
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MTDH-MAL2 (FusionGDB2 ID:55604) |
Fusion Gene Summary for MTDH-MAL2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MTDH-MAL2 | Fusion gene ID: 55604 | Hgene | Tgene | Gene symbol | MTDH | MAL2 | Gene ID | 92140 | 114569 |
| Gene name | metadherin | mal, T cell differentiation protein 2 (gene/pseudogene) | |
| Synonyms | 3D3|AEG-1|AEG1|LYRIC|LYRIC/3D3 | - | |
| Cytomap | 8q22.1 | 8q24.12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein LYRIC3D3/LYRICastrocyte elevated gene 1astrocyte elevated gene-1 proteinlysine-rich CEACAM1 co-isolated proteinmetastasis adhesion protein | protein MAL2MAL proteolipid protein 2MAL2 proteolipid proteinmal, T-cell differentiation protein 2myelin and lymphocyte protein 2 | |
| Modification date | 20200313 | 20200326 | |
| UniProtAcc | Q86UE4 | Q969L2 | |
| Ensembl transtripts involved in fusion gene | ENST00000336273, ENST00000519934, | ENST00000521748, ENST00000276681, | |
| Fusion gene scores | * DoF score | 15 X 10 X 9=1350 | 2 X 2 X 2=8 |
| # samples | 20 | 2 | |
| ** MAII score | log2(20/1350*10)=-2.75488750216347 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(2/8*10)=1.32192809488736 | |
| Context | PubMed: MTDH [Title/Abstract] AND MAL2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MTDH(98673401)-MAL2(120233827), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | MTDH | GO:0000122 | negative regulation of transcription by RNA polymerase II | 15927426 |
| Hgene | MTDH | GO:0010508 | positive regulation of autophagy | 21127263 |
| Hgene | MTDH | GO:0043066 | negative regulation of apoptotic process | 17704808 |
| Hgene | MTDH | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 16452207|18316612 |
| Hgene | MTDH | GO:0045766 | positive regulation of angiogenesis | 19940250 |
| Hgene | MTDH | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 16452207|18316612 |
| Hgene | MTDH | GO:0051897 | positive regulation of protein kinase B signaling | 17704808 |
 Fusion gene breakpoints across MTDH (5'-gene) Fusion gene breakpoints across MTDH (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
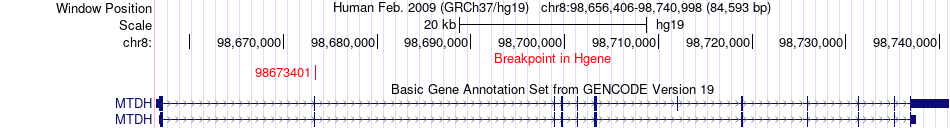 |
