
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MRS2-CTSC (FusionGDB2 ID:55240) |
Fusion Gene Summary for MRS2-CTSC |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MRS2-CTSC | Fusion gene ID: 55240 | Hgene | Tgene | Gene symbol | MRS2 | CTSC | Gene ID | 57380 | 1075 |
| Gene name | magnesium transporter MRS2 | cathepsin C | |
| Synonyms | HPT|MRS2L | CPPI|DPP-I|DPP1|DPPI|HMS|JP|JPD|PALS|PDON1|PLS | |
| Cytomap | 6p22.3 | 11q14.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | magnesium transporter MRS2 homolog, mitochondrialMRS2, magnesium transporterMRS2-like proteinMRS2-like, magnesium homeostasis factorputative magnesium transporter | dipeptidyl peptidase 1cathepsin Jdipeptidyl transferasedipeptidyl-peptidase I | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9HD23 | P53634 | |
| Ensembl transtripts involved in fusion gene | ENST00000483634, ENST00000274747, ENST00000378353, ENST00000378386, ENST00000443868, ENST00000535061, ENST00000543597, | ENST00000227266, ENST00000393301, ENST00000524463, ENST00000529974, | |
| Fusion gene scores | * DoF score | 7 X 6 X 6=252 | 6 X 4 X 4=96 |
| # samples | 7 | 6 | |
| ** MAII score | log2(7/252*10)=-1.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/96*10)=-0.678071905112638 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MRS2 [Title/Abstract] AND CTSC [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MRS2(24405469)-CTSC(88045722), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | CTSC | GO:0006508 | proteolysis | 8811434 |
 Fusion gene breakpoints across MRS2 (5'-gene) Fusion gene breakpoints across MRS2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
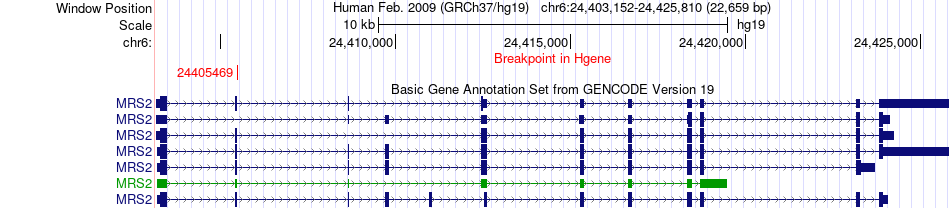 |
 Fusion gene breakpoints across CTSC (3'-gene) Fusion gene breakpoints across CTSC (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
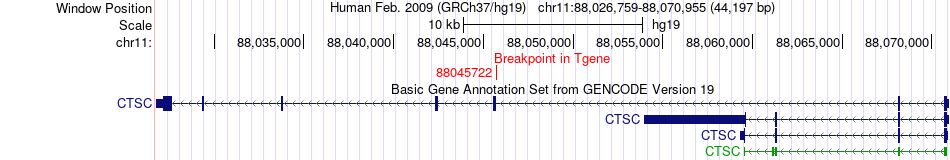 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | Non-Cancer | 257N | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
Top |
Fusion Gene ORF analysis for MRS2-CTSC |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000483634 | ENST00000227266 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| 3UTR-intron | ENST00000483634 | ENST00000393301 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| 3UTR-intron | ENST00000483634 | ENST00000524463 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| 3UTR-intron | ENST00000483634 | ENST00000529974 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| 5CDS-intron | ENST00000274747 | ENST00000393301 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| 5CDS-intron | ENST00000274747 | ENST00000524463 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| 5CDS-intron | ENST00000274747 | ENST00000529974 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| 5CDS-intron | ENST00000378353 | ENST00000393301 