
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MROH2B-MRPS30 (FusionGDB2 ID:54890) |
Fusion Gene Summary for MROH2B-MRPS30 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MROH2B-MRPS30 | Fusion gene ID: 54890 | Hgene | Tgene | Gene symbol | MROH2B | MRPS30 | Gene ID | 133558 | 10884 |
| Gene name | maestro heat like repeat family member 2B | mitochondrial ribosomal protein S30 | |
| Synonyms | HEATR7B2|SPIF | MRP-S30|PAP|PDCD9|S30mt | |
| Cytomap | 5p13.1 | 5p12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | maestro heat-like repeat-containing protein family member 2BHEAT repeat family member 7B2HEAT repeat family member B2HEAT repeat-containing protein 7B2sperm PKA-interacting factor | 39S ribosomal protein S30, mitochondrial28S ribosomal protein S30, mitochondrialmitochondrial large ribosomal subunit protein mL65mitochondrial large ribosomal subunit protein mS30programmed cell death 9 | |
| Modification date | 20200320 | 20200313 | |
| UniProtAcc | Q7Z745 | Q9NP92 | |
| Ensembl transtripts involved in fusion gene | ENST00000399564, ENST00000506092, | ENST00000230914, ENST00000507110, | |
| Fusion gene scores | * DoF score | 1 X 1 X 1=1 | 5 X 6 X 3=90 |
| # samples | 1 | 6 | |
| ** MAII score | log2(1/1*10)=3.32192809488736 | log2(6/90*10)=-0.584962500721156 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MROH2B [Title/Abstract] AND MRPS30 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MROH2B(41017952)-MRPS30(44813208), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across MROH2B (5'-gene) Fusion gene breakpoints across MROH2B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
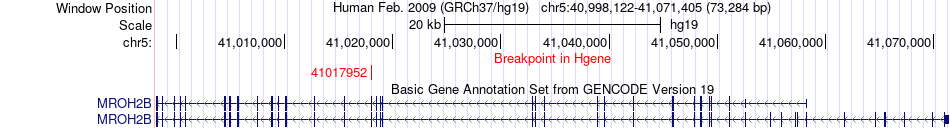 |
 Fusion gene breakpoints across MRPS30 (3'-gene) Fusion gene breakpoints across MRPS30 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
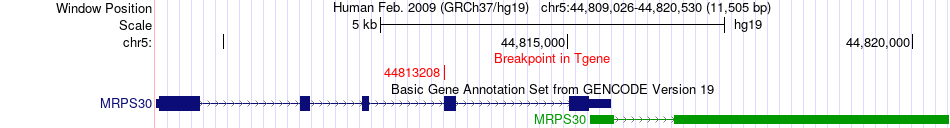 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-55-1596-01A | MROH2B | chr5 | 41017952 | - | MRPS30 | chr5 | 44813208 | + |
Top |
Fusion Gene ORF analysis for MROH2B-MRPS30 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000399564 | ENST00000230914 | MROH2B | chr5 | 41017952 | - | MRPS30 | chr5 | 44813208 | + |
