
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MTG2-AGR2 (FusionGDB2 ID:55671) |
Fusion Gene Summary for MTG2-AGR2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MTG2-AGR2 | Fusion gene ID: 55671 | Hgene | Tgene | Gene symbol | MTG2 | AGR2 | Gene ID | 26164 | 10551 |
| Gene name | mitochondrial ribosome associated GTPase 2 | anterior gradient 2, protein disulphide isomerase family member | |
| Synonyms | GTPBP5|ObgH1|dJ1005F21.2 | AG-2|AG2|GOB-4|HAG-2|HEL-S-116|HPC8|PDIA17|XAG-2 | |
| Cytomap | 20q13.33 | 7p21.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | mitochondrial ribosome-associated GTPase 2GTP binding protein 5 (putative)GTP-binding protein 5protein obg homolog 1 | anterior gradient protein 2 homologanterior gradient homolog 2epididymis secretory protein Li 116protein disulfide isomerase family A, member 17secreted cement gland homologsecreted cement gland protein XAG-2 homolog | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9H4K7 | O95994 | |
| Ensembl transtripts involved in fusion gene | ENST00000370823, ENST00000436421, ENST00000536470, ENST00000461411, | ENST00000486219, ENST00000401412, ENST00000419304, ENST00000419572, | |
| Fusion gene scores | * DoF score | 3 X 3 X 2=18 | 13 X 9 X 3=351 |
| # samples | 3 | 13 | |
| ** MAII score | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(13/351*10)=-1.43295940727611 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MTG2 [Title/Abstract] AND AGR2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MTG2(60776043)-AGR2(16841341), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | MTG2-AGR2 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | AGR2 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response | 23220234 |
| Tgene | AGR2 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response | 23220234 |
 Fusion gene breakpoints across MTG2 (5'-gene) Fusion gene breakpoints across MTG2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
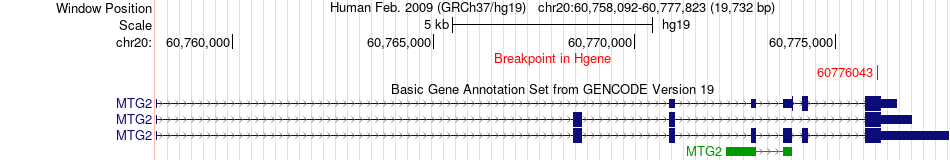 |
 Fusion gene breakpoints across AGR2 (3'-gene) Fusion gene breakpoints across AGR2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
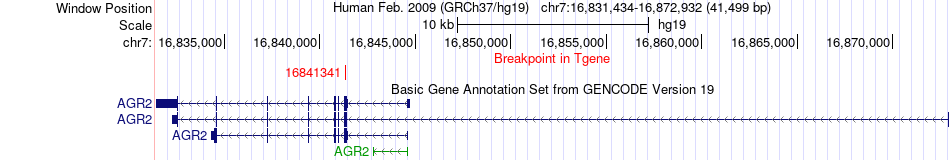 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-BR-A4J5-01A | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - |
Top |
