
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MTDH-LAPTM4B (FusionGDB2 ID:55602) |
Fusion Gene Summary for MTDH-LAPTM4B |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MTDH-LAPTM4B | Fusion gene ID: 55602 | Hgene | Tgene | Gene symbol | MTDH | LAPTM4B | Gene ID | 92140 | 55353 |
| Gene name | metadherin | lysosomal protein transmembrane 4 beta | |
| Synonyms | 3D3|AEG-1|AEG1|LYRIC|LYRIC/3D3 | LAPTM4beta|LC27 | |
| Cytomap | 8q22.1 | 8q22.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein LYRIC3D3/LYRICastrocyte elevated gene 1astrocyte elevated gene-1 proteinlysine-rich CEACAM1 co-isolated proteinmetastasis adhesion protein | lysosomal-associated transmembrane protein 4Blysosomal associated protein transmembrane 4 betalysosome-associated transmembrane protein 4-beta | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q86UE4 | Q86VI4 | |
| Ensembl transtripts involved in fusion gene | ENST00000336273, ENST00000519934, | ENST00000445593, ENST00000521545, | |
| Fusion gene scores | * DoF score | 15 X 10 X 9=1350 | 14 X 8 X 8=896 |
| # samples | 20 | 16 | |
| ** MAII score | log2(20/1350*10)=-2.75488750216347 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(16/896*10)=-2.48542682717024 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MTDH [Title/Abstract] AND LAPTM4B [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MTDH(98673401)-LAPTM4B(98817581), # samples:2 LAPTM4B(98817692)-MTDH(98731276), # samples:1 LAPTM4B(98837381)-MTDH(98673300), # samples:1 LAPTM4B(98817692)-MTDH(98731277), # samples:1 LAPTM4B(98788336)-MTDH(98736828), # samples:1 LAPTM4B(98837381)-MTDH(98735107), # samples:1 LAPTM4B(98837381)-MTDH(98736828), # samples:1 LAPTM4B(98837381)-MTDH(98698896), # samples:1 LAPTM4B(98837381)-MTDH(98731277), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | MTDH | GO:0000122 | negative regulation of transcription by RNA polymerase II | 15927426 |
| Hgene | MTDH | GO:0010508 | positive regulation of autophagy | 21127263 |
| Hgene | MTDH | GO:0043066 | negative regulation of apoptotic process | 17704808 |
| Hgene | MTDH | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 16452207|18316612 |
| Hgene | MTDH | GO:0045766 | positive regulation of angiogenesis | 19940250 |
| Hgene | MTDH | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 16452207|18316612 |
| Hgene | MTDH | GO:0051897 | positive regulation of protein kinase B signaling | 17704808 |
| Tgene | LAPTM4B | GO:0032509 | endosome transport via multivesicular body sorting pathway | 25588945 |