| 5CDS-intron | ENST00000506092 | ENST00000230914 | MROH2B | chr5 | 41017952 | - | MRPS30 | chr5 | 44813208 | + |
| In-frame | ENST00000399564 | ENST00000507110 | MROH2B | chr5 | 41017952 | - | MRPS30 | chr5 | 44813208 | + |
| In-frame | ENST00000506092 | ENST00000507110 | MROH2B | chr5 | 41017952 | - | MRPS30 | chr5 | 44813208 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000506092 | MROH2B | chr5 | 41017952 | - | ENST00000507110 | MRPS30 | chr5 | 44813208 | + | 2627 | 1840 | 6 | 2306 | 766 |
| ENST00000399564 | MROH2B | chr5 | 41017952 | - | ENST00000507110 | MRPS30 | chr5 | 44813208 | + | 4122 | 3335 | 451 | 3801 | 1116 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000506092 | ENST00000507110 | MROH2B | chr5 | 41017952 | - | MRPS30 | chr5 | 44813208 | + | 0.001045286 | 0.99895465 |
| ENST00000399564 | ENST00000507110 | MROH2B | chr5 | 41017952 | - | MRPS30 | chr5 | 44813208 | + | 0.00058043 | 0.99941957 |
Top |
Fusion Genomic Features for MROH2B-MRPS30 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
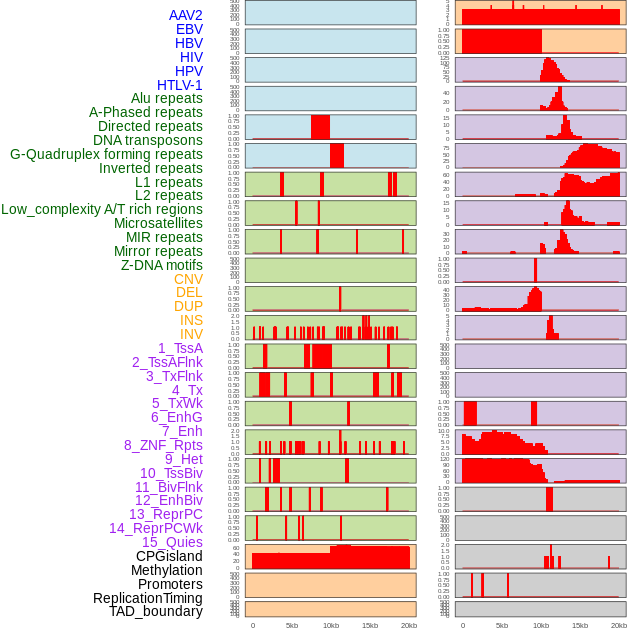 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
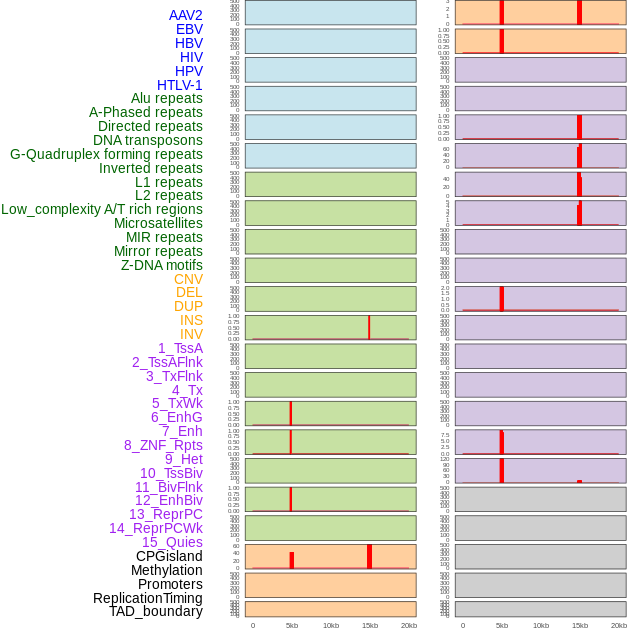 |
Top |
Fusion Protein Features for MROH2B-MRPS30 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:41017952/chr5:44813208) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MROH2B | MRPS30 |
| FUNCTION: May play a role in the process of sperm capacitation. {ECO:0000250|UniProtKB:Q7M6Y6}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MROH2B | chr5:41017952 | chr5:44813208 | ENST00000399564 | - | 28 | 42 | 228_263 | 961 | 1586.0 | Repeat | Note=HEAT 2 |
| Hgene | MROH2B | chr5:41017952 | chr5:44813208 | ENST00000399564 | - | 28 | 42 | 272_309 | 961 | 1586.0 | Repeat | Note=HEAT 3 |
| Hgene | MROH2B | chr5:41017952 | chr5:44813208 | ENST00000399564 | - | 28 | 42 | 28_65 | 961 | 1586.0 | Repeat | Note=HEAT 1 |
| Hgene | MROH2B | chr5:41017952 | chr5:44813208 | ENST00000399564 | - | 28 | 42 | 310_346 | 961 | 1586.0 | Repeat | Note=HEAT 4 |
| Hgene | MROH2B | chr5:41017952 | chr5:44813208 | ENST00000399564 | - | 28 | 42 | 405_445 | 961 | 1586.0 | Repeat | Note=HEAT 5 |