Fusion Gene ORF analysis for MTG2-AGR2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000370823 | ENST00000486219 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - |
| 5CDS-intron | ENST00000436421 | ENST00000486219 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - |
| 5CDS-intron | ENST00000536470 | ENST00000486219 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - |
| Frame-shift | ENST00000370823 | ENST00000401412 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - |
| Frame-shift | ENST00000370823 | ENST00000419304 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - |
| Frame-shift | ENST00000370823 | ENST00000419572 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - |
| In-frame | ENST00000436421 | ENST00000401412 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - |
| In-frame | ENST00000436421 | ENST00000419304 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - |
| In-frame | ENST00000436421 | ENST00000419572 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - |
| In-frame | ENST00000536470 | ENST00000401412 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - |
| In-frame | ENST00000536470 | ENST00000419304 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - |
| In-frame | ENST00000536470 | ENST00000419572 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - |
| intron-3CDS | ENST00000461411 | ENST00000401412 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - |
| intron-3CDS | ENST00000461411 | ENST00000419304 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - |
| intron-3CDS | ENST00000461411 | ENST00000419572 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - |
| intron-intron | ENST00000461411 | ENST00000486219 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000536470 | MTG2 | chr20 | 60776043 | + | ENST00000419304 | AGR2 | chr7 | 16841341 | - | 2668 | 1122 | 162 | 680 | 172 |
| ENST00000536470 | MTG2 | chr20 | 60776043 | + | ENST00000419572 | AGR2 | chr7 | 16841341 | - | 1837 | 1122 | 162 | 680 | 172 |
| ENST00000536470 | MTG2 | chr20 | 60776043 | + | ENST00000401412 | AGR2 | chr7 | 16841341 | - | 1783 | 1122 | 162 | 680 | 172 |
| ENST00000436421 | MTG2 | chr20 | 60776043 | + | ENST00000419304 | AGR2 | chr7 | 16841341 | - | 2788 | 1242 | 1138 | 638 | 166 |
| ENST00000436421 | MTG2 | chr20 | 60776043 | + | ENST00000419572 | AGR2 | chr7 | 16841341 | - | 1957 | 1242 | 1138 | 638 | 166 |