 Fusion gene breakpoints across MAL2 (3'-gene) Fusion gene breakpoints across MAL2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
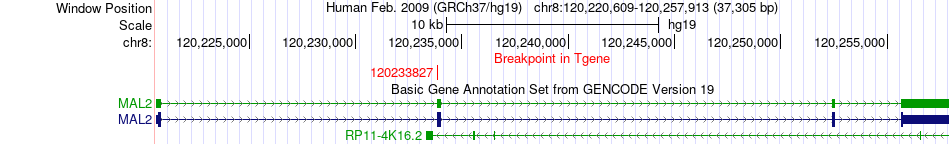 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-JV-A5VE-01A | MTDH | chr8 | 98673401 | + | MAL2 | chr8 | 120233827 | + |
Top |
Fusion Gene ORF analysis for MTDH-MAL2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000336273 | ENST00000521748 | MTDH | chr8 | 98673401 | + | MAL2 | chr8 | 120233827 | + |
| 5CDS-3UTR | ENST00000519934 | ENST00000521748 | MTDH | chr8 | 98673401 | + | MAL2 | chr8 | 120233827 | + |
| In-frame | ENST00000336273 | ENST00000276681 | MTDH | chr8 | 98673401 | + | MAL2 | chr8 | 120233827 | + |
| In-frame | ENST00000519934 | ENST00000276681 | MTDH | chr8 | 98673401 | + | MAL2 | chr8 | 120233827 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000336273 | MTDH | chr8 | 98673401 | + | ENST00000276681 | MAL2 | chr8 | 120233827 | + | 3391 | 811 | 301 | 1209 | 302 |
| ENST00000519934 | MTDH | chr8 | 98673401 | + | ENST00000276681 | MAL2 | chr8 | 120233827 | + | 3024 | 444 | 21 | 842 | 273 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000336273 | ENST00000276681 | MTDH | chr8 | 98673401 | + | MAL2 | chr8 | 120233827 | + | 0.000692461 | 0.9993075 |
| ENST00000519934 | ENST00000276681 | MTDH | chr8 | 98673401 | + | MAL2 | chr8 | 120233827 | + | 0.000821438 | 0.9991786 |
Top |
Fusion Genomic Features for MTDH-MAL2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
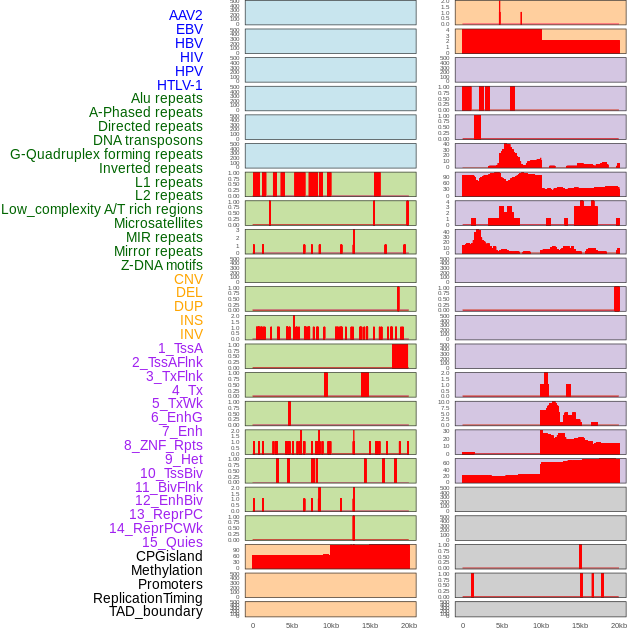 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
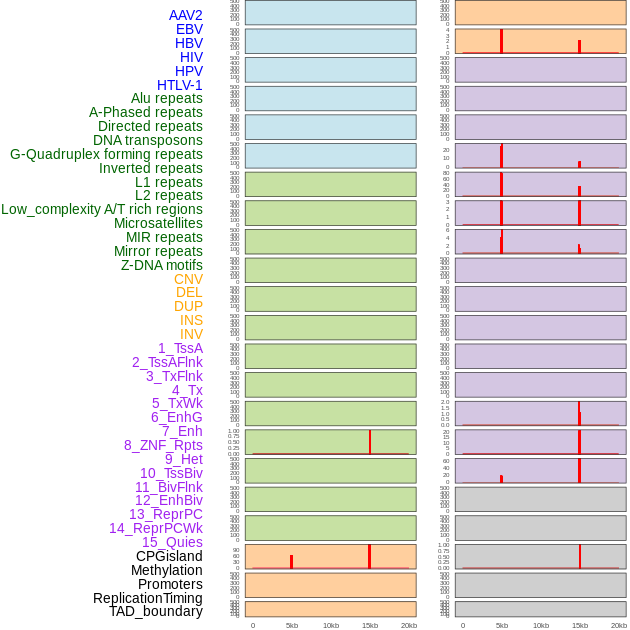 |
Top |
Fusion Protein Features for MTDH-MAL2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:98673401/chr8:120233827) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MTDH | MAL2 |
| FUNCTION: Downregulates SLC1A2/EAAT2 promoter activity when expressed ectopically. Activates the nuclear factor kappa-B (NF-kappa-B) transcription factor. Promotes anchorage-independent growth of immortalized melanocytes and astrocytes which is a key component in tumor cell expansion. Promotes lung metastasis and also has an effect on bone and brain metastasis, possibly by enhancing the seeding of