| Tgene | LAPTM4B | GO:0032911 | negative regulation of transforming growth factor beta1 production | 26126825 |
 Fusion gene breakpoints across MTDH (5'-gene) Fusion gene breakpoints across MTDH (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
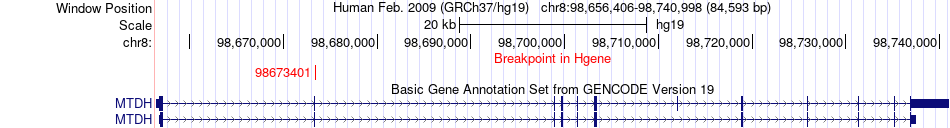 |
 Fusion gene breakpoints across LAPTM4B (3'-gene) Fusion gene breakpoints across LAPTM4B (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
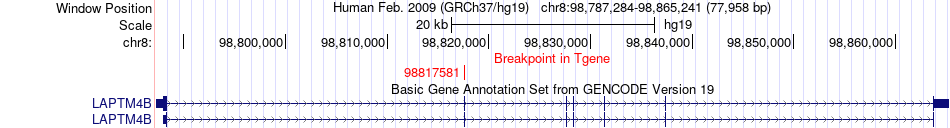 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | COAD | TCGA-A6-6782-01A | MTDH | chr8 | 98673401 | + | LAPTM4B | chr8 | 98817581 | + |
| ChimerDB4 | OV | TCGA-61-2088-01A | MTDH | chr8 | 98673401 | + | LAPTM4B | chr8 | 98817581 | + |
Top |
Fusion Gene ORF analysis for MTDH-LAPTM4B |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000336273 | ENST00000445593 | MTDH | chr8 | 98673401 | + | LAPTM4B | chr8 | 98817581 | + |
| In-frame | ENST00000336273 | ENST00000521545 | MTDH | chr8 | 98673401 | + | LAPTM4B | chr8 | 98817581 | + |
| In-frame | ENST00000519934 | ENST00000445593 | MTDH | chr8 | 98673401 | + | LAPTM4B | chr8 | 98817581 | + |
| In-frame | ENST00000519934 | ENST00000521545 | MTDH | chr8 | 98673401 | + | LAPTM4B | chr8 | 98817581 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000336273 | MTDH | chr8 | 98673401 | + | ENST00000445593 | LAPTM4B | chr8 | 98817581 | + | 2932 | 811 | 301 | 1392 | 363 |
| ENST00000336273 | MTDH | chr8 | 98673401 | + | ENST00000521545 | LAPTM4B | chr8 | 98817581 | + | 1393 | 811 | 301 | 1392 | 364 |
| ENST00000519934 | MTDH | chr8 | 98673401 | + | ENST00000445593 | LAPTM4B | chr8 | 98817581 | + | 2565 | 444 | 21 | 1025 | 334 |
| ENST00000519934 | MTDH | chr8 | 98673401 | + | ENST00000521545 | LAPTM4B | chr8 | 98817581 | + | 1026 | 444 | 21 | 1025 | 334 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000336273 | ENST00000445593 | MTDH | chr8 | 98673401 | + | LAPTM4B | chr8 | 98817581 | + | 0.000620664 | 0.9993793 |
| ENST00000336273 | ENST00000521545 | MTDH | chr8 | 98673401 | + | LAPTM4B | chr8 | 98817581 | + | 0.003426064 | 0.9965739 |
| ENST00000519934 | ENST00000445593 | MTDH | chr8 | 98673401 | + | LAPTM4B | chr8 | 98817581 | + | 0.000409979 | 0.99959 |
| ENST00000519934 | ENST00000521545 | MTDH | chr8 | 98673401 | + | LAPTM4B | chr8 | 98817581 | + | 0.001560712 | 0.99843925 |
Top |
Fusion Genomic Features for MTDH-LAPTM4B |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