| ENST00000436421 | MTG2 | chr20 | 60776043 | + | ENST00000401412 | AGR2 | chr7 | 16841341 | - | 1903 | 1242 | 1138 | 638 | 166 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000536470 | ENST00000419304 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - | 0.60100013 | 0.39899984 |
| ENST00000536470 | ENST00000419572 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - | 0.29829496 | 0.701705 |
| ENST00000536470 | ENST00000401412 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - | 0.85797083 | 0.14202921 |
| ENST00000436421 | ENST00000419304 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - | 0.50875825 | 0.4912418 |
| ENST00000436421 | ENST00000419572 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - | 0.6747124 | 0.32528758 |
| ENST00000436421 | ENST00000401412 | MTG2 | chr20 | 60776043 | + | AGR2 | chr7 | 16841341 | - | 0.47542778 | 0.52457225 |
Top |
Fusion Genomic Features for MTG2-AGR2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
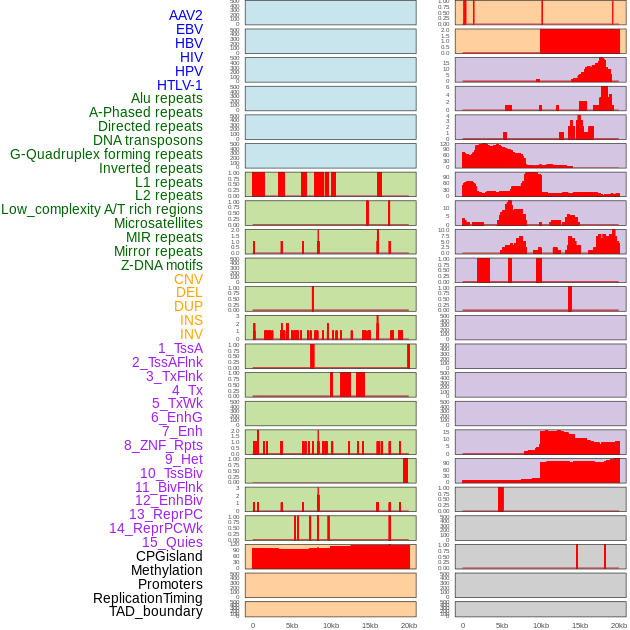 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for MTG2-AGR2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr20:60776043/chr7:16841341) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MTG2 | AGR2 |
| FUNCTION: Plays a role in the regulation of the mitochondrial ribosome assembly and of translational activity. Displays GTPase activity. Involved in the ribosome maturation process. {ECO:0000269|PubMed:17054726, ECO:0000269|PubMed:23396448}. | FUNCTION: Required for MUC2 post-transcriptional synthesis and secretion. May play a role in the production of mucus by intestinal cells (By similarity). Proto-oncogene that may play a role in cell migration, cell differentiation and cell growth. Promotes cell adhesion (PubMed:23274113). {ECO:0000250, ECO:0000269|PubMed:18199544, ECO:0000269|PubMed:23274113}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | AGR2 | chr20:60776043 | chr7:16841341 | ENST00000419304 | 0 | 8 | 45_54 | 0 | 176.0 | Motif | Homodimer stabilization%3B interchain | |
| Tgene | AGR2 | chr20:60776043 | chr7:16841341 | ENST00000419304 | 0 | 8 | 60_67 | 0 | 176.0 | Motif | Homodimer stabilization%3B interchain | |
| Tgene | AGR2 | chr20:60776043 | chr7:16841341 | ENST00000419304 | 0 | 8 | 21_40 | 0 | 176.0 | Region | Required to promote cell adhesion |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MTG2 | chr20:60776043 | chr7:16841341 | ENST00000370823 | + | 1 | 7 | 225_390 | 0 | 407.0 | Domain | Note=OBG-type G |