| Hgene | MROH2B | chr5:41017952 | chr5:44813208 | ENST00000399564 | - | 28 | 42 | 531_569 | 961 | 1586.0 | Repeat | Note=HEAT 6 |
| Hgene | MROH2B | chr5:41017952 | chr5:44813208 | ENST00000399564 | - | 28 | 42 | 572_611 | 961 | 1586.0 | Repeat | Note=HEAT 7 |
| Hgene | MROH2B | chr5:41017952 | chr5:44813208 | ENST00000399564 | - | 28 | 42 | 662_699 | 961 | 1586.0 | Repeat | Note=HEAT 8 |
| Hgene | MROH2B | chr5:41017952 | chr5:44813208 | ENST00000399564 | - | 28 | 42 | 777_819 | 961 | 1586.0 | Repeat | Note=HEAT 9 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MROH2B | chr5:41017952 | chr5:44813208 | ENST00000399564 | - | 28 | 42 | 1021_1059 | 961 | 1586.0 | Repeat | Note=HEAT 11 |
| Hgene | MROH2B | chr5:41017952 | chr5:44813208 | ENST00000399564 | - | 28 | 42 | 1112_1151 | 961 | 1586.0 | Repeat | Note=HEAT 12 |
| Hgene | MROH2B | chr5:41017952 | chr5:44813208 | ENST00000399564 | - | 28 | 42 | 1157_1195 | 961 | 1586.0 | Repeat | Note=HEAT 13 |
| Hgene | MROH2B | chr5:41017952 | chr5:44813208 | ENST00000399564 | - | 28 | 42 | 1258_1295 | 961 | 1586.0 | Repeat | Note=HEAT 14 |
| Hgene | MROH2B | chr5:41017952 | chr5:44813208 | ENST00000399564 | - | 28 | 42 | 1363_1402 | 961 | 1586.0 | Repeat | Note=HEAT 15 |
| Hgene | MROH2B | chr5:41017952 | chr5:44813208 | ENST00000399564 | - | 28 | 42 | 964_1001 | 961 | 1586.0 | Repeat | Note=HEAT 10 |
Top |
Fusion Gene Sequence for MROH2B-MRPS30 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >54890_54890_1_MROH2B-MRPS30_MROH2B_chr5_41017952_ENST00000399564_MRPS30_chr5_44813208_ENST00000507110_length(transcript)=4122nt_BP=3335nt GAGTTCACCATAAAGCCTTTCCCAACATCAGGACAGTAAAGCAACTACTGAAAACCATTCCTGAATTTGTGATCAATGGACTCAAGTTTA GGGAGAGGAGCACCTTTGGTCTGCAACAATGGTTTGAATCCCAGTCTCTAATTACTTCTTTCCCAAATACTCTGGTCACCAAAGGGACAA TATTTGGTGAGATGCAGCCATTTTGTTGGTTAGCAGGAGTTCAGACGCAAGACAATGTCATAGTGGTAGAATACAGTATCAACTTCAACA GCAAGCCCATCTCTGGAAGCTCCCAGACCATTGTGCCAATCAAGTGGAGGAGGCACTTTCATTTAGAGAGGCTTATGTGAACCTTATATG AGAAACTTGCCAACCTGCCTCTTTGATTTTTTAGCCTTACTATTTCTAATACTTAGGTGCTATCTTTTAGAGACCCTGAAACAGCCAAGA CATGACACTTAGTACAGAGGAATCCATAGAGATGTTTGGGGATATTAACCTCACTCTTGGCATGCTGAACAAGGAAGATATTGTTAACAA