tumor cells to the target organ endothelium. Induces chemoresistance. {ECO:0000269|PubMed:15927426, ECO:0000269|PubMed:16452207, ECO:0000269|PubMed:18316612, ECO:0000269|PubMed:19111877}. | FUNCTION: Member of the machinery of polarized transport. Required for the indirect transcytotic route at the step of the egress of the transcytosing cargo from perinuclear endosomes in order for it to travel to the apical surface via a raft-dependent pathway. {ECO:0000269|PubMed:12370246}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MTDH | chr8:98673401 | chr8:120233827 | ENST00000336273 | + | 2 | 12 | 59_67 | 161 | 583.0 | Compositional bias | Note=Poly-Leu |
| Hgene | MTDH | chr8:98673401 | chr8:120233827 | ENST00000336273 | + | 2 | 12 | 1_71 | 161 | 583.0 | Region | Note=Activation of NF-kappa-B |
| Hgene | MTDH | chr8:98673401 | chr8:120233827 | ENST00000336273 | + | 2 | 12 | 1_48 | 161 | 583.0 | Topological domain | Lumenal |
| Hgene | MTDH | chr8:98673401 | chr8:120233827 | ENST00000336273 | + | 2 | 12 | 49_69 | 161 | 583.0 | Transmembrane | Helical |
| Tgene | MAL2 | chr8:98673401 | chr8:120233827 | ENST00000276681 | 1 | 5 | 124_149 | 44 | 177.0 | Topological domain | Lumenal | |
| Tgene | MAL2 | chr8:98673401 | chr8:120233827 | ENST00000276681 | 1 | 5 | 171_176 | 44 | 177.0 | Topological domain | Cytoplasmic | |
| Tgene | MAL2 | chr8:98673401 | chr8:120233827 | ENST00000276681 | 1 | 5 | 56_66 | 44 | 177.0 | Topological domain | Lumenal | |
| Tgene | MAL2 | chr8:98673401 | chr8:120233827 | ENST00000276681 | 1 | 5 | 88_102 | 44 | 177.0 | Topological domain | Cytoplasmic | |
| Tgene | MAL2 | chr8:98673401 | chr8:120233827 | ENST00000276681 | 1 | 5 | 103_123 | 44 | 177.0 | Transmembrane | Helical | |
| Tgene | MAL2 | chr8:98673401 | chr8:120233827 | ENST00000276681 | 1 | 5 | 150_170 | 44 | 177.0 | Transmembrane | Helical | |
| Tgene | MAL2 | chr8:98673401 | chr8:120233827 | ENST00000276681 | 1 | 5 | 67_87 | 44 | 177.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MTDH | chr8:98673401 | chr8:120233827 | ENST00000336273 | + | 2 | 12 | 441_451 | 161 | 583.0 | Compositional bias | Note=Poly-Lys |
| Hgene | MTDH | chr8:98673401 | chr8:120233827 | ENST00000336273 | + | 2 | 12 | 573_577 | 161 | 583.0 | Compositional bias | Note=Poly-Lys |