| Hgene | MTG2 | chr20:60776043 | chr7:16841341 | ENST00000370823 | + | 1 | 7 | 70_224 | 0 | 407.0 | Domain | Obg |
| Hgene | MTG2 | chr20:60776043 | chr7:16841341 | ENST00000370823 | + | 1 | 7 | 231_238 | 0 | 407.0 | Nucleotide binding | GTP |
| Hgene | MTG2 | chr20:60776043 | chr7:16841341 | ENST00000370823 | + | 1 | 7 | 256_260 | 0 | 407.0 | Nucleotide binding | GTP |
| Hgene | MTG2 | chr20:60776043 | chr7:16841341 | ENST00000370823 | + | 1 | 7 | 278_281 | 0 | 407.0 | Nucleotide binding | GTP |
| Hgene | MTG2 | chr20:60776043 | chr7:16841341 | ENST00000370823 | + | 1 | 7 | 345_348 | 0 | 407.0 | Nucleotide binding | GTP |
| Hgene | MTG2 | chr20:60776043 | chr7:16841341 | ENST00000370823 | + | 1 | 7 | 371_373 | 0 | 407.0 | Nucleotide binding | GTP |
| Hgene | MTG2 | chr20:60776043 | chr7:16841341 | ENST00000370823 | + | 1 | 7 | 15_406 | 0 | 407.0 | Region | Note=Localized in the mitochondria |
| Hgene | MTG2 | chr20:60776043 | chr7:16841341 | ENST00000370823 | + | 1 | 7 | 30_406 | 0 | 407.0 | Region | Note=Not localized in the mitochondria |
Top |
Fusion Gene Sequence for MTG2-AGR2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >55671_55671_1_MTG2-AGR2_MTG2_chr20_60776043_ENST00000436421_AGR2_chr7_16841341_ENST00000401412_length(transcript)=1903nt_BP=1242nt GTGCTTTCCCGAGCCGGGGCCATGGCACCTGCAAGGTGTTTCTCAGCAAGATTGAGGACCGTGTTTCAGGGCGTGGGGCATTGGGCTTTG TCCACATGGGCTGGCCTGAAGCCCAGCCGGCTACTGCCACAGCGGGCTTCTCCCAGGCTGCTCTCGGTCGGCCGTGCGGACCTCGCCAAG CATCAGGAACTCCCGGGGAAGAAGCTGCTCTCTGAGAAAAAGCTGAAAAGGTACTTTGTGGACTATCGGAGAGTGCTTGTCTGTGGAGGA AACGGAGGCGCTGGGGCAAGCTGCTTCCACAGTGAGCCCCGCAAGGAGTTTGGAGGCCCTGATGGAGGGGACGGAGGCAACGGTGGACAC GTCATTCTGAGAGCTGGAGCAGCTGCTGTTGCACCTGAAGGTGCTGTATGACGCCTACGCGGAGGCCGAGCTGGGCCAGGGCCGCCAGCC GCTCAGGTGGTAGCCACGCCAGAGCGGGGTCGCCTCTGGGCCTCTGTCTGAGCAAACCTGGGTGTGAATTCGGTGGTTTTGAATGCATAA AGTGCCTTGTGGACACGGGGGAGTTGTGGTGCTTCTGGGTCTCTGGGCCCCGCCTGCTGGCCTGAGATGCCCTCATGTTGGGAAGCATTC CGTGCCCCCTACCCCGCCTGCCCTCCGTATTTCCTGCACCTGTCAGCCTGCACCGACTGATGAGCCAGTTGCTCATTTGTGCTGATTAAC ACCCCTAATAAGGGGTTGGGGTGCCCATAACGGGGTGGCCCTGCCGCTGACTCAGGTCTCCGCCATGCACGCGTGGACTCTCGGATGAGC TCAGCAGAACCGCACAGCCAGAGCCCCAGGTCAGAAGTGCAGACCAGGGTTCTCAGCACAGTGCCCGTCGTGCTTCCATGGCTTGCTACG GAGAGAGACCTCTGGATCCACACTGGGGCTGCGTCTGGCCCGTTGTCCAGCAGCCCTGCGGTACCGCAAGCCCAGGCACCAGTGTCTCGG GGGGCCTCACTGCTGCGCAAGGGGTGGGGCCGAGGATGCAAGTCCAGGCAGAGCGGCGCAGGCAGCTGTGAGCTTTTCTCCATCAGCCGT