GGAAGACATTTACAGTCATCTCACTTCTGTTATTCAGAATACTGACATCTTGGATGATGCAATTGTCCAACGATTGATTTATTATGCTTC TAAGGATATGAGAGACAACAATATGCTCAGAGAAATCAGAATGCTAGCTGGTGAGGTTTTGGTGTCTCTTGCTGCACATGATTTCAACTC TGTGATGTATGAAGTACAAAGTAACTTCAGGATCTTAGAGCTACCAGATGAATTCGTTGTGCTTGCCCTGGCTGAATTGGCAACCAGCTA TGTGTCCCAGAGTATTCCTTTCATGATGATGACCCTGCTCACCATGCAAACCATGCTCAGGCTGGCCGAGGATGAAAGGATGAAGGGGAC TTTCTGTATTGCCCTTGAGAAATTCAGCAAAGCCATTTACAAATATGTCAACCACTGGAGAGATTTTCCCTACCCCAGACTGGATGCCAA CCGACTGTCTGACAAGATCTTCATGCTGTTCTGGTATATAATGGAGAAGTGGGCCCCTTTGGCCTCACCCATGCAGACTTTGAGCATCGT TAAGGCCCACGGGCCCACGGTGAGCTTGCTGCTGCATCGGGAAGACTTCCGTGGATACGCCCTGGGCCAGGTGCCCTGGCTCCTGAACCA GTATAAAGACAAAGAGATTGATTTCCATGTCACTCAGAGCCTAAAACAAATACTGACTGCAGCAGTTCTTTATGACATTGGCCTTCCAAG GAGCTTGAGAAGATCCATCTTTATCAATTTACTCCAGCAGATCTGCAGAGCTCCAGAGCCTCCAGTAAAGGAAAATGAAATGAAAGCTTC AAGCTGTTTTCTCATTCTAGCCCATTCCAATCCTGGAGAGCTGATGGAATTTTTTGATGAACAAGTAAGAAGTAATAATGAAGCCATTCG AGTGGGAATCTTGACTTTGTTAAGATTGGCTGTCAATGCTGATGAGCCCAGGTTGAGGGATCACATCATTTCAATAGAAAGAACAGTCAA AATCGTCATGGGTGATCTCAGTACAAAAGTAAGAAATTCTGTTCTTCTCCTCATCCAAACCATGTGTGAAAAGTCCTATATTGAAGCTCG GGAAGGATGGCCATTGATTGATTATGTCTTCTCCCAGTTTGCAACGTTGAACAGGAATCTGGAGAAACCAGTGAAAACTAACTTTCATGA GAATGAAAAGGAAGAGGAATCTGTCCGAGAAACAAGCCTTGAGGTCTTAAAAACCCTGGACCCACTGGTCATTGGAATGCCTCAGGTGCT ATGGCCAAGGATCCTGACTTTTGTGGTACCTGCAGAATACACAGAAGCTCTGGAGCCCCTGTTTAGTATCATCAGAATTCTGATTATGGC AGAGGAGAAGAAGCAGCACAGTGCCAAGGAGTCGACAGCACTTGTCGTCTCTACAGGAGCTGTGAAACTTCCTTCACCACAGCAGCTACT GGCCAGACTTCTGGTAATATCTATGCCTGCCAGTTTAGGGGAGTTACGTGGGGCTGGTGCAATAGGGCTTTTGAAGATACTGCCTGAGAT AATTCACCCAAAATTGGTAGACCTATGGAAAACACGTTTACCTGAGCTTCTGCAGCCTCTGGAAGGAAAGAACATCAGTACCGTTCTATG GGAAACCATGCTGCTTCAGTTGCTCAAAGAATCCTTATGGAAGATCAGTGATGTGGCCTGGACCATTCAGCTGACTCAGGATTTCAAACA GCAAATGGGCAGTTACAGCAATAACTCCACTGAGAAGAAATTCCTTTGGAAAGCCTTGGGAACCACCTTAGCATGCTGCCAAGATTCAGA CTTTGTAAACTCACAGATTAAGGAGTTTCTGACTGCTCCCAACCAACTGGGGGATCAAAGACAGGGAATAACATCTATTTTAGGATACTG TGCCGAGAACCATTTGGATATTGTTTTAAAAGTTCTTAAAACATTCCAAAATCAGGAAAAGTTTTTCATGAATCGATGTAAGAGCCTTTT TTCTGGGAAAAAGAGCCTGACCAAGACAGATGTCATGGTCATCTATGGAGCAGTGGCCCTCCATGCTCCCAAGAAGCAACTTCTCTCCAG ACTTAATCAAGATATCATATCCCAAGTCCTGTCTCTTCATGGCCAGTGCTCTCAGGTTCTGGGCATGTCTGTGATGAACAAGGACATGGA