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| 5CDS-intron | ENST00000378353 | ENST00000524463 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| 5CDS-intron | ENST00000378353 | ENST00000529974 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| 5CDS-intron | ENST00000378386 | ENST00000393301 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| 5CDS-intron | ENST00000378386 | ENST00000524463 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| 5CDS-intron | ENST00000378386 | ENST00000529974 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| 5CDS-intron | ENST00000443868 | ENST00000393301 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| 5CDS-intron | ENST00000443868 | ENST00000524463 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| 5CDS-intron | ENST00000443868 | ENST00000529974 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| 5CDS-intron | ENST00000535061 | ENST00000393301 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| 5CDS-intron | ENST00000535061 | ENST00000524463 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| 5CDS-intron | ENST00000535061 | ENST00000529974 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| In-frame | ENST00000274747 | ENST00000227266 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| In-frame | ENST00000378353 | ENST00000227266 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| In-frame | ENST00000378386 | ENST00000227266 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| In-frame | ENST00000443868 | ENST00000227266 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| In-frame | ENST00000535061 | ENST00000227266 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| intron-3CDS | ENST00000543597 | ENST00000227266 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| intron-intron | ENST00000543597 | ENST00000393301 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| intron-intron | ENST00000543597 | ENST00000524463 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
| intron-intron | ENST00000543597 | ENST00000529974 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000274747 | MRS2 | chr6 | 24405469 | + | ENST00000227266 | CTSC | chr11 | 88045722 | - | 1874 | 386 | 95 | 1459 | 454 |
| ENST00000535061 | MRS2 | chr6 | 24405469 | + | ENST00000227266 | CTSC | chr11 | 88045722 | - | 1863 | 375 | 84 | 1448 | 454 |
| ENST00000378353 | MRS2 | chr6 | 24405469 | + | ENST00000227266 | CTSC | chr11 | 88045722 | - | 1845 | 357 | 66 | 1430 | 454 |
| ENST00000378386 | MRS2 | chr6 | 24405469 | + | ENST00000227266 | CTSC | chr11 | 88045722 | - | 1845 | 357 | 66 | 1430 | 454 |
| ENST00000443868 | MRS2 | chr6 | 24405469 | + | ENST00000227266 | CTSC | chr11 | 88045722 | - | 1840 | 352 | 61 | 1425 | 454 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000274747 | ENST00000227266 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - | 0.003955589 | 0.9960444 |