CTGAGAAGAGCAGTGAGGCCAGCTGCTTCCTGTCCTTCAGAACACTTCTCTGTGCTCAGTGGGAGCCAGGAAGCCTCAGGCTTCACGACT GAATGCACCCAATATCCGACCTGGCTGCGTGTTTCTGGCTGGGCTGCCGTGTGCACAGCAAGTTAACTAGAGACCCATCTCTGACAGTTA GAGCCGATATCACTGGAAGATATTCAAATCGTCTCTATGCTTACGAACCTGCAGATACAGCTCTGTGTAAGTGTGACCTTCAATTCGGAT GTTGCCCAAGAGTTGAACACCTCTATTGCTGTAGTGATGGGACTAGAGGCTGCATGGTGAGGTGAGAGAGAGCTAGGCTTAGGGACGCCT CGGACTCAGTTTGACCCTAACTAGCCTTGCATCTTCAGGCAAATCACACTCTTTCCATCTCAGGTTTGTTTGTTTTTAATCTCAATTGAA GATGAATTATTTAGATTAGGTATCTTTTTCTCTAGGAGGATTCTATGACCACATTACCTATGAAACAACTGACAAACACCTTTCTCCTGA TGGCCAGTATGTCCCCAGGATTATGTTTGTTGCTTTAAAGAAAGTGTTTGCTGAAAATAAAGAAATCCAGAAATTGGCAGAGCAGTTTGT CCTCCTCAATCTGGTTCAACAAACCCTTGATGATTATTCATCACTTGGATGAGTGCCCACACAGTCAAGGTTGGGGTGACCAACTCATCT GGACTCAGACATATGAAGAAGCTCTATATAAATCCAAGACAAGCTGGAGCCAAAAAGGACACAAAGGACTCTCGACCCAAACTGCCCCAG >55671_55671_1_MTG2-AGR2_MTG2_chr20_60776043_ENST00000436421_AGR2_chr7_16841341_ENST00000401412_length(amino acids)=166AA_BP= MSTEKCSEGQEAAGLTALLRRLMEKSSQLPAPLCLDLHPRPHPLRSSEAPRDTGAWACGTAGLLDNGPDAAPVWIQRSLSVASHGSTTGT -------------------------------------------------------------- >55671_55671_2_MTG2-AGR2_MTG2_chr20_60776043_ENST00000436421_AGR2_chr7_16841341_ENST00000419304_length(transcript)=2788nt_BP=1242nt GTGCTTTCCCGAGCCGGGGCCATGGCACCTGCAAGGTGTTTCTCAGCAAGATTGAGGACCGTGTTTCAGGGCGTGGGGCATTGGGCTTTG TCCACATGGGCTGGCCTGAAGCCCAGCCGGCTACTGCCACAGCGGGCTTCTCCCAGGCTGCTCTCGGTCGGCCGTGCGGACCTCGCCAAG CATCAGGAACTCCCGGGGAAGAAGCTGCTCTCTGAGAAAAAGCTGAAAAGGTACTTTGTGGACTATCGGAGAGTGCTTGTCTGTGGAGGA AACGGAGGCGCTGGGGCAAGCTGCTTCCACAGTGAGCCCCGCAAGGAGTTTGGAGGCCCTGATGGAGGGGACGGAGGCAACGGTGGACAC GTCATTCTGAGAGCTGGAGCAGCTGCTGTTGCACCTGAAGGTGCTGTATGACGCCTACGCGGAGGCCGAGCTGGGCCAGGGCCGCCAGCC GCTCAGGTGGTAGCCACGCCAGAGCGGGGTCGCCTCTGGGCCTCTGTCTGAGCAAACCTGGGTGTGAATTCGGTGGTTTTGAATGCATAA AGTGCCTTGTGGACACGGGGGAGTTGTGGTGCTTCTGGGTCTCTGGGCCCCGCCTGCTGGCCTGAGATGCCCTCATGTTGGGAAGCATTC CGTGCCCCCTACCCCGCCTGCCCTCCGTATTTCCTGCACCTGTCAGCCTGCACCGACTGATGAGCCAGTTGCTCATTTGTGCTGATTAAC ACCCCTAATAAGGGGTTGGGGTGCCCATAACGGGGTGGCCCTGCCGCTGACTCAGGTCTCCGCCATGCACGCGTGGACTCTCGGATGAGC TCAGCAGAACCGCACAGCCAGAGCCCCAGGTCAGAAGTGCAGACCAGGGTTCTCAGCACAGTGCCCGTCGTGCTTCCATGGCTTGCTACG GAGAGAGACCTCTGGATCCACACTGGGGCTGCGTCTGGCCCGTTGTCCAGCAGCCCTGCGGTACCGCAAGCCCAGGCACCAGTGTCTCGG GGGGCCTCACTGCTGCGCAAGGGGTGGGGCCGAGGATGCAAGTCCAGGCAGAGCGGCGCAGGCAGCTGTGAGCTTTTCTCCATCAGCCGT CTGAGAAGAGCAGTGAGGCCAGCTGCTTCCTGTCCTTCAGAACACTTCTCTGTGCTCAGTGGGAGCCAGGAAGCCTCAGGCTTCACGACT