TCTGCAAATGAGTTTCACAAGAAGCATCACTGAGATTGGCATTGCTGTCCAAGATGCTGAGGATCAGGGGTTCCAGTTTTCCTACAAGGA GATGCTGATTGGTTACATGCTGGACTTCATTAGAGACGAGCCCCTGGATTCCTTAGCTAGCCCTATTCGGTGGAAAGCCTTAATCGCCAT TAGGTATCTCAGTAAACTGAAACCTCAGCTCTCACTACAAGACCACCTTAACATTCTTGAGGAGAATATTCGGAGGCTGCTGCCCCTTCC ACCTCTGGAAAATCTGAAAAGTGAAGGCCAGACAGACAAGGACAAGGAGCACATTCAATTTCTCTATGAACGATCCATGGACGCCCTAGG AAAACTTCTGAAGACCATGATGTGGGATAATGTGAATGCAGAGGACTGTCAAGAAATGTTTAATCTTCTCCAAATGTGGCTTGTTTCACA AAAAGAGTGGGAAAGAGAAAGAGCCTTCCAGATCACTGCGAAAGTGCTGACAAATGATATTGAGGCACCAGAGAACTTTAAAATTGGTTC ACTGCTTGGACTTCTGGCTCCTCACTCCTGTGATACCCTGCCCACCATCCGTCAGGCGGCTGCTAGCTCAACTATTGGTCTGTTCTATAT AAAAGGCTCAAAAACTGCAGATCCTTGCTGTTACGGTCACACCCAGTTTCATCTGTTACCTGACAAATTAAGAAGGGAAAGGCTTTTGAG ACAAAACTGTGCTGATCAGATAGAAGTTGTTTTTAGAGCTAATGCTATTGCAAGCCTTTTTGCTTGGACTGGAGCACAAGCTATGTATCA AGGATTCTGGAGTGAAGCAGATGTTACTCGACCTTTTGTCTCCCAGGCTGTGATCACAGATGGAAAATACTTTTCCTTTTTCTGCTACCA GCTAAATACTTTGGCACTGACTACACAAGCTGATCAAAATAACCCTCGTAAAAATATATGTTGGGGTACACAAAGTAAGCCTCTTTATGA AACAATTGAGGATAATGATGTGAAAGGTTTTAATGATGATGTTCTACTTCAGATAGTTCACTTTCTACTGAATAGACCAAAAGAAGAAAA ATCACAGCTGTTGGAAAACTGAAAAAGCATATTTGATTGAGAACTGTGGGAATATTTAAATTTTACTGAAGGAACAATAATGATGAGATT TGTAACTGTCAACTATTAAATACATTGATTTTTGAGACAAATATTTCTTATGTCAACCTGTTATTAGATCTCTTACTCTGCTCAAATTCA TCACTGAAAGATTTAATTTTAGTTACCTTTTGTTGATTTAAAAATAATTGCATTTGTATATTGCTAACTGATAAGACAAATTGAGTTATT >54890_54890_1_MROH2B-MRPS30_MROH2B_chr5_41017952_ENST00000399564_MRPS30_chr5_44813208_ENST00000507110_length(amino acids)=1116AA_BP=951 MTLSTEESIEMFGDINLTLGMLNKEDIVNKEDIYSHLTSVIQNTDILDDAIVQRLIYYASKDMRDNNMLREIRMLAGEVLVSLAAHDFNS VMYEVQSNFRILELPDEFVVLALAELATSYVSQSIPFMMMTLLTMQTMLRLAEDERMKGTFCIALEKFSKAIYKYVNHWRDFPYPRLDAN RLSDKIFMLFWYIMEKWAPLASPMQTLSIVKAHGPTVSLLLHREDFRGYALGQVPWLLNQYKDKEIDFHVTQSLKQILTAAVLYDIGLPR SLRRSIFINLLQQICRAPEPPVKENEMKASSCFLILAHSNPGELMEFFDEQVRSNNEAIRVGILTLLRLAVNADEPRLRDHIISIERTVK IVMGDLSTKVRNSVLLLIQTMCEKSYIEAREGWPLIDYVFSQFATLNRNLEKPVKTNFHENEKEEESVRETSLEVLKTLDPLVIGMPQVL WPRILTFVVPAEYTEALEPLFSIIRILIMAEEKKQHSAKESTALVVSTGAVKLPSPQQLLARLLVISMPASLGELRGAGAIGLLKILPEI IHPKLVDLWKTRLPELLQPLEGKNISTVLWETMLLQLLKESLWKISDVAWTIQLTQDFKQQMGSYSNNSTEKKFLWKALGTTLACCQDSD FVNSQIKEFLTAPNQLGDQRQGITSILGYCAENHLDIVLKVLKTFQNQEKFFMNRCKSLFSGKKSLTKTDVMVIYGAVALHAPKKQLLSR