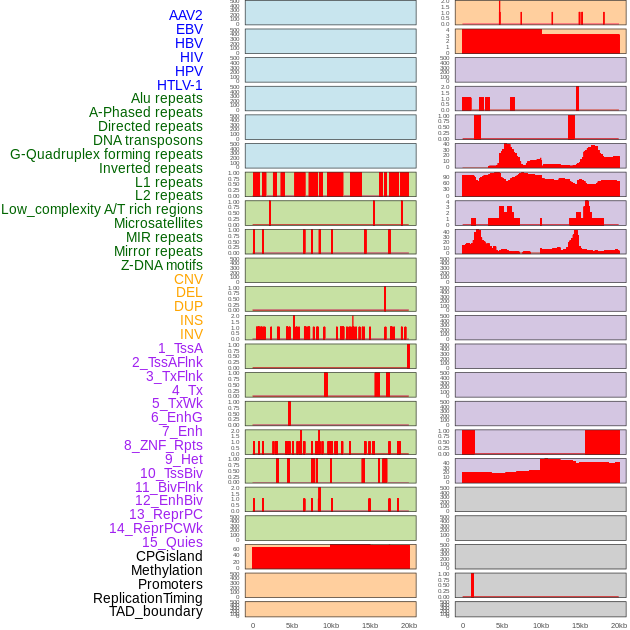 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
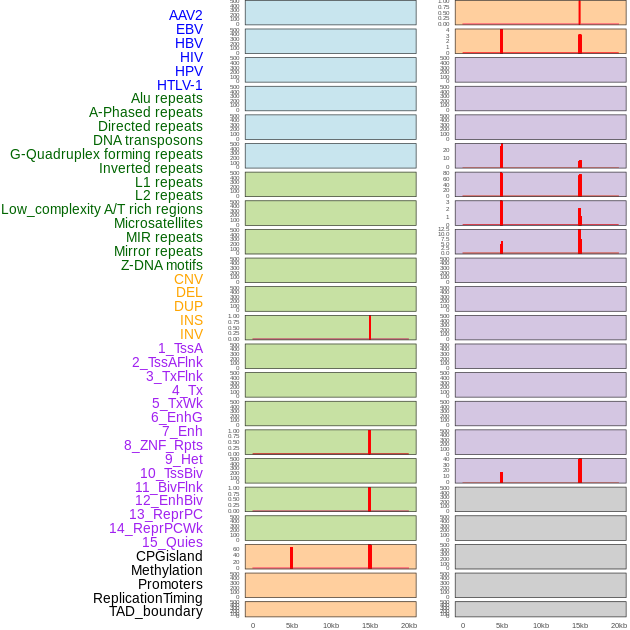 |
Top |
Fusion Protein Features for MTDH-LAPTM4B |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:98673401/chr8:98817581) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MTDH | LAPTM4B |
| FUNCTION: Downregulates SLC1A2/EAAT2 promoter activity when expressed ectopically. Activates the nuclear factor kappa-B (NF-kappa-B) transcription factor. Promotes anchorage-independent growth of immortalized melanocytes and astrocytes which is a key component in tumor cell expansion. Promotes lung metastasis and also has an effect on bone and brain metastasis, possibly by enhancing the seeding of tumor cells to the target organ endothelium. Induces chemoresistance. {ECO:0000269|PubMed:15927426, ECO:0000269|PubMed:16452207, ECO:0000269|PubMed:18316612, ECO:0000269|PubMed:19111877}. | FUNCTION: Required for optimal lysosomal function (PubMed:21224396). Blocks EGF-stimulated EGFR intraluminal sorting and degradation. Conversely by binding with the phosphatidylinositol 4,5-bisphosphate, regulates its PIP5K1C interaction, inhibits HGS ubiquitination and relieves LAPTM4B inhibition of EGFR degradation (PubMed:25588945). Recruits SLC3A2 and SLC7A5 (the Leu transporter) to the lysosome, promoting entry of leucine and other essential amino acid (EAA) into the lysosome, stimulating activation of proton-transporting vacuolar (V)-ATPase protein pump (V-ATPase) and hence mTORC1 activation (PubMed:25998567). Plays a role as negative regulator of TGFB1 production in regulatory T cells (PubMed:26126825). Binds ceramide and facilitates its exit from late endosome in order to control cell death pathways (PubMed:26280656). {ECO:0000269|PubMed:21224396, ECO:0000269|PubMed:25588945, ECO:0000269|PubMed:25998567, ECO:0000269|PubMed:26126825, ECO:0000269|PubMed:26280656}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MTDH | chr8:98673401 | chr8:98817581 | ENST00000336273 | + | 2 | 12 | 59_67 | 161 | 583.0 | Compositional bias | Note=Poly-Leu |
| Hgene | MTDH | chr8:98673401 | chr8:98817581 | ENST00000336273 | + | 2 | 12 | 1_71 | 161 | 583.0 | Region | Note=Activation of NF-kappa-B |
| Hgene | MTDH | chr8:98673401 | chr8:98817581 | ENST00000336273 | + | 2 | 12 | 1_48 | 161 | 583.0 | Topological domain | Lumenal |
| Hgene | MTDH | chr8:98673401 | chr8:98817581 | ENST00000336273 | + | 2 | 12 | 49_69 | 161 | 583.0 | Transmembrane | Helical |
