAAAGACTTCGGGCATTATTGGCCAAAGAAGGCAAGAATCCAAAGATGGATCC TGAGCTTGAAAAAAAACTGAAAGAGAATAAAATATCTTTAGAGTCGGAATATGAGAAAATCAAAGACTCCAAGTTTGATGACTGGAAGAA TATTCGAGGACCCAGGCCTTGGGAAGATCCTGACCTCCTCCAAGGAAGAAATCCAGAAAGCCTTAAGACTAAGACAACTTGACTCTGCTG ATTCTTTTTTCCTTTTTTTTTTTTTTAAATAAAAATATTATTAACTGGACTTCCTAATATATACTTCTATCAAGTGGAAAGGAAATTCCA GGCCCATGGAAACTTGGATATGGGTAATTTGATGACAAATAATCTTCACTAAAGGTCATGTACAGGTTTTTATACTTCCCAGCTATTCCA TCTGTGGATGAAAGTAACAATGTTGGCCACGTATATTTTACACCTCGAAATAAAAAATGTGAATACTGCTCCAAAAACAGAGTCACGTAT TCCACTCTCCAACTACCCACATATTCCTTTTGCAATAGCCATTAGGGCATCATTTTGATATTTCATTCTGATTTCTGATTCTCTGATTTC TGATTCCTAATGAGGACAGTAGGTCTGGATCCAAATTCTCACAGTAAAATCAAGCAGTAATTTTCTCTCATATCTATTAGGGAAAGAAAA ATGATCACAGTCTGCTAAGAGTCTTGATTTTCTTTGTAATGCCTCACATAGTATGATAATCAGTCTCCAAAGCATCACATGATAATTACA ATGATACCATTAACATGTCAAGGAAATTATATTATTTATGGTTGTCAAAAATTATGAAGTAGTGTATGATTATAAGCAGATATGGCAAAT TTGTTCAGTAAATCCATAGATGACTACATTTTGAGAAATACTAAGATAATACTAAAAATTATGCCTTAGCATAATTTGCATGCAAAATTG CCCTCTAGTGTTTTTGTTTTGTTTTGAGACATAGTCTCGCTCTGTTCGCCCAGGCTGGAGTGCAGGGGCACGATCTCTGCTCACTGCAAG CTCTGCTTCCCGGGTTCACACCATTCTCCTGCCTCAGCATCCTGAGTAGCTGGGACTACAGGCACATGCTGTCACACCCGGCTAATTTTT TGTATTTAGTAGAGATGGGGTTTCACCACGTTAGCCAGGATGGTCTCCATCTCCTGACCTTGTGATCCGCCCACCTCGGCCTCACAAATT GCTGGGATTACAGGTGTGAGCCACCACGCCTGGCCGAAAGTTAGTTGTTTTGAACTCATTTTCTCCTTTTTACCCCTGCCTTTATCAGCT TCTACCTTCTGGTTTCATGCTGAGTTCTAGTTGCCTAAAGTCTCCATTGAAACACTCCTGTTATAATTGCCATAAAATCATAAATCCACT >54745_54745_1_MPP5-COX16_MPP5_chr14_67790529_ENST00000261681_COX16_chr14_70795953_ENST00000389912_length(amino acids)=676AA_BP=617 MTTSHMNGHVTEESDSEVKNVDLASPEEHQKHREMAVDCPGDLGTRMMPIRRSAQLERIRQQQEDMRRRREEEGKKQELDLNSSMRLKKL AQIPPKTGIDNPMFDTEEGIVLESPHYAVKILEIEDLFSSLKHIQHTLVDSQSQEDISLLLQLVQNKDFQNAFKIHNAITVHMNKASPPF PLISNAQDLAQEVQTVLKPVHHKEGQELTALLNTPHIQALLLAHDKVAEQEMQLEPITDERVYESIGQYGGETVKIVRIEKARDIPLGAT VRNEMDSVIISRIVKGGAAEKSGLLHEGDEVLEINGIEIRGKDVNEVFDLLSDMHGTLTFVLIPSQQIKPPPAKETVIHVKAHFDYDPSD DPYVPCRELGLSFQKGDILHVISQEDPNWWQAYREGDEDNQPLAGLVPGKSFQQQREAMKQTIEEDKEPEKSGKLWCAKKNKKKRKKVLY NANKNDDYDNEEILTYEEMSLYHQPANRKRPIILIGPQNCGQNELRQRLMNKEKDRFASAVPHTTRSRRDQEVAGRDYHFVSRQAFEADI AAGKFIEHGEFEKNLYGTSIDSVRQVINSGKICLLSLRTQSLKTLRNSDLKPYIIFIAPPSQERLRALLAKEGKNPKMDPELEKKLKENK -------------------------------------------------------------- >54745_54745_2_MPP5-COX16_MPP5_chr14_67790529_ENST00000555925_COX16_chr14_70795953_ENST00000389912_length(transcript)=3398nt_BP=1966nt