LNQDIISQVLSLHGQCSQVLGMSVMNKDMDLQMSFTRSITEIGIAVQDAEDQGFQFSYKEMLIGYMLDFIRDEPLDSLASPIRWKALIAI RYLSKLKPQLSLQDHLNILEENIRRLLPLPPLENLKSEGQTDKDKEHIQFLYERSMDALGKLLKTMMWDNVNAEDCQEMFNLLQMWLVSQ KEWERERAFQITAKVLTNDIEAPENFKIGSLLGLLAPHSCDTLPTIRQAAASSTIGLFYIKGSKTADPCCYGHTQFHLLPDKLRRERLLR QNCADQIEVVFRANAIASLFAWTGAQAMYQGFWSEADVTRPFVSQAVITDGKYFSFFCYQLNTLALTTQADQNNPRKNICWGTQSKPLYE -------------------------------------------------------------- >54890_54890_2_MROH2B-MRPS30_MROH2B_chr5_41017952_ENST00000506092_MRPS30_chr5_44813208_ENST00000507110_length(transcript)=2627nt_BP=1840nt ATTTGTTTGCTGCCTGTACAGACTTTGAGCATCGTTAAGGCCCACGGGCCCACGGTGAGCTTGCTGCTGCATCGGGAAGACTTCCGTGGA TACGCCCTGGGCCAGGTGCCCTGGCTCCTGAACCAGTATAAAGACAAAGAGATTGATTTCCATGTCACTCAGTTTGCAACGTTGAACAGG AATCTGGAGAAACCAGTGAAAACTAACTTTCATGAGAATGAAAAGGAAGAGGAATCTGTCCGAGAAACAAGCCTTGAGGTCTTAAAAACC CTGGACCCACTGGTCATTGGAATGCCTCAGGTGCTATGGCCAAGGATCCTGACTTTTGTGGTACCTGCAGAATACACAGAAGCTCTGGAG CCCCTGTTTAGTATCATCAGAATTCTGATTATGGCAGAGGAGAAGAAGCAGCACAGTGCCAAGGAGTCGACAGCACTTGTCGTCTCTACA GGAGCTGTGAAACTTCCTTCACCACAGCAGCTACTGGCCAGACTTCTGGTAATATCTATGCCTGCCAGTTTAGGGGAGTTACGTGGGGCT GGTGCAATAGGGCTTTTGAAGATACTGCCTGAGATAATTCACCCAAAATTGGTAGACCTATGGAAAACACGTTTACCTGAGCTTCTGCAG CCTCTGGAAGGAAAGAACATCAGTACCGTTCTATGGGAAACCATGCTGCTTCAGTTGCTCAAAGAATCCTTATGGAAGATCAGTGATGTG GCCTGGACCATTCAGCTGACTCAGGATTTCAAACAGCAAATGGGCAGTTACAGCAATAACTCCACTGAGAAGAAATTCCTTTGGAAAGCC TTGGGAACCACCTTAGCATGCTGCCAAGATTCAGACTTTGTAAACTCACAGATTAAGGAGTTTCTGACTGCTCCCAACCAACTGGGGGAT CAAAGACAGGGAATAACATCTATTTTAGGATACTGTGCCGAGAACCATTTGGATATTGTTTTAAAAGTTCTTAAAACATTCCAAAATCAG GAAAAGTTTTTCATGAATCGATGTAAGAGCCTTTTTTCTGGGAAAAAGAGCCTGACCAAGACAGATGTCATGGTCATCTATGGAGCAGTG GCCCTCCATGCTCCCAAGAAGCAACTTCTCTCCAGACTTAATCAAGATATCATATCCCAAGTCCTGTCTCTTCATGGCCAGTGCTCTCAG GTTCTGGGCATGTCTGTGATGAACAAGGACATGGATCTGCAAATGAGTTTCACAAGAAGCATCACTGAGATTGGCATTGCTGTCCAAGAT GCTGAGGATCAGGGGTTCCAGTTTTCCTACAAGGAGATGCTGATTGGTTACATGCTGGACTTCATTAGAGACGAGCCCCTGGATTCCTTA GCTAGCCCTATTCGGTGGAAAGCCTTAATCGCCATTAGGTATCTCAGTAAACTGAAACCTCAGCTCTCACTACAAGACCACCTTAACATT CTTGAGGAGAATATTCGGAGGCTGCTGCCCCTTCCACCTCTGGAAAATCTGAAAAGTGAAGGCCAGACAGACAAGGACAAGGAGCACATT