| Hgene | MTDH | chr8:98673401 | chr8:120233827 | ENST00000336273 | + | 2 | 12 | 381_443 | 161 | 583.0 | Region | Lung-homing for mammary tumors |
| Hgene | MTDH | chr8:98673401 | chr8:120233827 | ENST00000336273 | + | 2 | 12 | 70_582 | 161 | 583.0 | Topological domain | Cytoplasmic |
| Tgene | MAL2 | chr8:98673401 | chr8:120233827 | ENST00000276681 | 1 | 5 | 31_175 | 44 | 177.0 | Domain | MARVEL | |
| Tgene | MAL2 | chr8:98673401 | chr8:120233827 | ENST00000276681 | 1 | 5 | 1_34 | 44 | 177.0 | Topological domain | Cytoplasmic | |
| Tgene | MAL2 | chr8:98673401 | chr8:120233827 | ENST00000276681 | 1 | 5 | 35_55 | 44 | 177.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for MTDH-MAL2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >55604_55604_1_MTDH-MAL2_MTDH_chr8_98673401_ENST00000336273_MAL2_chr8_120233827_ENST00000276681_length(transcript)=3391nt_BP=811nt GGCCCAAGCCGCCATTGTTCCGCCGAGGGAGGACAGCGGGGCCTGGCGCTGGCGCCGAGACGCCGCTTAGCGGCCGCCACTGGAGACACT CCCTCCCGCCTCCCGGGTCTCCTGGCGGCGGCGGAGTGAGGCTGACAGCGGGGAACCTGGGAGACCCCTCCGCCCTCCCCGCGGTGGCAG CGGCCGATCCCCGGCTCCGGCGCGAGGGACGGCCGCGATGCGCTCGGCCTGAGGTTACCCGGCCCGGCCCTTCCTCGCTTCCCTCGACTA TTCCACTGCGTCTCCGCGCCCCGGCGTCATCCTGCGAGTCCCTCTGACGGGAGGGAAGATGGCTGCACGGAGCTGGCAGGACGAGCTGGC CCAGCAGGCCGAGGAGGGCTCGGCCCGGCTGCGGGAAATGCTCTCGGTCGGCCTAGGCTTTCTGCGCACCGAGCTGGGCCTCGACCTGGG GCTGGAGCCGAAACGGTACCCCGGCTGGGTGATCCTGGTGGGCACTGGCGCGCTCGGGCTGCTGCTGCTGTTTCTGCTGGGCTACGGCTG GGCCGCGGCTTGCGCCGGCGCCCGCAAAAAGCGGAGGAGCCCGCCCCGCAAGCGGGAGGAGGCGGCGGCCGTGCCGGCCGCGGCCCCCGA CGACCTGGCCTTGCTGAAGAATCTCCGGAGCGAGGAACAGAAGAAGAAGAACCGGAAGAAACTGTCCGAGAAGCCCAAACCAAATGGGCG GACTGTTGAAGTGGCTGAGGGTGAAGCTGTTCGAACACCTCAAAGTGTAACAGCAAAGCAGCCACCAGAGATTGACAAGAAAAATGAAAA GCTGTTCGGGGGTCTTGTCTGGATTTTGGTTGCCTCCTCCAATGTTCCTCTACCTCTACTACAAGGATGGGTCATGTTTGTGTCCGTGAC AGCGTTTTTCTTTTCGCTCCTCTTTCTGGGCATGTTCCTCTCTGGCATGGTGGCTCAAATTGATGCTAACTGGAACTTCCTGGATTTTGC CTACCATTTTACAGTATTTGTCTTCTATTTTGGAGCCTTTTTATTGGAAGCAGCAGCCACATCCCTGCATGATTTGCATTGCAATACAAC CATAACCGGGCAGCCACTCCTGAGTGATAACCAGTATAACATAAACGTAGCAGCCTCAATTTTTGCCTTTATGACGACAGCTTGTTATGG TTGCAGTTTGGGTCTGGCTTTACGAAGATGGCGACCGTAACACTCCTTAGAAACTGGCAGTCGTATGTTAGTTTCACTTGTCTACTTTAT