GAATGCACCCAATATCCGACCTGGCTGCGTGTTTCTGGCTGGGCTGCCGTGTGCACAGCAAGTTAACTAGAGTGCTTGACAACATGAAGA AAGCTCTCAAGTTGCTGAAGACTGAATTGTAAAGAAAAAAAATCTCCAAGCCCTTCTGTCTGTCAGGCCTTGAGACTTGAAACCAGAAGA AGTGTGAGAAGACTGGCTAGTGTGGAAGCATAGTGAACACACTGATTAGGTTATGGTTTAATGTTACAACAACTATTTTTTAAGAAAAAC AAGTTTTAGAAATTTGGTTTCAAGTGTACATGTGTGAAAACAATATTGTATACTACCATAGTGAGCCATGATTTTCTAAAAAAAAAAATA AATGTTTTGGGGGTGTTCTGTTTTCTCCAACTTGGTCTTTCACAGTGGTTCGTTTACCAAATAGGATTAAACACACACAAAATGCTCAAG GAAGGGACAAGACAAAACCAAAACTAGTTCAAATGATGAAGACCAAAGACCAAGTTATCATCTCACCACACCACAGGTTCTCACTAGATG ACTGTAAGTAGACACGAGCTTAATCAACAGAAGTATCAAGCCATGTGCTTTAGCATAAAAGAATATTTAGAAAAACATCCCAAGAAAATC ACATCACTACCTAGAGTCAACTCTGGCCAGGAACTCTAAGGTACACACTTTCATTTAGTAATTAAATTTTAGTCAGATTTTGCCCAACCT AATGCTCTCAGGGAAAGCCTCTGGCAAGTAGCTTTCTCCTTCAGAGGTCTAATTTAGTAGAAAGGTCATCCAAAGAACATCTGCACTCCT GAACACACCCTGAAGAAATCCTGGGAATTGACCTTGTAATCGATTTGTCTGTCAAGGTCCTAAAGTACTGGAGTGAAATAAATTCAGCCA ACATGTGACTAATTGGAAGAAGAGCAAAGGGTGGTGACGTGTTGATGAGGCAGATGGAGATCAGAGGTTACTAGGGTTTAGGAAACGTGA AAGGCTGTGGCATCAGGGTAGGGGAGCATTCTGCCTAACAGAAATTAGAATTGTGTGTTAATGTCTTCACTCTATACTTAATCTCACATT CATTAATATATGGAATTCCTCTACTGCCCAGCCCCTCCTGATTTCTTTGGCCCCTGGACTATGGTGCTGTATATAATGCTTTGCAGTATC TGTTGCTTGTCTTGATTAACTTTTTTGGATAAAACCTTTTTTGAACAGAACCCATCTCTGACAGTTAGAGCCGATATCACTGGAAGATAT TCAAATCGTCTCTATGCTTACGAACCTGCAGATACAGCTCTGTTATGAAACAACTGACAAACACCTTTCTCCTGATGGCCAGTATGTCCC CAGGATTATGTTTGTTGCTTTAAAGAAAGTGTTTGCTGAAAATAAAGAAATCCAGAAATTGGCAGAGCAGTTTGTCCTCCTCAATCTGGT TCAACAAACCCTTGATGATTATTCATCACTTGGATGAGTGCCCACACAGTCAAGGTTGGGGTGACCAACTCATCTGGACTCAGACATATG >55671_55671_2_MTG2-AGR2_MTG2_chr20_60776043_ENST00000436421_AGR2_chr7_16841341_ENST00000419304_length(amino acids)=166AA_BP= MSTEKCSEGQEAAGLTALLRRLMEKSSQLPAPLCLDLHPRPHPLRSSEAPRDTGAWACGTAGLLDNGPDAAPVWIQRSLSVASHGSTTGT -------------------------------------------------------------- >55671_55671_3_MTG2-AGR2_MTG2_chr20_60776043_ENST00000436421_AGR2_chr7_16841341_ENST00000419572_length(transcript)=1957nt_BP=1242nt GTGCTTTCCCGAGCCGGGGCCATGGCACCTGCAAGGTGTTTCTCAGCAAGATTGAGGACCGTGTTTCAGGGCGTGGGGCATTGGGCTTTG TCCACATGGGCTGGCCTGAAGCCCAGCCGGCTACTGCCACAGCGGGCTTCTCCCAGGCTGCTCTCGGTCGGCCGTGCGGACCTCGCCAAG CATCAGGAACTCCCGGGGAAGAAGCTGCTCTCTGAGAAAAAGCTGAAAAGGTACTTTGTGGACTATCGGAGAGTGCTTGTCTGTGGAGGA AACGGAGGCGCTGGGGCAAGCTGCTTCCACAGTGAGCCCCGCAAGGAGTTTGGAGGCCCTGATGGAGGGGACGGAGGCAACGGTGGACAC GTCATTCTGAGAGCTGGAGCAGCTGCTGTTGCACCTGAAGGTGCTGTATGACGCCTACGCGGAGGCCGAGCTGGGCCAGGGCCGCCAGCC