| Tgene | LAPTM4B | chr8:98673401 | chr8:98817581 | ENST00000445593 | 0 | 7 | 163_183 | 124 | 318.0 | Transmembrane | Helical | |
| Tgene | LAPTM4B | chr8:98673401 | chr8:98817581 | ENST00000445593 | 0 | 7 | 191_211 | 124 | 318.0 | Transmembrane | Helical | |
| Tgene | LAPTM4B | chr8:98673401 | chr8:98817581 | ENST00000445593 | 0 | 7 | 244_264 | 124 | 318.0 | Transmembrane | Helical | |
| Tgene | LAPTM4B | chr8:98673401 | chr8:98817581 | ENST00000521545 | 0 | 7 | 117_137 | 33 | 227.0 | Transmembrane | Helical | |
| Tgene | LAPTM4B | chr8:98673401 | chr8:98817581 | ENST00000521545 | 0 | 7 | 163_183 | 33 | 227.0 | Transmembrane | Helical | |
| Tgene | LAPTM4B | chr8:98673401 | chr8:98817581 | ENST00000521545 | 0 | 7 | 191_211 | 33 | 227.0 | Transmembrane | Helical | |
| Tgene | LAPTM4B | chr8:98673401 | chr8:98817581 | ENST00000521545 | 0 | 7 | 244_264 | 33 | 227.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MTDH | chr8:98673401 | chr8:98817581 | ENST00000336273 | + | 2 | 12 | 441_451 | 161 | 583.0 | Compositional bias | Note=Poly-Lys |
| Hgene | MTDH | chr8:98673401 | chr8:98817581 | ENST00000336273 | + | 2 | 12 | 573_577 | 161 | 583.0 | Compositional bias | Note=Poly-Lys |
| Hgene | MTDH | chr8:98673401 | chr8:98817581 | ENST00000336273 | + | 2 | 12 | 381_443 | 161 | 583.0 | Region | Lung-homing for mammary tumors |
| Hgene | MTDH | chr8:98673401 | chr8:98817581 | ENST00000336273 | + | 2 | 12 | 70_582 | 161 | 583.0 | Topological domain | Cytoplasmic |
| Tgene | LAPTM4B | chr8:98673401 | chr8:98817581 | ENST00000445593 | 0 | 7 | 117_137 | 124 | 318.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for MTDH-LAPTM4B |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >55602_55602_1_MTDH-LAPTM4B_MTDH_chr8_98673401_ENST00000336273_LAPTM4B_chr8_98817581_ENST00000445593_length(transcript)=2932nt_BP=811nt GGCCCAAGCCGCCATTGTTCCGCCGAGGGAGGACAGCGGGGCCTGGCGCTGGCGCCGAGACGCCGCTTAGCGGCCGCCACTGGAGACACT CCCTCCCGCCTCCCGGGTCTCCTGGCGGCGGCGGAGTGAGGCTGACAGCGGGGAACCTGGGAGACCCCTCCGCCCTCCCCGCGGTGGCAG CGGCCGATCCCCGGCTCCGGCGCGAGGGACGGCCGCGATGCGCTCGGCCTGAGGTTACCCGGCCCGGCCCTTCCTCGCTTCCCTCGACTA TTCCACTGCGTCTCCGCGCCCCGGCGTCATCCTGCGAGTCCCTCTGACGGGAGGGAAGATGGCTGCACGGAGCTGGCAGGACGAGCTGGC CCAGCAGGCCGAGGAGGGCTCGGCCCGGCTGCGGGAAATGCTCTCGGTCGGCCTAGGCTTTCTGCGCACCGAGCTGGGCCTCGACCTGGG GCTGGAGCCGAAACGGTACCCCGGCTGGGTGATCCTGGTGGGCACTGGCGCGCTCGGGCTGCTGCTGCTGTTTCTGCTGGGCTACGGCTG GGCCGCGGCTTGCGCCGGCGCCCGCAAAAAGCGGAGGAGCCCGCCCCGCAAGCGGGAGGAGGCGGCGGCCGTGCCGGCCGCGGCCCCCGA CGACCTGGCCTTGCTGAAGAATCTCCGGAGCGAGGAACAGAAGAAGAAGAACCGGAAGAAACTGTCCGAGAAGCCCAAACCAAATGGGCG