ATGTCTGATCAATTTGGATACCATTTTGTCCAGATGCAAAAACATTCCAAAAGTAATGTGTTTAGTAGAGAGAGACTCTAAGCTCAAGTT CTGGTTTATTTCATGGATGGAATGTTAATTTTATTATGATATTAAAGAAATGGCCTTTTATTTTACATCTCTCCCCTTTTTCCCTTTCCC CCTTTATTTTCCTCCTTTTCTTTCTGAAAGTTTCCTTTTATGTCCATAAAATACAAATATATTGTTCATAAAAAATTAGTATCCCTTTTG TTTGGTTGCTGAGTCACCTGAACCTTAATTTTAATTGGTAATTACAGCCCCTAAAAAAAACACATTTCAAATAGGCTTCCCACTAAACTC TATATTTTAGTGTAAACCAGGAATTGGCACACTTTTTTTAGAATGGGCCAGATGGTAAATATTTATGCTTCACGGTCCATACAGTCTCTG TCACAACTATTCAGTTCTGCTAGTATAGCGTGAAAGCAGCTATACACAATACAGAAATGAATGAGTGTGGTTATGTTCTAATAAAACTTA TTTATAAAAACAAGGGGAGGCTGGGTTTAGCCTGTGGGCCATAGTTTGTCAACCACTGGTGTAAAACCTTAGTTATATATGATCTGCATT TTCTTGAACTGATCATTGAAAACTTATAAACCTAACAGAAAAGCCACATAATATTTAGTGTCATTATGCAATAATCACATTGCCTTTGTG TTAATAGTCAAATACTTACCTTTGGAGAATACTTACCTTTGGAGGAATGTATAAAATTTCTCAGGCAGAGTCCTGGATATAGGAAAAAGT AATTTATGAAGTAAACTTCAGTTGCTTAATCAAACTAATGATAGTCTAACAACTGAGCAAGATCCTCATCTGAGAGTGCTTAAAATGGGA TCCCCAGAGACCATTAACCAATACTGGAACTGGTATCTAGCTACTGATGTCTTACTTTGAGTTTATTTATGCTTCAGAATACAGTTGTTT GCCCTGTGCATGAATATACCCATATTTGTGTGTGGATATGTGAAGCTTTTCCAAATAGAGCTCTCAGAAGAATTAAGTTTTTACTTCTAA TTATTTTGCATTACTTTGAGTTAAATTTGAATAGAGTATTAAATATAAAGTTGTAGATTCTTATGTGTTTTTGTATTAGCCCAGACATCT GTAATGTTTTTGCACTGGTGACAGACAAAATCTGTTTTAAAATCATATCCAGCACAAAAACTATTTCTGGCTGAATAGCACAGAAAAGTA TTTTAACCTACCTGTAGAGATCCTCGTCATGGAAAGGTGCCAAACTGTTTTGAATGGAAGGACAAGTAAGAGTGAGGCCACAGTTCCCAC CACACGAGGGCTTTTGTATTGTTCTACTTTTTCAGCCCTTTACTTTCTGGCTGAAGCATCCCCTTGGAGTGCCATGTATAAGTTGGGCTA TTAGAGTTCATGGAACATAGAACAACCATGAATGAGTGGCATGATCCGTGCTTAATGATCAAGTGTTACTTATCTAATAATCCTCTAGAA AGAACCCTGTTAGATCTTGGTTTGTGATAAAAATATAAAGACAGAAGACATGAGGAAAAACAAAAGGTTTGAGGAAATCAGGCATATGAC TTTATACTTAACATCAGATCTTTTCTATAATATCCTACTACTTTGGTTTTCCTAGCTCCATACCACACACCTAAACCTGTATTATGAATT ACATATTACAAAGTCATAAATGTGCCATATGGATATACAGTACATTCTAGTTGGAATCGTTTACTCTGCTAGAATTTAGGTGTGAGATTT TTTGTTTCCCAGGTATAGCAGGCTTATGTTTGGTGGCATTAAATTGGTTTCTTTAAAATGCTTTGGTGGCACTTTTGTAAACAGATTGCT TCTAGATTGTTACAAACCAAGCCTAAGACACATCTGTGAATACTTAGATTTGTAGCTTAATCACATTCTAGACTTGTGAGTTGAATGACA AAGCAGTTGAACAAAAATTATGGCATTTAAGAATTTAACATGTCTTAGCTGTAAAAATGAGAAAGTGTTGGTTGGTTTTAAAATCTGGTA >55604_55604_1_MTDH-MAL2_MTDH_chr8_98673401_ENST00000336273_MAL2_chr8_120233827_ENST00000276681_length(amino