GGGAGTTTCTGGATTCTTTATCCGGAATTTCAAGGGCCGCCGGAGGGCTGTCGCTTCTGCAGTGCGTAGGAGCGGCCGGGGCGGGAGGCT CCGCGGAGCCGAGGCGTGGAGAATAAGAAAGCCTTGAGTTTTGTGAAAAACCGGAGAAGAGAAACTAAAAGGACAGTAGAAAAGGCTTTT CCAGTTTGCATAATAGGAACATCAGAAGCACCGAGAGATGGCTGTTGACTGCCCTGGAGATTTGGGCACCAGGATGATGCCAATACGTCG AAGTGCACAGTTGGAGCGTATTCGGCAACAACAGGAGGACATGAGGCGTAGGAGAGAGGAAGAAGGGAAAAAGCAAGAACTTGACCTTAA TTCTTCCATGAGACTTAAGAAACTAGCCCAAATTCCTCCAAAGACCGGAATAGATAACCCTATGTTTGATACAGAGGAAGGAATTGTCTT AGAAAGTCCTCATTATGCTGTGAAAATATTAGAAATAGAAGACTTGTTTTCTTCACTTAAACATATCCAACATACTTTGGTAGATTCTCA GAGCCAGGAGGATATTTCACTGCTTTTACAACTTGTTCAAAATAAGGATTTCCAGAATGCATTTAAGATACACAATGCCATCACAGTACA CATGAACAAGGCCAGTCCTCCATTTCCTCTTATCTCCAACGCACAAGATCTTGCTCAAGAGGTACAAACTGTTTTGAAGCCAGTTCATCA TAAGGAAGGACAAGAACTAACTGCTTTGCTGAATACTCCACATATTCAGGCACTTTTACTGGCCCACGATAAGGTTGCTGAGCAGGAAAT GCAGCTAGAGCCCATTACAGATGAGAGAGTTTATGAAAGTATTGGCCAGTATGGAGGAGAAACTGTAAAAATAGTTCGTATAGAAAAGGC TCGTGATATTCCGTTGGGTGCTACAGTTCGTAATGAAATGGACTCTGTCATCATTAGCCGGATAGTAAAAGGGGGTGCTGCAGAGAAAAG TGGTCTGTTGCATGAAGGAGATGAAGTTCTAGAGATTAATGGCATTGAAATTCGGGGGAAAGATGTCAATGAGGTTTTTGACTTGTTGTC TGATATGCATGGTACTTTGACTTTTGTCCTGATTCCCAGTCAACAGATCAAGCCGCCTCCTGCCAAGGAAACAGTAATCCATGTAAAAGC TCATTTTGACTATGACCCCTCAGATGACCCTTATGTTCCATGTCGAGAGTTAGGTCTGTCTTTTCAAAAAGGTGATATACTTCATGTGAT CAGTCAAGAAGATCCAAACTGGTGGCAGGCCTACAGGGAAGGGGACGAAGATAATCAACCTCTAGCCGGGCTTGTTCCAGGGAAAAGCTT TCAGCAGCAAAGGGAAGCCATGAAACAAACCATAGAAGAAGATAAGGAGCCAGAAAAATCAGGAAAACTGTGGTGTGCAAAGAAGAATAA AAAGAAGAGGAAAAAGGTTTTATATAATGCCAATAAAAATGATGATTATGACAACGAGGAGATCTTAACCTATGAGGAAATGTCACTTTA TCATCAGCCAGCAAATAGGAAGAGACCTATCATCTTGATTGGTCCACAGAACTGTGGCCAGAATGAATTGCGTCAGAGGCTCATGAACAA AGAAAAGGACCGCTTTGCATCTGCAGTTCCTCATACAACCCGGAGTAGGCGAGACCAAGAAGTAGCCGGTAGAGATTACCACTTTGTTTC GCGGCAAGCATTCGAGGCAGACATAGCAGCTGGAAAGTTCATTGAGCATGGTGAATTTGAGAAGAATTTGTATGGAACTAGCATAGATTC TGTACGGCAAGTGATCAACTCTGGCAAAATATGTCTTTTAAGTCTTCGTACACAGTCATTGAAGACTCTCCGGAATTCAGATTTGAAACC ATATATTATCTTCATTGCACCCCCTTCACAAGAAAGACTTCGGGCATTATTGGCCAAAGAAGGCAAGAATCCAAAGATGGATCCTGAGCT TGAAAAAAAACTGAAAGAGAATAAAATATCTTTAGAGTCGGAATATGAGAAAATCAAAGACTCCAAGTTTGATGACTGGAAGAATATTCG AGGACCCAGGCCTTGGGAAGATCCTGACCTCCTCCAAGGAAGAAATCCAGAAAGCCTTAAGACTAAGACAACTTGACTCTGCTGATTCTT TTTTCCTTTTTTTTTTTTTTAAATAAAAATATTATTAACTGGACTTCCTAATATATACTTCTATCAAGTGGAAAGGAAATTCCAGGCCCA TGGAAACTTGGATATGGGTAATTTGATGACAAATAATCTTCACTAAAGGTCATGTACAGGTTTTTATACTTCCCAGCTATTCCATCTGTG GATGAAAGTAACAATGTTGGCCACGTATATTTTACACCTCGAAATAAAAAATGTGAATACTGCTCCAAAAACAGAGTCACGTATTCCACT