GACTGTTGAAGTGGCTGAGGGTGAAGCTGTTCGAACACCTCAAAGTGTAACAGCAAAGCAGCCACCAGAGATTGACAAGAAAAATGAAAA GATCATCAATGCTGTGGTACTGTTGATTTTATTGAGTGCCCTGGCTGATCCGGATCAGTATAACTTTTCAAGTTCTGAACTGGGAGGTGA CTTTGAGTTCATGGATGATGCCAACATGTGCATTGCCATTGCGATTTCTCTTCTCATGATCCTGATATGTGCTATGGCTACTTACGGAGC GTACAAGCAACGCGCAGCCTGGATCATCCCATTCTTCTGTTACCAGATCTTTGACTTTGCCCTGAACATGTTGGTTGCAATCACTGTGCT TATTTATCCAAACTCCATTCAGGAATACATACGGCAACTGCCTCCTAATTTTCCCTACAGAGATGATGTCATGTCAGTGAATCCTACCTG TTTGGTCCTTATTATTCTTCTGTTTATTAGCATTATCTTGACTTTTAAGGGTTACTTGATTAGCTGTGTTTGGAACTGCTACCGATACAT CAATGGTAGGAACTCCTCTGATGTCCTGGTTTATGTTACCAGCAATGACACTACGGTGCTGCTACCCCCGTATGATGATGCCACTGTGAA TGGTGCTGCCAAGGAGCCACCGCCACCTTACGTGTCTGCCTAAGCCTTCAAGTGGGCGGAGCTGAGGGCAGCAGCTTGACTTTGCAGACA TCTGAGCAATAGTTCTGTTATTTCACTTTTGCCATGAGCCTCTCTGAGCTTGTTTGTTGCTGAAATGCTACTTTTTAAAATTTAGATGTT AGATTGAAAACTGTAGTTTTCAACATATGCTTTGCTGGAACACTGTGATAGATTAACTGTAGAATTCTTCCTGTACGATTGGGGATATAA TGGGCTTCACTAACCTTCCCTAGGCATTGAAACTTCCCCCAAATCTGATGGACCTAGAAGTCTGCTTTTGTACCTGCTGGGCCCCAAAGT TGGGCATTTTTCTCTCTGTTCCCTCTCTTTTGAAAATGTAAAATAAAACCAAAAATAGACAACTTTTTCTTCAGCCATTCCAGCATAGAG AACAAAACCTTATGGAAACAGGAATGTCAATTGTGTAATCATTGTTCTAATTAGGTAAATAGAAGTCCTTATGTATGTGTTACAAGAATT TCCCCCACAACATCCTTTATGACTGAAGTTCAATGACAGTTTGTGTTTGGTGGTAAAGGATTTTCTCCATGGCCTGAATTAAGACCATTA GAAAGCACCAGGCCGTGGGAGCAGTGACCATCTGCTGACTGTTCTTGTGGATCTTGTGTCCAGGGACATGGGGTGACATGCCTCGTATGT GTTAGAGGGTGGAATGGATGTGTTTGGCGCTGCATGGGATCTGGTGCCCCTCTTCTCCTGGATTCACATCCCCACCCAGGGCCCGCTTTT ACTAAGTGTTCTGCCCTAGATTGGTTCAAGGAGGTCATCCAACTGACTTTATCAAGTGGAATTGGGATATATTTGATATACTTCTGCCTA ACAACATGGAAAAGGGTTTTCTTTTCCCTGCAAGCTACATCCTACTGCTTTGAACTTCCAAGTATGTCTAGTCACCTTTTAAAATGTAAA CATTTTCAGAAAAATGAGGATTGCCTTCCTTGTATGCGCTTTTTACCTTGACTACCTGAATTGCAAGGGATTTTTATATATTCATATGTT ACAAAGTCAGCAACTCTCCTGTTGGTTCATTATTGAATGTGCTGTAAATTAAGTTGTTTGCAATTAAAACAAGGTTTGCCCACATCCAAG ATGACCTTGTGATTTTGTGCTGATTGTGTCTGAGGACCTTTCCCTCCACATATGGTCTGGCAGATGCACCCAGTTCAGCCTAAGGAGTAG GCTTTTTTTTGGGGGGGGGAGGTCGGGTGGGGGGGATTTTTAATCTTTTAATTTTCAAGATGGTTAAAATATTGAAATGTTTAAGTGGAT AACTATATTATTCATAAAATGACTGAGTGAAAACTAATACACTATGAGATGAAAAGTACTTGTCAGGGATTTTCAAGCTTTTGCTCAAGT AATTACTATGAAATAACAATTTCTAGAAATGAAGAACAATCCGTGGAGAATAAATTACCATTGGTGTGGGGGAAAAAAGCCAAACAGAAG >55602_55602_1_MTDH-LAPTM4B_MTDH_chr8_98673401_ENST00000336273_LAPTM4B_chr8_98817581_ENST00000445593_length(amino acids)=363AA_BP=1 MRVPLTGGKMAARSWQDELAQQAEEGSARLREMLSVGLGFLRTELGLDLGLEPKRYPGWVILVGTGALGLLLLFLLGYGWAAACAGARKK RRSPPRKREEAAAVPAAAPDDLALLKNLRSEEQKKKNRKKLSEKPKPNGRTVEVAEGEAVRTPQSVTAKQPPEIDKKNEKIINAVVLLIL