acids)=302AA_BP=170 MRVPLTGGKMAARSWQDELAQQAEEGSARLREMLSVGLGFLRTELGLDLGLEPKRYPGWVILVGTGALGLLLLFLLGYGWAAACAGARKK RRSPPRKREEAAAVPAAAPDDLALLKNLRSEEQKKKNRKKLSEKPKPNGRTVEVAEGEAVRTPQSVTAKQPPEIDKKNEKLFGGLVWILV ASSNVPLPLLQGWVMFVSVTAFFFSLLFLGMFLSGMVAQIDANWNFLDFAYHFTVFVFYFGAFLLEAAATSLHDLHCNTTITGQPLLSDN -------------------------------------------------------------- >55604_55604_2_MTDH-MAL2_MTDH_chr8_98673401_ENST00000519934_MAL2_chr8_120233827_ENST00000276681_length(transcript)=3024nt_BP=444nt GCCGAGGAGGGCTCGGCCCGGCTGCGGGAAATGCTCTCGGTCGGCCTAGGCTTTCTGCGCACCGAGCTGGGCCTCGACCTGGGGCTGGAG CCGAAACGGTACCCCGGCTGGGTGATCCTGGTGGGCACTGGCGCGCTCGGGCTGCTGCTGCTGTTTCTGCTGGGCTACGGCTGGGCCGCG GCTTGCGCCGGCGCCCGCAAAAAGCGGAGGAGCCCGCCCCGCAAGCGGGAGGAGGCGGCGGCCGTGCCGGCCGCGGCCCCCGACGACCTG GCCTTGCTGAAGAATCTCCGGAGCGAGGAACAGAAGAAGAAGAACCGGAAGAAACTGTCCGAGAAGCCCAAACCAAATGGGCGGACTGTT GAAGTGGCTGAGGGTGAAGCTGTTCGAACACCTCAAAGTGTAACAGCAAAGCAGCCACCAGAGATTGACAAGAAAAATGAAAAGCTGTTC GGGGGTCTTGTCTGGATTTTGGTTGCCTCCTCCAATGTTCCTCTACCTCTACTACAAGGATGGGTCATGTTTGTGTCCGTGACAGCGTTT TTCTTTTCGCTCCTCTTTCTGGGCATGTTCCTCTCTGGCATGGTGGCTCAAATTGATGCTAACTGGAACTTCCTGGATTTTGCCTACCAT TTTACAGTATTTGTCTTCTATTTTGGAGCCTTTTTATTGGAAGCAGCAGCCACATCCCTGCATGATTTGCATTGCAATACAACCATAACC GGGCAGCCACTCCTGAGTGATAACCAGTATAACATAAACGTAGCAGCCTCAATTTTTGCCTTTATGACGACAGCTTGTTATGGTTGCAGT TTGGGTCTGGCTTTACGAAGATGGCGACCGTAACACTCCTTAGAAACTGGCAGTCGTATGTTAGTTTCACTTGTCTACTTTATATGTCTG ATCAATTTGGATACCATTTTGTCCAGATGCAAAAACATTCCAAAAGTAATGTGTTTAGTAGAGAGAGACTCTAAGCTCAAGTTCTGGTTT ATTTCATGGATGGAATGTTAATTTTATTATGATATTAAAGAAATGGCCTTTTATTTTACATCTCTCCCCTTTTTCCCTTTCCCCCTTTAT TTTCCTCCTTTTCTTTCTGAAAGTTTCCTTTTATGTCCATAAAATACAAATATATTGTTCATAAAAAATTAGTATCCCTTTTGTTTGGTT GCTGAGTCACCTGAACCTTAATTTTAATTGGTAATTACAGCCCCTAAAAAAAACACATTTCAAATAGGCTTCCCACTAAACTCTATATTT TAGTGTAAACCAGGAATTGGCACACTTTTTTTAGAATGGGCCAGATGGTAAATATTTATGCTTCACGGTCCATACAGTCTCTGTCACAAC TATTCAGTTCTGCTAGTATAGCGTGAAAGCAGCTATACACAATACAGAAATGAATGAGTGTGGTTATGTTCTAATAAAACTTATTTATAA AAACAAGGGGAGGCTGGGTTTAGCCTGTGGGCCATAGTTTGTCAACCACTGGTGTAAAACCTTAGTTATATATGATCTGCATTTTCTTGA ACTGATCATTGAAAACTTATAAACCTAACAGAAAAGCCACATAATATTTAGTGTCATTATGCAATAATCACATTGCCTTTGTGTTAATAG TCAAATACTTACCTTTGGAGAATACTTACCTTTGGAGGAATGTATAAAATTTCTCAGGCAGAGTCCTGGATATAGGAAAAAGTAATTTAT