| ENST00000535061 | ENST00000227266 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - | 0.003821709 | 0.99617827 |
| ENST00000378353 | ENST00000227266 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - | 0.003798486 | 0.99620146 |
| ENST00000378386 | ENST00000227266 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - | 0.003798486 | 0.99620146 |
| ENST00000443868 | ENST00000227266 | MRS2 | chr6 | 24405469 | + | CTSC | chr11 | 88045722 | - | 0.003802447 | 0.9961976 |
Top |
Fusion Genomic Features for MRS2-CTSC |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
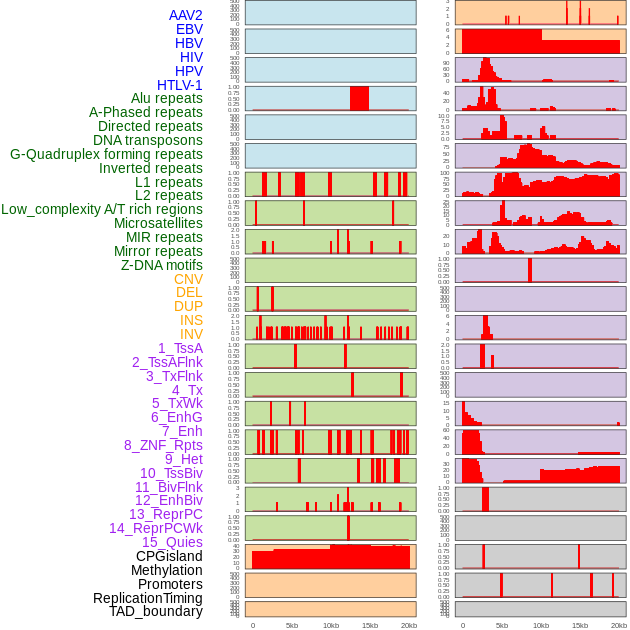 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for MRS2-CTSC |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr6:24405469/chr11:88045722) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MRS2 | CTSC |
| FUNCTION: Magnesium transporter that mediates the influx of magnesium into the mitochondrial matrix (PubMed:11401429, PubMed:18384665). Required for normal expression of the mitochondrial respiratory complex I subunits (PubMed:18384665). {ECO:0000269|PubMed:11401429, ECO:0000269|PubMed:18384665}. | FUNCTION: Thiol protease. Has dipeptidylpeptidase activity. Active against a broad range of dipeptide substrates composed of both polar and hydrophobic amino acids. Proline cannot occupy the P1 position and arginine cannot occupy the P2 position of the substrate. Can act as both an exopeptidase and endopeptidase. Activates serine proteases such as elastase, cathepsin G and granzymes A and B. Can also activate neuraminidase and factor XIII. {ECO:0000269|PubMed:1586157}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MRS2 | chr6:24405469 | chr11:88045722 | ENST00000274747 | + | 2 | 10 | 361_372 | 88 | 713.3333333333334 | Topological domain | Mitochondrial intermembrane |
| Hgene | MRS2 | chr6:24405469 | chr11:88045722 | ENST00000274747 | + | 2 | 10 | 394_443 | 88 | 713.3333333333334 | Topological domain | Mitochondrial matrix |
| Hgene | MRS2 | chr6:24405469 | chr11:88045722 | ENST00000274747 | + | 2 | 10 | 50_339 | 88 | 713.3333333333334 | Topological domain | Mitochondrial matrix |
| Hgene | MRS2 | chr6:24405469 | chr11:88045722 | ENST00000378353 | + | 2 | 10 | 361_372 | 88 | 409.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | MRS2 | chr6:24405469 | chr11:88045722 | ENST00000378353 | + | 2 | 10 | 394_443 | 88 | 409.0 | Topological domain | Mitochondrial matrix |
| Hgene | MRS2 | chr6:24405469 | chr11:88045722 | ENST00000378353 | + | 2 | 10 | 50_339 | 88 | 409.0 | Topological domain | Mitochondrial matrix |
| Hgene | MRS2 | chr6:24405469 | chr11:88045722 | ENST00000378386 | + | 2 | 11 | 361_372 | 88 | 444.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | MRS2 | chr6:24405469 | chr11:88045722 | ENST00000378386 | + | 2 | 11 | 394_443 | 88 | 444.0 | Topological domain | Mitochondrial matrix |