LSALADPDQYNFSSSELGGDFEFMDDANMCIAIAISLLMILICAMATYGAYKQRAAWIIPFFCYQIFDFALNMLVAITVLIYPNSIQEYI RQLPPNFPYRDDVMSVNPTCLVLIILLFISIILTFKGYLISCVWNCYRYINGRNSSDVLVYVTSNDTTVLLPPYDDATVNGAAKEPPPPY -------------------------------------------------------------- >55602_55602_2_MTDH-LAPTM4B_MTDH_chr8_98673401_ENST00000336273_LAPTM4B_chr8_98817581_ENST00000521545_length(transcript)=1393nt_BP=811nt GGCCCAAGCCGCCATTGTTCCGCCGAGGGAGGACAGCGGGGCCTGGCGCTGGCGCCGAGACGCCGCTTAGCGGCCGCCACTGGAGACACT CCCTCCCGCCTCCCGGGTCTCCTGGCGGCGGCGGAGTGAGGCTGACAGCGGGGAACCTGGGAGACCCCTCCGCCCTCCCCGCGGTGGCAG CGGCCGATCCCCGGCTCCGGCGCGAGGGACGGCCGCGATGCGCTCGGCCTGAGGTTACCCGGCCCGGCCCTTCCTCGCTTCCCTCGACTA TTCCACTGCGTCTCCGCGCCCCGGCGTCATCCTGCGAGTCCCTCTGACGGGAGGGAAGATGGCTGCACGGAGCTGGCAGGACGAGCTGGC CCAGCAGGCCGAGGAGGGCTCGGCCCGGCTGCGGGAAATGCTCTCGGTCGGCCTAGGCTTTCTGCGCACCGAGCTGGGCCTCGACCTGGG GCTGGAGCCGAAACGGTACCCCGGCTGGGTGATCCTGGTGGGCACTGGCGCGCTCGGGCTGCTGCTGCTGTTTCTGCTGGGCTACGGCTG GGCCGCGGCTTGCGCCGGCGCCCGCAAAAAGCGGAGGAGCCCGCCCCGCAAGCGGGAGGAGGCGGCGGCCGTGCCGGCCGCGGCCCCCGA CGACCTGGCCTTGCTGAAGAATCTCCGGAGCGAGGAACAGAAGAAGAAGAACCGGAAGAAACTGTCCGAGAAGCCCAAACCAAATGGGCG GACTGTTGAAGTGGCTGAGGGTGAAGCTGTTCGAACACCTCAAAGTGTAACAGCAAAGCAGCCACCAGAGATTGACAAGAAAAATGAAAA GATCATCAATGCTGTGGTACTGTTGATTTTATTGAGTGCCCTGGCTGATCCGGATCAGTATAACTTTTCAAGTTCTGAACTGGGAGGTGA CTTTGAGTTCATGGATGATGCCAACATGTGCATTGCCATTGCGATTTCTCTTCTCATGATCCTGATATGTGCTATGGCTACTTACGGAGC GTACAAGCAACGCGCAGCCTGGATCATCCCATTCTTCTGTTACCAGATCTTTGACTTTGCCCTGAACATGTTGGTTGCAATCACTGTGCT TATTTATCCAAACTCCATTCAGGAATACATACGGCAACTGCCTCCTAATTTTCCCTACAGAGATGATGTCATGTCAGTGAATCCTACCTG TTTGGTCCTTATTATTCTTCTGTTTATTAGCATTATCTTGACTTTTAAGGGTTACTTGATTAGCTGTGTTTGGAACTGCTACCGATACAT CAATGGTAGGAACTCCTCTGATGTCCTGGTTTATGTTACCAGCAATGACACTACGGTGCTGCTACCCCCGTATGATGATGCCACTGTGAA >55602_55602_2_MTDH-LAPTM4B_MTDH_chr8_98673401_ENST00000336273_LAPTM4B_chr8_98817581_ENST00000521545_length(amino acids)=364AA_BP=1 MRVPLTGGKMAARSWQDELAQQAEEGSARLREMLSVGLGFLRTELGLDLGLEPKRYPGWVILVGTGALGLLLLFLLGYGWAAACAGARKK RRSPPRKREEAAAVPAAAPDDLALLKNLRSEEQKKKNRKKLSEKPKPNGRTVEVAEGEAVRTPQSVTAKQPPEIDKKNEKIINAVVLLIL LSALADPDQYNFSSSELGGDFEFMDDANMCIAIAISLLMILICAMATYGAYKQRAAWIIPFFCYQIFDFALNMLVAITVLIYPNSIQEYI RQLPPNFPYRDDVMSVNPTCLVLIILLFISIILTFKGYLISCVWNCYRYINGRNSSDVLVYVTSNDTTVLLPPYDDATVNGAAKEPPPPY -------------------------------------------------------------- >55602_55602_3_MTDH-LAPTM4B_MTDH_chr8_98673401_ENST00000519934_LAPTM4B_chr8_98817581_ENST00000445593_length(transcript)=2565nt_BP=444nt GCCGAGGAGGGCTCGGCCCGGCTGCGGGAAATGCTCTCGGTCGGCCTAGGCTTTCTGCGCACCGAGCTGGGCCTCGACCTGGGGCTGGAG