GAAGTAAACTTCAGTTGCTTAATCAAACTAATGATAGTCTAACAACTGAGCAAGATCCTCATCTGAGAGTGCTTAAAATGGGATCCCCAG AGACCATTAACCAATACTGGAACTGGTATCTAGCTACTGATGTCTTACTTTGAGTTTATTTATGCTTCAGAATACAGTTGTTTGCCCTGT GCATGAATATACCCATATTTGTGTGTGGATATGTGAAGCTTTTCCAAATAGAGCTCTCAGAAGAATTAAGTTTTTACTTCTAATTATTTT GCATTACTTTGAGTTAAATTTGAATAGAGTATTAAATATAAAGTTGTAGATTCTTATGTGTTTTTGTATTAGCCCAGACATCTGTAATGT TTTTGCACTGGTGACAGACAAAATCTGTTTTAAAATCATATCCAGCACAAAAACTATTTCTGGCTGAATAGCACAGAAAAGTATTTTAAC CTACCTGTAGAGATCCTCGTCATGGAAAGGTGCCAAACTGTTTTGAATGGAAGGACAAGTAAGAGTGAGGCCACAGTTCCCACCACACGA GGGCTTTTGTATTGTTCTACTTTTTCAGCCCTTTACTTTCTGGCTGAAGCATCCCCTTGGAGTGCCATGTATAAGTTGGGCTATTAGAGT TCATGGAACATAGAACAACCATGAATGAGTGGCATGATCCGTGCTTAATGATCAAGTGTTACTTATCTAATAATCCTCTAGAAAGAACCC TGTTAGATCTTGGTTTGTGATAAAAATATAAAGACAGAAGACATGAGGAAAAACAAAAGGTTTGAGGAAATCAGGCATATGACTTTATAC TTAACATCAGATCTTTTCTATAATATCCTACTACTTTGGTTTTCCTAGCTCCATACCACACACCTAAACCTGTATTATGAATTACATATT ACAAAGTCATAAATGTGCCATATGGATATACAGTACATTCTAGTTGGAATCGTTTACTCTGCTAGAATTTAGGTGTGAGATTTTTTGTTT CCCAGGTATAGCAGGCTTATGTTTGGTGGCATTAAATTGGTTTCTTTAAAATGCTTTGGTGGCACTTTTGTAAACAGATTGCTTCTAGAT TGTTACAAACCAAGCCTAAGACACATCTGTGAATACTTAGATTTGTAGCTTAATCACATTCTAGACTTGTGAGTTGAATGACAAAGCAGT TGAACAAAAATTATGGCATTTAAGAATTTAACATGTCTTAGCTGTAAAAATGAGAAAGTGTTGGTTGGTTTTAAAATCTGGTAACTCCAT >55604_55604_2_MTDH-MAL2_MTDH_chr8_98673401_ENST00000519934_MAL2_chr8_120233827_ENST00000276681_length(amino acids)=273AA_BP=141 MREMLSVGLGFLRTELGLDLGLEPKRYPGWVILVGTGALGLLLLFLLGYGWAAACAGARKKRRSPPRKREEAAAVPAAAPDDLALLKNLR SEEQKKKNRKKLSEKPKPNGRTVEVAEGEAVRTPQSVTAKQPPEIDKKNEKLFGGLVWILVASSNVPLPLLQGWVMFVSVTAFFFSLLFL GMFLSGMVAQIDANWNFLDFAYHFTVFVFYFGAFLLEAAATSLHDLHCNTTITGQPLLSDNQYNINVAASIFAFMTTACYGCSLGLALRR -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MTDH-MAL2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | MTDH | chr8:98673401 | chr8:120233827 | ENST00000336273 | + | 2 | 12 | 72_169 | 161.0 | 583.0 | BCCIP |
| Hgene | MTDH | chr8:98673401 | chr8:120233827 | ENST00000336273 | + | 2 | 12 | 101_205 | 161.0 | 583.0 | RELA |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MTDH-MAL2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MTDH-MAL2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