| Hgene | MRS2 | chr6:24405469 | chr11:88045722 | ENST00000378386 | + | 2 | 11 | 50_339 | 88 | 444.0 | Topological domain | Mitochondrial matrix |
| Hgene | MRS2 | chr6:24405469 | chr11:88045722 | ENST00000274747 | + | 2 | 10 | 340_360 | 88 | 713.3333333333334 | Transmembrane | Helical |
| Hgene | MRS2 | chr6:24405469 | chr11:88045722 | ENST00000274747 | + | 2 | 10 | 373_393 | 88 | 713.3333333333334 | Transmembrane | Helical |
| Hgene | MRS2 | chr6:24405469 | chr11:88045722 | ENST00000378353 | + | 2 | 10 | 340_360 | 88 | 409.0 | Transmembrane | Helical |
| Hgene | MRS2 | chr6:24405469 | chr11:88045722 | ENST00000378353 | + | 2 | 10 | 373_393 | 88 | 409.0 | Transmembrane | Helical |
| Hgene | MRS2 | chr6:24405469 | chr11:88045722 | ENST00000378386 | + | 2 | 11 | 340_360 | 88 | 444.0 | Transmembrane | Helical |
| Hgene | MRS2 | chr6:24405469 | chr11:88045722 | ENST00000378386 | + | 2 | 11 | 373_393 | 88 | 444.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for MRS2-CTSC |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >55240_55240_1_MRS2-CTSC_MRS2_chr6_24405469_ENST00000274747_CTSC_chr11_88045722_ENST00000227266_length(transcript)=1874nt_BP=386nt ATGCCTGGTGCACAGAGTCTGCAGGTCGGGCGGTAGCGACAGGTCAGAGCTGCGGCCTGAGCAGCCAGCGTCCGGCATGAAGGTCTGGGG TCTGGCTGCTGCCTGCTTCTTGCTCCAGCACCATGGAATGCCTGCGCAGTTTACCCTGCCTCCTGCCCCGCGCGATGAGACTTCCCCGGC GGACGCTGTGTGCCCTGGCCTTGGACGTGACCTCTGTGGGTCCTCCCGTTGCTGCCTGCGGCCGCCGAGCCAACCTGATTGGAAGGAGCC GAGCGGCGCAGCTTTGCGGGCCCGACCGGCTCCGCGTGGCAGGTGAAGTGCACCGGTTTAGAACCTCTGACGTCTCTCAAGCCACTTTAG CCAGTGTAGCCCCAGTATTTACTGTGTATAAAGAAGAGGGCAGCAAGGTGACCACTTACTGCAACGAGACAATGACTGGGTGGGTGCATG ATGTGTTGGGCCGGAACTGGGCTTGTTTCACCGGAAAGAAGGTGGGAACTGCCTCTGAGAATGTGTATGTCAACATAGCACACCTTAAGA ATTCTCAGGAAAAGTATTCTAATAGGCTCTACAAGTATGATCACAACTTTGTGAAAGCTATCAATGCCATTCAGAAGTCTTGGACTGCAA CTACATACATGGAATATGAGACTCTTACCCTGGGAGATATGATTAGGAGAAGTGGTGGCCACAGTCGAAAAATCCCAAGGCCCAAACCTG CACCACTGACTGCTGAAATACAGCAAAAGATTTTGCATTTGCCAACATCTTGGGACTGGAGAAATGTTCATGGTATCAATTTTGTCAGTC CTGTTCGAAACCAAGCATCCTGTGGCAGCTGCTACTCATTTGCTTCTATGGGTATGCTAGAAGCGAGAATCCGTATACTAACCAACAATT CTCAGACCCCAATCCTAAGCCCTCAGGAGGTTGTGTCTTGTAGCCAGTATGCTCAAGGCTGTGAAGGCGGCTTCCCATACCTTATTGCAG GAAAGTACGCCCAAGATTTTGGGCTGGTGGAAGAAGCTTGCTTCCCCTACACAGGCACTGATTCTCCATGCAAAATGAAGGAAGACTGCT TTCGTTATTACTCCTCTGAGTACCACTATGTAGGAGGTTTCTATGGAGGCTGCAATGAAGCCCTGATGAAGCTTGAGTTGGTCCATCATG GGCCCATGGCAGTTGCTTTTGAAGTATATGATGACTTCCTCCACTACAAAAAGGGGATCTACCACCACACTGGTCTAAGAGACCCTTTCA ACCCCTTTGAGCTGACTAATCATGCTGTTCTGCTTGTGGGCTATGGCACTGACTCAGCCTCTGGGATGGATTACTGGATTGTTAAAAACA GCTGGGGCACCGGCTGGGGTGAGAATGGCTACTTCCGGATCCGCAGAGGAACTGATGAGTGTGCAATTGAGAGCATAGCAGTGGCAGCCA CACCAATTCCTAAATTGTAGGGTATGCCTTCCAGTATTTCATAATGATCTGCATCAGTTGTAAAGGGGAATTGGTATATTCACAGACTGT AGACTTTCAGCAGCAATCTCAGAAGCTTACAAATAGATTTCCATGAAGATATTTGTCTTCAGAATTAAAACTGCCCTTAATTTTAATATA