CCGAAACGGTACCCCGGCTGGGTGATCCTGGTGGGCACTGGCGCGCTCGGGCTGCTGCTGCTGTTTCTGCTGGGCTACGGCTGGGCCGCG GCTTGCGCCGGCGCCCGCAAAAAGCGGAGGAGCCCGCCCCGCAAGCGGGAGGAGGCGGCGGCCGTGCCGGCCGCGGCCCCCGACGACCTG GCCTTGCTGAAGAATCTCCGGAGCGAGGAACAGAAGAAGAAGAACCGGAAGAAACTGTCCGAGAAGCCCAAACCAAATGGGCGGACTGTT GAAGTGGCTGAGGGTGAAGCTGTTCGAACACCTCAAAGTGTAACAGCAAAGCAGCCACCAGAGATTGACAAGAAAAATGAAAAGATCATC AATGCTGTGGTACTGTTGATTTTATTGAGTGCCCTGGCTGATCCGGATCAGTATAACTTTTCAAGTTCTGAACTGGGAGGTGACTTTGAG TTCATGGATGATGCCAACATGTGCATTGCCATTGCGATTTCTCTTCTCATGATCCTGATATGTGCTATGGCTACTTACGGAGCGTACAAG CAACGCGCAGCCTGGATCATCCCATTCTTCTGTTACCAGATCTTTGACTTTGCCCTGAACATGTTGGTTGCAATCACTGTGCTTATTTAT CCAAACTCCATTCAGGAATACATACGGCAACTGCCTCCTAATTTTCCCTACAGAGATGATGTCATGTCAGTGAATCCTACCTGTTTGGTC CTTATTATTCTTCTGTTTATTAGCATTATCTTGACTTTTAAGGGTTACTTGATTAGCTGTGTTTGGAACTGCTACCGATACATCAATGGT AGGAACTCCTCTGATGTCCTGGTTTATGTTACCAGCAATGACACTACGGTGCTGCTACCCCCGTATGATGATGCCACTGTGAATGGTGCT GCCAAGGAGCCACCGCCACCTTACGTGTCTGCCTAAGCCTTCAAGTGGGCGGAGCTGAGGGCAGCAGCTTGACTTTGCAGACATCTGAGC AATAGTTCTGTTATTTCACTTTTGCCATGAGCCTCTCTGAGCTTGTTTGTTGCTGAAATGCTACTTTTTAAAATTTAGATGTTAGATTGA AAACTGTAGTTTTCAACATATGCTTTGCTGGAACACTGTGATAGATTAACTGTAGAATTCTTCCTGTACGATTGGGGATATAATGGGCTT CACTAACCTTCCCTAGGCATTGAAACTTCCCCCAAATCTGATGGACCTAGAAGTCTGCTTTTGTACCTGCTGGGCCCCAAAGTTGGGCAT TTTTCTCTCTGTTCCCTCTCTTTTGAAAATGTAAAATAAAACCAAAAATAGACAACTTTTTCTTCAGCCATTCCAGCATAGAGAACAAAA CCTTATGGAAACAGGAATGTCAATTGTGTAATCATTGTTCTAATTAGGTAAATAGAAGTCCTTATGTATGTGTTACAAGAATTTCCCCCA CAACATCCTTTATGACTGAAGTTCAATGACAGTTTGTGTTTGGTGGTAAAGGATTTTCTCCATGGCCTGAATTAAGACCATTAGAAAGCA CCAGGCCGTGGGAGCAGTGACCATCTGCTGACTGTTCTTGTGGATCTTGTGTCCAGGGACATGGGGTGACATGCCTCGTATGTGTTAGAG GGTGGAATGGATGTGTTTGGCGCTGCATGGGATCTGGTGCCCCTCTTCTCCTGGATTCACATCCCCACCCAGGGCCCGCTTTTACTAAGT GTTCTGCCCTAGATTGGTTCAAGGAGGTCATCCAACTGACTTTATCAAGTGGAATTGGGATATATTTGATATACTTCTGCCTAACAACAT GGAAAAGGGTTTTCTTTTCCCTGCAAGCTACATCCTACTGCTTTGAACTTCCAAGTATGTCTAGTCACCTTTTAAAATGTAAACATTTTC AGAAAAATGAGGATTGCCTTCCTTGTATGCGCTTTTTACCTTGACTACCTGAATTGCAAGGGATTTTTATATATTCATATGTTACAAAGT CAGCAACTCTCCTGTTGGTTCATTATTGAATGTGCTGTAAATTAAGTTGTTTGCAATTAAAACAAGGTTTGCCCACATCCAAGATGACCT TGTGATTTTGTGCTGATTGTGTCTGAGGACCTTTCCCTCCACATATGGTCTGGCAGATGCACCCAGTTCAGCCTAAGGAGTAGGCTTTTT TTTGGGGGGGGGAGGTCGGGTGGGGGGGATTTTTAATCTTTTAATTTTCAAGATGGTTAAAATATTGAAATGTTTAAGTGGATAACTATA TTATTCATAAAATGACTGAGTGAAAACTAATACACTATGAGATGAAAAGTACTTGTCAGGGATTTTCAAGCTTTTGCTCAAGTAATTACT ATGAAATAACAATTTCTAGAAATGAAGAACAATCCGTGGAGAATAAATTACCATTGGTGTGGGGGAAAAAAGCCAAACAGAAGTAGAAAA >55602_55602_3_MTDH-LAPTM4B_MTDH_chr8_98673401_ENST00000519934_LAPTM4B_chr8_98817581_ENST00000445593_length(amino acids)=334AA_BP=1 MREMLSVGLGFLRTELGLDLGLEPKRYPGWVILVGTGALGLLLLFLLGYGWAAACAGARKKRRSPPRKREEAAAVPAAAPDDLALLKNLR SEEQKKKNRKKLSEKPKPNGRTVEVAEGEAVRTPQSVTAKQPPEIDKKNEKIINAVVLLILLSALADPDQYNFSSSELGGDFEFMDDANM CIAIAISLLMILICAMATYGAYKQRAAWIIPFFCYQIFDFALNMLVAITVLIYPNSIQEYIRQLPPNFPYRDDVMSVNPTCLVLIILLFI -------------------------------------------------------------- >55602_55602_4_MTDH-LAPTM4B_MTDH_chr8_98673401_ENST00000519934_LAPTM4B_chr8_98817581_ENST00000521545_length(transcript)=1026nt_BP=444nt GCCGAGGAGGGCTCGGCCCGGCTGCGGGAAATGCTCTCGGTCGGCCTAGGCTTTCTGCGCACCGAGCTGGGCCTCGACCTGGGGCTGGAG CCGAAACGGTACCCCGGCTGGGTGATCCTGGTGGGCACTGGCGCGCTCGGGCTGCTGCTGCTGTTTCTGCTGGGCTACGGCTGGGCCGCG GCTTGCGCCGGCGCCCGCAAAAAGCGGAGGAGCCCGCCCCGCAAGCGGGAGGAGGCGGCGGCCGTGCCGGCCGCGGCCCCCGACGACCTG GCCTTGCTGAAGAATCTCCGGAGCGAGGAACAGAAGAAGAAGAACCGGAAGAAACTGTCCGAGAAGCCCAAACCAAATGGGCGGACTGTT GAAGTGGCTGAGGGTGAAGCTGTTCGAACACCTCAAAGTGTAACAGCAAAGCAGCCACCAGAGATTGACAAGAAAAATGAAAAGATCATC AATGCTGTGGTACTGTTGATTTTATTGAGTGCCCTGGCTGATCCGGATCAGTATAACTTTTCAAGTTCTGAACTGGGAGGTGACTTTGAG TTCATGGATGATGCCAACATGTGCATTGCCATTGCGATTTCTCTTCTCATGATCCTGATATGTGCTATGGCTACTTACGGAGCGTACAAG CAACGCGCAGCCTGGATCATCCCATTCTTCTGTTACCAGATCTTTGACTTTGCCCTGAACATGTTGGTTGCAATCACTGTGCTTATTTAT CCAAACTCCATTCAGGAATACATACGGCAACTGCCTCCTAATTTTCCCTACAGAGATGATGTCATGTCAGTGAATCCTACCTGTTTGGTC CTTATTATTCTTCTGTTTATTAGCATTATCTTGACTTTTAAGGGTTACTTGATTAGCTGTGTTTGGAACTGCTACCGATACATCAATGGT AGGAACTCCTCTGATGTCCTGGTTTATGTTACCAGCAATGACACTACGGTGCTGCTACCCCCGTATGATGATGCCACTGTGAATGGTGCT >55602_55602_4_MTDH-LAPTM4B_MTDH_chr8_98673401_ENST00000519934_LAPTM4B_chr8_98817581_ENST00000521545_length(amino acids)=334AA_BP=1 MREMLSVGLGFLRTELGLDLGLEPKRYPGWVILVGTGALGLLLLFLLGYGWAAACAGARKKRRSPPRKREEAAAVPAAAPDDLALLKNLR SEEQKKKNRKKLSEKPKPNGRTVEVAEGEAVRTPQSVTAKQPPEIDKKNEKIINAVVLLILLSALADPDQYNFSSSELGGDFEFMDDANM CIAIAISLLMILICAMATYGAYKQRAAWIIPFFCYQIFDFALNMLVAITVLIYPNSIQEYIRQLPPNFPYRDDVMSVNPTCLVLIILLFI -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MTDH-LAPTM4B |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | MTDH | chr8:98673401 | chr8:98817581 | ENST00000336273 | + | 2 | 12 | 72_169 | 161.0 | 583.0 | BCCIP |
| Hgene | MTDH | chr8:98673401 | chr8:98817581 | ENST00000336273 | + | 2 | 12 | 101_205 | 161.0 | 583.0 | RELA |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MTDH-LAPTM4B |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MTDH-LAPTM4B |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