CAATTTCTCTATGAACGATCCATGGACGCCCTAGGAAAACTTCTGAAGACCATGATGTGGGATAATGTGAATGCAGAGGACTGTCAAGAA ATGTTTAATCTTCTCCAAATGTGGCTTGTTTCACAAAAAGAGTGGGAAAGAGAAAGAGCCTTCCAGATCACTGCGAAAGTGCTGACAAAT GATATTGAGGCACCAGAGAACTTTAAAATTGGTTCACTGCTTGGACTTCTGGCTCCTCACTCCTGTGATACCCTGCCCACCATCCGTCAG GCGGCTGCTAGCTCAACTATTGGTCTGTTCTATATAAAAGGCTCAAAAACTGCAGATCCTTGCTGTTACGGTCACACCCAGTTTCATCTG TTACCTGACAAATTAAGAAGGGAAAGGCTTTTGAGACAAAACTGTGCTGATCAGATAGAAGTTGTTTTTAGAGCTAATGCTATTGCAAGC CTTTTTGCTTGGACTGGAGCACAAGCTATGTATCAAGGATTCTGGAGTGAAGCAGATGTTACTCGACCTTTTGTCTCCCAGGCTGTGATC ACAGATGGAAAATACTTTTCCTTTTTCTGCTACCAGCTAAATACTTTGGCACTGACTACACAAGCTGATCAAAATAACCCTCGTAAAAAT ATATGTTGGGGTACACAAAGTAAGCCTCTTTATGAAACAATTGAGGATAATGATGTGAAAGGTTTTAATGATGATGTTCTACTTCAGATA GTTCACTTTCTACTGAATAGACCAAAAGAAGAAAAATCACAGCTGTTGGAAAACTGAAAAAGCATATTTGATTGAGAACTGTGGGAATAT TTAAATTTTACTGAAGGAACAATAATGATGAGATTTGTAACTGTCAACTATTAAATACATTGATTTTTGAGACAAATATTTCTTATGTCA ACCTGTTATTAGATCTCTTACTCTGCTCAAATTCATCACTGAAAGATTTAATTTTAGTTACCTTTTGTTGATTTAAAAATAATTGCATTT GTATATTGCTAACTGATAAGACAAATTGAGTTATTGAGCTATTAAATGCACATTTTAATATAAATGCAGAAATCCCAAATAAAATGCTAA >54890_54890_2_MROH2B-MRPS30_MROH2B_chr5_41017952_ENST00000506092_MRPS30_chr5_44813208_ENST00000507110_length(amino acids)=766AA_BP=601 MLPVQTLSIVKAHGPTVSLLLHREDFRGYALGQVPWLLNQYKDKEIDFHVTQFATLNRNLEKPVKTNFHENEKEEESVRETSLEVLKTLD PLVIGMPQVLWPRILTFVVPAEYTEALEPLFSIIRILIMAEEKKQHSAKESTALVVSTGAVKLPSPQQLLARLLVISMPASLGELRGAGA IGLLKILPEIIHPKLVDLWKTRLPELLQPLEGKNISTVLWETMLLQLLKESLWKISDVAWTIQLTQDFKQQMGSYSNNSTEKKFLWKALG TTLACCQDSDFVNSQIKEFLTAPNQLGDQRQGITSILGYCAENHLDIVLKVLKTFQNQEKFFMNRCKSLFSGKKSLTKTDVMVIYGAVAL HAPKKQLLSRLNQDIISQVLSLHGQCSQVLGMSVMNKDMDLQMSFTRSITEIGIAVQDAEDQGFQFSYKEMLIGYMLDFIRDEPLDSLAS PIRWKALIAIRYLSKLKPQLSLQDHLNILEENIRRLLPLPPLENLKSEGQTDKDKEHIQFLYERSMDALGKLLKTMMWDNVNAEDCQEMF NLLQMWLVSQKEWERERAFQITAKVLTNDIEAPENFKIGSLLGLLAPHSCDTLPTIRQAAASSTIGLFYIKGSKTADPCCYGHTQFHLLP DKLRRERLLRQNCADQIEVVFRANAIASLFAWTGAQAMYQGFWSEADVTRPFVSQAVITDGKYFSFFCYQLNTLALTTQADQNNPRKNIC -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MROH2B-MRPS30 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MROH2B-MRPS30 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MROH2B-MRPS30 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