CCTTTCAATCGGCCACTGGCCATTTTTTTCTAAGTATTCAATTAAGTGGGAATTTTCTGGAAGATGGTCAGCTATGAAGTAATAGAGTTT GCTTAATCATTTGTAATTCAAACATGCTATATTTTTTAAAATCAATGTGAAAACATAGACTTATTTTTAAATTGTACCAATCACAAGAAA >55240_55240_1_MRS2-CTSC_MRS2_chr6_24405469_ENST00000274747_CTSC_chr11_88045722_ENST00000227266_length(amino acids)=454AA_BP=97 MLPASCSSTMECLRSLPCLLPRAMRLPRRTLCALALDVTSVGPPVAACGRRANLIGRSRAAQLCGPDRLRVAGEVHRFRTSDVSQATLAS VAPVFTVYKEEGSKVTTYCNETMTGWVHDVLGRNWACFTGKKVGTASENVYVNIAHLKNSQEKYSNRLYKYDHNFVKAINAIQKSWTATT YMEYETLTLGDMIRRSGGHSRKIPRPKPAPLTAEIQQKILHLPTSWDWRNVHGINFVSPVRNQASCGSCYSFASMGMLEARIRILTNNSQ TPILSPQEVVSCSQYAQGCEGGFPYLIAGKYAQDFGLVEEACFPYTGTDSPCKMKEDCFRYYSSEYHYVGGFYGGCNEALMKLELVHHGP MAVAFEVYDDFLHYKKGIYHHTGLRDPFNPFELTNHAVLLVGYGTDSASGMDYWIVKNSWGTGWGENGYFRIRRGTDECAIESIAVAATP -------------------------------------------------------------- >55240_55240_2_MRS2-CTSC_MRS2_chr6_24405469_ENST00000378353_CTSC_chr11_88045722_ENST00000227266_length(transcript)=1845nt_BP=357nt GCGGTAGCGACAGGTCAGAGCTGCGGCCTGAGCAGCCAGCGTCCGGCATGAAGGTCTGGGGTCTGGCTGCTGCCTGCTTCTTGCTCCAGC ACCATGGAATGCCTGCGCAGTTTACCCTGCCTCCTGCCCCGCGCGATGAGACTTCCCCGGCGGACGCTGTGTGCCCTGGCCTTGGACGTG ACCTCTGTGGGTCCTCCCGTTGCTGCCTGCGGCCGCCGAGCCAACCTGATTGGAAGGAGCCGAGCGGCGCAGCTTTGCGGGCCCGACCGG CTCCGCGTGGCAGGTGAAGTGCACCGGTTTAGAACCTCTGACGTCTCTCAAGCCACTTTAGCCAGTGTAGCCCCAGTATTTACTGTGTAT AAAGAAGAGGGCAGCAAGGTGACCACTTACTGCAACGAGACAATGACTGGGTGGGTGCATGATGTGTTGGGCCGGAACTGGGCTTGTTTC ACCGGAAAGAAGGTGGGAACTGCCTCTGAGAATGTGTATGTCAACATAGCACACCTTAAGAATTCTCAGGAAAAGTATTCTAATAGGCTC TACAAGTATGATCACAACTTTGTGAAAGCTATCAATGCCATTCAGAAGTCTTGGACTGCAACTACATACATGGAATATGAGACTCTTACC CTGGGAGATATGATTAGGAGAAGTGGTGGCCACAGTCGAAAAATCCCAAGGCCCAAACCTGCACCACTGACTGCTGAAATACAGCAAAAG ATTTTGCATTTGCCAACATCTTGGGACTGGAGAAATGTTCATGGTATCAATTTTGTCAGTCCTGTTCGAAACCAAGCATCCTGTGGCAGC TGCTACTCATTTGCTTCTATGGGTATGCTAGAAGCGAGAATCCGTATACTAACCAACAATTCTCAGACCCCAATCCTAAGCCCTCAGGAG GTTGTGTCTTGTAGCCAGTATGCTCAAGGCTGTGAAGGCGGCTTCCCATACCTTATTGCAGGAAAGTACGCCCAAGATTTTGGGCTGGTG GAAGAAGCTTGCTTCCCCTACACAGGCACTGATTCTCCATGCAAAATGAAGGAAGACTGCTTTCGTTATTACTCCTCTGAGTACCACTAT GTAGGAGGTTTCTATGGAGGCTGCAATGAAGCCCTGATGAAGCTTGAGTTGGTCCATCATGGGCCCATGGCAGTTGCTTTTGAAGTATAT GATGACTTCCTCCACTACAAAAAGGGGATCTACCACCACACTGGTCTAAGAGACCCTTTCAACCCCTTTGAGCTGACTAATCATGCTGTT CTGCTTGTGGGCTATGGCACTGACTCAGCCTCTGGGATGGATTACTGGATTGTTAAAAACAGCTGGGGCACCGGCTGGGGTGAGAATGGC TACTTCCGGATCCGCAGAGGAACTGATGAGTGTGCAATTGAGAGCATAGCAGTGGCAGCCACACCAATTCCTAAATTGTAGGGTATGCCT TCCAGTATTTCATAATGATCTGCATCAGTTGTAAAGGGGAATTGGTATATTCACAGACTGTAGACTTTCAGCAGCAATCTCAGAAGCTTA CAAATAGATTTCCATGAAGATATTTGTCTTCAGAATTAAAACTGCCCTTAATTTTAATATACCTTTCAATCGGCCACTGGCCATTTTTTT CTAAGTATTCAATTAAGTGGGAATTTTCTGGAAGATGGTCAGCTATGAAGTAATAGAGTTTGCTTAATCATTTGTAATTCAAACATGCTA TATTTTTTAAAATCAATGTGAAAACATAGACTTATTTTTAAATTGTACCAATCACAAGAAAATAATGGCAATAATTATCAAAACTTTTAA >55240_55240_2_MRS2-CTSC_MRS2_chr6_24405469_ENST00000378353_CTSC_chr11_88045722_ENST00000227266_length(amino acids)=454AA_BP=97 MLPASCSSTMECLRSLPCLLPRAMRLPRRTLCALALDVTSVGPPVAACGRRANLIGRSRAAQLCGPDRLRVAGEVHRFRTSDVSQATLAS VAPVFTVYKEEGSKVTTYCNETMTGWVHDVLGRNWACFTGKKVGTASENVYVNIAHLKNSQEKYSNRLYKYDHNFVKAINAIQKSWTATT YMEYETLTLGDMIRRSGGHSRKIPRPKPAPLTAEIQQKILHLPTSWDWRNVHGINFVSPVRNQASCGSCYSFASMGMLEARIRILTNNSQ TPILSPQEVVSCSQYAQGCEGGFPYLIAGKYAQDFGLVEEACFPYTGTDSPCKMKEDCFRYYSSEYHYVGGFYGGCNEALMKLELVHHGP MAVAFEVYDDFLHYKKGIYHHTGLRDPFNPFELTNHAVLLVGYGTDSASGMDYWIVKNSWGTGWGENGYFRIRRGTDECAIESIAVAATP -------------------------------------------------------------- >55240_55240_3_MRS2-CTSC_MRS2_chr6_24405469_ENST00000378386_CTSC_chr11_88045722_ENST00000227266_length(transcript)=1845nt_BP=357nt GCGGTAGCGACAGGTCAGAGCTGCGGCCTGAGCAGCCAGCGTCCGGCATGAAGGTCTGGGGTCTGGCTGCTGCCTGCTTCTTGCTCCAGC ACCATGGAATGCCTGCGCAGTTTACCCTGCCTCCTGCCCCGCGCGATGAGACTTCCCCGGCGGACGCTGTGTGCCCTGGCCTTGGACGTG ACCTCTGTGGGTCCTCCCGTTGCTGCCTGCGGCCGCCGAGCCAACCTGATTGGAAGGAGCCGAGCGGCGCAGCTTTGCGGGCCCGACCGG CTCCGCGTGGCAGGTGAAGTGCACCGGTTTAGAACCTCTGACGTCTCTCAAGCCACTTTAGCCAGTGTAGCCCCAGTATTTACTGTGTAT AAAGAAGAGGGCAGCAAGGTGACCACTTACTGCAACGAGACAATGACTGGGTGGGTGCATGATGTGTTGGGCCGGAACTGGGCTTGTTTC ACCGGAAAGAAGGTGGGAACTGCCTCTGAGAATGTGTATGTCAACATAGCACACCTTAAGAATTCTCAGGAAAAGTATTCTAATAGGCTC TACAAGTATGATCACAACTTTGTGAAAGCTATCAATGCCATTCAGAAGTCTTGGACTGCAACTACATACATGGAATATGAGACTCTTACC CTGGGAGATATGATTAGGAGAAGTGGTGGCCACAGTCGAAAAATCCCAAGGCCCAAACCTGCACCACTGACTGCTGAAATACAGCAAAAG ATTTTGCATTTGCCAACATCTTGGGACTGGAGAAATGTTCATGGTATCAATTTTGTCAGTCCTGTTCGAAACCAAGCATCCTGTGGCAGC TGCTACTCATTTGCTTCTATGGGTATGCTAGAAGCGAGAATCCGTATACTAACCAACAATTCTCAGACCCCAATCCTAAGCCCTCAGGAG GTTGTGTCTTGTAGCCAGTATGCTCAAGGCTGTGAAGGCGGCTTCCCATACCTTATTGCAGGAAAGTACGCCCAAGATTTTGGGCTGGTG GAAGAAGCTTGCTTCCCCTACACAGGCACTGATTCTCCATGCAAAATGAAGGAAGACTGCTTTCGTTATTACTCCTCTGAGTACCACTAT GTAGGAGGTTTCTATGGAGGCTGCAATGAAGCCCTGATGAAGCTTGAGTTGGTCCATCATGGGCCCATGGCAGTTGCTTTTGAAGTATAT GATGACTTCCTCCACTACAAAAAGGGGATCTACCACCACACTGGTCTAAGAGACCCTTTCAACCCCTTTGAGCTGACTAATCATGCTGTT CTGCTTGTGGGCTATGGCACTGACTCAGCCTCTGGGATGGATTACTGGATTGTTAAAAACAGCTGGGGCACCGGCTGGGGTGAGAATGGC TACTTCCGGATCCGCAGAGGAACTGATGAGTGTGCAATTGAGAGCATAGCAGTGGCAGCCACACCAATTCCTAAATTGTAGGGTATGCCT TCCAGTATTTCATAATGATCTGCATCAGTTGTAAAGGGGAATTGGTATATTCACAGACTGTAGACTTTCAGCAGCAATCTCAGAAGCTTA CAAATAGATTTCCATGAAGATATTTGTCTTCAGAATTAAAACTGCCCTTAATTTTAATATACCTTTCAATCGGCCACTGGCCATTTTTTT CTAAGTATTCAATTAAGTGGGAATTTTCTGGAAGATGGTCAGCTATGAAGTAATAGAGTTTGCTTAATCATTTGTAATTCAAACATGCTA TATTTTTTAAAATCAATGTGAAAACATAGACTTATTTTTAAATTGTACCAATCACAAGAAAATAATGGCAATAATTATCAAAACTTTTAA >55240_55240_3_MRS2-CTSC_MRS2_chr6_24405469_ENST00000378386_CTSC_chr11_88045722_ENST00000227266_length(amino acids)=454AA_BP=97 MLPASCSSTMECLRSLPCLLPRAMRLPRRTLCALALDVTSVGPPVAACGRRANLIGRSRAAQLCGPDRLRVAGEVHRFRTSDVSQATLAS VAPVFTVYKEEGSKVTTYCNETMTGWVHDVLGRNWACFTGKKVGTASENVYVNIAHLKNSQEKYSNRLYKYDHNFVKAINAIQKSWTATT