CTCCAACTACCCACATATTCCTTTTGCAATAGCCATTAGGGCATCATTTTGATATTTCATTCTGATTTCTGATTCTCTGATTTCTGATTC CTAATGAGGACAGTAGGTCTGGATCCAAATTCTCACAGTAAAATCAAGCAGTAATTTTCTCTCATATCTATTAGGGAAAGAAAAATGATC ACAGTCTGCTAAGAGTCTTGATTTTCTTTGTAATGCCTCACATAGTATGATAATCAGTCTCCAAAGCATCACATGATAATTACAATGATA CCATTAACATGTCAAGGAAATTATATTATTTATGGTTGTCAAAAATTATGAAGTAGTGTATGATTATAAGCAGATATGGCAAATTTGTTC AGTAAATCCATAGATGACTACATTTTGAGAAATACTAAGATAATACTAAAAATTATGCCTTAGCATAATTTGCATGCAAAATTGCCCTCT AGTGTTTTTGTTTTGTTTTGAGACATAGTCTCGCTCTGTTCGCCCAGGCTGGAGTGCAGGGGCACGATCTCTGCTCACTGCAAGCTCTGC TTCCCGGGTTCACACCATTCTCCTGCCTCAGCATCCTGAGTAGCTGGGACTACAGGCACATGCTGTCACACCCGGCTAATTTTTTGTATT TAGTAGAGATGGGGTTTCACCACGTTAGCCAGGATGGTCTCCATCTCCTGACCTTGTGATCCGCCCACCTCGGCCTCACAAATTGCTGGG ATTACAGGTGTGAGCCACCACGCCTGGCCGAAAGTTAGTTGTTTTGAACTCATTTTCTCCTTTTTACCCCTGCCTTTATCAGCTTCTACC TTCTGGTTTCATGCTGAGTTCTAGTTGCCTAAAGTCTCCATTGAAACACTCCTGTTATAATTGCCATAAAATCATAAATCCACTCCCCCT >54745_54745_2_MPP5-COX16_MPP5_chr14_67790529_ENST00000555925_COX16_chr14_70795953_ENST00000389912_length(amino acids)=642AA_BP=583 MAVDCPGDLGTRMMPIRRSAQLERIRQQQEDMRRRREEEGKKQELDLNSSMRLKKLAQIPPKTGIDNPMFDTEEGIVLESPHYAVKILEI EDLFSSLKHIQHTLVDSQSQEDISLLLQLVQNKDFQNAFKIHNAITVHMNKASPPFPLISNAQDLAQEVQTVLKPVHHKEGQELTALLNT PHIQALLLAHDKVAEQEMQLEPITDERVYESIGQYGGETVKIVRIEKARDIPLGATVRNEMDSVIISRIVKGGAAEKSGLLHEGDEVLEI NGIEIRGKDVNEVFDLLSDMHGTLTFVLIPSQQIKPPPAKETVIHVKAHFDYDPSDDPYVPCRELGLSFQKGDILHVISQEDPNWWQAYR EGDEDNQPLAGLVPGKSFQQQREAMKQTIEEDKEPEKSGKLWCAKKNKKKRKKVLYNANKNDDYDNEEILTYEEMSLYHQPANRKRPIIL IGPQNCGQNELRQRLMNKEKDRFASAVPHTTRSRRDQEVAGRDYHFVSRQAFEADIAAGKFIEHGEFEKNLYGTSIDSVRQVINSGKICL LSLRTQSLKTLRNSDLKPYIIFIAPPSQERLRALLAKEGKNPKMDPELEKKLKENKISLESEYEKIKDSKFDDWKNIRGPRPWEDPDLLQ -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MPP5-COX16 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Hgene | MPP5 | chr14:67790529 | chr14:70795953 | ENST00000261681 | + | 14 | 15 | 181_243 | 617.0 | 676.0 | LIN7C |
| Hgene | MPP5 | chr14:67790529 | chr14:70795953 | ENST00000555925 | + | 14 | 15 | 181_243 | 583.0 | 642.0 | LIN7C |
| Hgene | MPP5 | chr14:67790529 | chr14:70795953 | ENST00000261681 | + | 14 | 15 | 21_140 | 617.0 | 676.0 | PARD6B |
| Hgene | MPP5 | chr14:67790529 | chr14:70795953 | ENST00000555925 | + | 14 | 15 | 21_140 | 583.0 | 642.0 | PARD6B |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MPP5-COX16 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MPP5-COX16 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