YMEYETLTLGDMIRRSGGHSRKIPRPKPAPLTAEIQQKILHLPTSWDWRNVHGINFVSPVRNQASCGSCYSFASMGMLEARIRILTNNSQ TPILSPQEVVSCSQYAQGCEGGFPYLIAGKYAQDFGLVEEACFPYTGTDSPCKMKEDCFRYYSSEYHYVGGFYGGCNEALMKLELVHHGP MAVAFEVYDDFLHYKKGIYHHTGLRDPFNPFELTNHAVLLVGYGTDSASGMDYWIVKNSWGTGWGENGYFRIRRGTDECAIESIAVAATP -------------------------------------------------------------- >55240_55240_4_MRS2-CTSC_MRS2_chr6_24405469_ENST00000443868_CTSC_chr11_88045722_ENST00000227266_length(transcript)=1840nt_BP=352nt AGCGACAGGTCAGAGCTGCGGCCTGAGCAGCCAGCGTCCGGCATGAAGGTCTGGGGTCTGGCTGCTGCCTGCTTCTTGCTCCAGCACCAT GGAATGCCTGCGCAGTTTACCCTGCCTCCTGCCCCGCGCGATGAGACTTCCCCGGCGGACGCTGTGTGCCCTGGCCTTGGACGTGACCTC TGTGGGTCCTCCCGTTGCTGCCTGCGGCCGCCGAGCCAACCTGATTGGAAGGAGCCGAGCGGCGCAGCTTTGCGGGCCCGACCGGCTCCG CGTGGCAGGTGAAGTGCACCGGTTTAGAACCTCTGACGTCTCTCAAGCCACTTTAGCCAGTGTAGCCCCAGTATTTACTGTGTATAAAGA AGAGGGCAGCAAGGTGACCACTTACTGCAACGAGACAATGACTGGGTGGGTGCATGATGTGTTGGGCCGGAACTGGGCTTGTTTCACCGG AAAGAAGGTGGGAACTGCCTCTGAGAATGTGTATGTCAACATAGCACACCTTAAGAATTCTCAGGAAAAGTATTCTAATAGGCTCTACAA GTATGATCACAACTTTGTGAAAGCTATCAATGCCATTCAGAAGTCTTGGACTGCAACTACATACATGGAATATGAGACTCTTACCCTGGG AGATATGATTAGGAGAAGTGGTGGCCACAGTCGAAAAATCCCAAGGCCCAAACCTGCACCACTGACTGCTGAAATACAGCAAAAGATTTT GCATTTGCCAACATCTTGGGACTGGAGAAATGTTCATGGTATCAATTTTGTCAGTCCTGTTCGAAACCAAGCATCCTGTGGCAGCTGCTA CTCATTTGCTTCTATGGGTATGCTAGAAGCGAGAATCCGTATACTAACCAACAATTCTCAGACCCCAATCCTAAGCCCTCAGGAGGTTGT GTCTTGTAGCCAGTATGCTCAAGGCTGTGAAGGCGGCTTCCCATACCTTATTGCAGGAAAGTACGCCCAAGATTTTGGGCTGGTGGAAGA AGCTTGCTTCCCCTACACAGGCACTGATTCTCCATGCAAAATGAAGGAAGACTGCTTTCGTTATTACTCCTCTGAGTACCACTATGTAGG AGGTTTCTATGGAGGCTGCAATGAAGCCCTGATGAAGCTTGAGTTGGTCCATCATGGGCCCATGGCAGTTGCTTTTGAAGTATATGATGA CTTCCTCCACTACAAAAAGGGGATCTACCACCACACTGGTCTAAGAGACCCTTTCAACCCCTTTGAGCTGACTAATCATGCTGTTCTGCT TGTGGGCTATGGCACTGACTCAGCCTCTGGGATGGATTACTGGATTGTTAAAAACAGCTGGGGCACCGGCTGGGGTGAGAATGGCTACTT CCGGATCCGCAGAGGAACTGATGAGTGTGCAATTGAGAGCATAGCAGTGGCAGCCACACCAATTCCTAAATTGTAGGGTATGCCTTCCAG TATTTCATAATGATCTGCATCAGTTGTAAAGGGGAATTGGTATATTCACAGACTGTAGACTTTCAGCAGCAATCTCAGAAGCTTACAAAT AGATTTCCATGAAGATATTTGTCTTCAGAATTAAAACTGCCCTTAATTTTAATATACCTTTCAATCGGCCACTGGCCATTTTTTTCTAAG TATTCAATTAAGTGGGAATTTTCTGGAAGATGGTCAGCTATGAAGTAATAGAGTTTGCTTAATCATTTGTAATTCAAACATGCTATATTT TTTAAAATCAATGTGAAAACATAGACTTATTTTTAAATTGTACCAATCACAAGAAAATAATGGCAATAATTATCAAAACTTTTAAAATAG >55240_55240_4_MRS2-CTSC_MRS2_chr6_24405469_ENST00000443868_CTSC_chr11_88045722_ENST00000227266_length(amino acids)=454AA_BP=97 MLPASCSSTMECLRSLPCLLPRAMRLPRRTLCALALDVTSVGPPVAACGRRANLIGRSRAAQLCGPDRLRVAGEVHRFRTSDVSQATLAS VAPVFTVYKEEGSKVTTYCNETMTGWVHDVLGRNWACFTGKKVGTASENVYVNIAHLKNSQEKYSNRLYKYDHNFVKAINAIQKSWTATT YMEYETLTLGDMIRRSGGHSRKIPRPKPAPLTAEIQQKILHLPTSWDWRNVHGINFVSPVRNQASCGSCYSFASMGMLEARIRILTNNSQ TPILSPQEVVSCSQYAQGCEGGFPYLIAGKYAQDFGLVEEACFPYTGTDSPCKMKEDCFRYYSSEYHYVGGFYGGCNEALMKLELVHHGP MAVAFEVYDDFLHYKKGIYHHTGLRDPFNPFELTNHAVLLVGYGTDSASGMDYWIVKNSWGTGWGENGYFRIRRGTDECAIESIAVAATP -------------------------------------------------------------- >55240_55240_5_MRS2-CTSC_MRS2_chr6_24405469_ENST00000535061_CTSC_chr11_88045722_ENST00000227266_length(transcript)=1863nt_BP=375nt ACAGAGTCTGCAGGTCGGGCGGTAGCGACAGGTCAGAGCTGCGGCCTGAGCAGCCAGCGTCCGGCATGAAGGTCTGGGGTCTGGCTGCTG CCTGCTTCTTGCTCCAGCACCATGGAATGCCTGCGCAGTTTACCCTGCCTCCTGCCCCGCGCGATGAGACTTCCCCGGCGGACGCTGTGT GCCCTGGCCTTGGACGTGACCTCTGTGGGTCCTCCCGTTGCTGCCTGCGGCCGCCGAGCCAACCTGATTGGAAGGAGCCGAGCGGCGCAG CTTTGCGGGCCCGACCGGCTCCGCGTGGCAGGTGAAGTGCACCGGTTTAGAACCTCTGACGTCTCTCAAGCCACTTTAGCCAGTGTAGCC CCAGTATTTACTGTGTATAAAGAAGAGGGCAGCAAGGTGACCACTTACTGCAACGAGACAATGACTGGGTGGGTGCATGATGTGTTGGGC CGGAACTGGGCTTGTTTCACCGGAAAGAAGGTGGGAACTGCCTCTGAGAATGTGTATGTCAACATAGCACACCTTAAGAATTCTCAGGAA AAGTATTCTAATAGGCTCTACAAGTATGATCACAACTTTGTGAAAGCTATCAATGCCATTCAGAAGTCTTGGACTGCAACTACATACATG GAATATGAGACTCTTACCCTGGGAGATATGATTAGGAGAAGTGGTGGCCACAGTCGAAAAATCCCAAGGCCCAAACCTGCACCACTGACT GCTGAAATACAGCAAAAGATTTTGCATTTGCCAACATCTTGGGACTGGAGAAATGTTCATGGTATCAATTTTGTCAGTCCTGTTCGAAAC CAAGCATCCTGTGGCAGCTGCTACTCATTTGCTTCTATGGGTATGCTAGAAGCGAGAATCCGTATACTAACCAACAATTCTCAGACCCCA ATCCTAAGCCCTCAGGAGGTTGTGTCTTGTAGCCAGTATGCTCAAGGCTGTGAAGGCGGCTTCCCATACCTTATTGCAGGAAAGTACGCC CAAGATTTTGGGCTGGTGGAAGAAGCTTGCTTCCCCTACACAGGCACTGATTCTCCATGCAAAATGAAGGAAGACTGCTTTCGTTATTAC TCCTCTGAGTACCACTATGTAGGAGGTTTCTATGGAGGCTGCAATGAAGCCCTGATGAAGCTTGAGTTGGTCCATCATGGGCCCATGGCA GTTGCTTTTGAAGTATATGATGACTTCCTCCACTACAAAAAGGGGATCTACCACCACACTGGTCTAAGAGACCCTTTCAACCCCTTTGAG CTGACTAATCATGCTGTTCTGCTTGTGGGCTATGGCACTGACTCAGCCTCTGGGATGGATTACTGGATTGTTAAAAACAGCTGGGGCACC GGCTGGGGTGAGAATGGCTACTTCCGGATCCGCAGAGGAACTGATGAGTGTGCAATTGAGAGCATAGCAGTGGCAGCCACACCAATTCCT AAATTGTAGGGTATGCCTTCCAGTATTTCATAATGATCTGCATCAGTTGTAAAGGGGAATTGGTATATTCACAGACTGTAGACTTTCAGC AGCAATCTCAGAAGCTTACAAATAGATTTCCATGAAGATATTTGTCTTCAGAATTAAAACTGCCCTTAATTTTAATATACCTTTCAATCG GCCACTGGCCATTTTTTTCTAAGTATTCAATTAAGTGGGAATTTTCTGGAAGATGGTCAGCTATGAAGTAATAGAGTTTGCTTAATCATT TGTAATTCAAACATGCTATATTTTTTAAAATCAATGTGAAAACATAGACTTATTTTTAAATTGTACCAATCACAAGAAAATAATGGCAAT >55240_55240_5_MRS2-CTSC_MRS2_chr6_24405469_ENST00000535061_CTSC_chr11_88045722_ENST00000227266_length(amino acids)=454AA_BP=97 MLPASCSSTMECLRSLPCLLPRAMRLPRRTLCALALDVTSVGPPVAACGRRANLIGRSRAAQLCGPDRLRVAGEVHRFRTSDVSQATLAS VAPVFTVYKEEGSKVTTYCNETMTGWVHDVLGRNWACFTGKKVGTASENVYVNIAHLKNSQEKYSNRLYKYDHNFVKAINAIQKSWTATT YMEYETLTLGDMIRRSGGHSRKIPRPKPAPLTAEIQQKILHLPTSWDWRNVHGINFVSPVRNQASCGSCYSFASMGMLEARIRILTNNSQ TPILSPQEVVSCSQYAQGCEGGFPYLIAGKYAQDFGLVEEACFPYTGTDSPCKMKEDCFRYYSSEYHYVGGFYGGCNEALMKLELVHHGP MAVAFEVYDDFLHYKKGIYHHTGLRDPFNPFELTNHAVLLVGYGTDSASGMDYWIVKNSWGTGWGENGYFRIRRGTDECAIESIAVAATP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MRS2-CTSC |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MRS2-CTSC |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MRS2-CTSC |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