GCTCAGGTGGTAGCCACGCCAGAGCGGGGTCGCCTCTGGGCCTCTGTCTGAGCAAACCTGGGTGTGAATTCGGTGGTTTTGAATGCATAA AGTGCCTTGTGGACACGGGGGAGTTGTGGTGCTTCTGGGTCTCTGGGCCCCGCCTGCTGGCCTGAGATGCCCTCATGTTGGGAAGCATTC CGTGCCCCCTACCCCGCCTGCCCTCCGTATTTCCTGCACCTGTCAGCCTGCACCGACTGATGAGCCAGTTGCTCATTTGTGCTGATTAAC ACCCCTAATAAGGGGTTGGGGTGCCCATAACGGGGTGGCCCTGCCGCTGACTCAGGTCTCCGCCATGCACGCGTGGACTCTCGGATGAGC TCAGCAGAACCGCACAGCCAGAGCCCCAGGTCAGAAGTGCAGACCAGGGTTCTCAGCACAGTGCCCGTCGTGCTTCCATGGCTTGCTACG GAGAGAGACCTCTGGATCCACACTGGGGCTGCGTCTGGCCCGTTGTCCAGCAGCCCTGCGGTACCGCAAGCCCAGGCACCAGTGTCTCGG GGGGCCTCACTGCTGCGCAAGGGGTGGGGCCGAGGATGCAAGTCCAGGCAGAGCGGCGCAGGCAGCTGTGAGCTTTTCTCCATCAGCCGT CTGAGAAGAGCAGTGAGGCCAGCTGCTTCCTGTCCTTCAGAACACTTCTCTGTGCTCAGTGGGAGCCAGGAAGCCTCAGGCTTCACGACT GAATGCACCCAATATCCGACCTGGCTGCGTGTTTCTGGCTGGGCTGCCGTGTGCACAGCAAGTTAACTAGAGTGCTTGACAACATGAAGA AAGCTCTCAAGTTGCTGAAGACTGAATTGTAAAGAAAAAAAATCTCCAAGCCCTTCTGTCTGTCAGGCCTTGAGACTTGAAACCAGAAGA AGTGTGAGAAGACTGGCTAGTGTGGAAGCATAGTGAACACACTGATTAGGTTATGGTTTAATGTTACAACAACTATTTTTTAAGAAAAAC AAGTTTTAGAAATTTGGTTTCAAGTGTACATGTGTGAAAACAATATTGTATACTACCATAGTGAGCCATGATTTTCTAAAAAAAAAAATA AATGTTTTGGGGGTGTTCTGTTTTCTCCACCCATCTCTGACAGTTAGAGCCGATATCACTGGAAGATATTCAAATCGTCTCTATGCTTAC GAACCTGCAGATACAGCTCTGTTATGAAACAACTGACAAACACCTTTCTCCTGATGGCCAGTATGTCCCCAGGATTATGTTTGTTGCTTT AAAGAAAGTGTTTGCTGAAAATAAAGAAATCCAGAAATTGGCAGAGCAGTTTGTCCTCCTCAATCTGGTTCAACAAACCCTTGATGATTA TTCATCACTTGGATGAGTGCCCACACAGTCAAGGTTGGGGTGACCAACTCATCTGGACTCAGACATATGAAGAAGCTCTATATAAATCCA >55671_55671_3_MTG2-AGR2_MTG2_chr20_60776043_ENST00000436421_AGR2_chr7_16841341_ENST00000419572_length(amino acids)=166AA_BP= MSTEKCSEGQEAAGLTALLRRLMEKSSQLPAPLCLDLHPRPHPLRSSEAPRDTGAWACGTAGLLDNGPDAAPVWIQRSLSVASHGSTTGT -------------------------------------------------------------- >55671_55671_4_MTG2-AGR2_MTG2_chr20_60776043_ENST00000536470_AGR2_chr7_16841341_ENST00000401412_length(transcript)=1783nt_BP=1122nt GTGACGTGCTTTCCCGAGCCGAAAAGGTACTTTGTGGACTATCGGAGAGTGCTTGTCTGTGGAGGAAACGGAGGCGCTGGGGCAAGCTGC TTCCACAGTGAGCCCCGCAAGGAGTTTGGAGGCCCTGATGGAGGGGACGGAGGCAACGGTGGACACGTCATTCTGAGAGTTGACCAGCAA GTCAAGTCCCTGTCGTCGGTCCTGTCGCGGTACCAGGGTTTCAGTGGAGAAGATGGAGGGAGTAAAAACTGCTTCGGGCGCAGTGGCGCC GTCCTCTACATCCGGGTCCCCGTGGGCACGCTGGTGAAGGAGGGAGGCAGAGTTGTGGCCGACCTGTCTTGCGTGGGAGATGAGTACATT GCCGCGCTGGGCGGGGCAGGAGGGAAAGGCAACCGCTTCTTCCTGGCCAACAACAACCGTGCCCCTGTGACCTGTACCCCTGGACAGCCA GGACAGCAGCGAGTTCTCCACCTGGAGCTCAAGACGGTGGCCCACGCCGGAATGGTGGGATTCCCCAACGCCGGGAAGTCCTCACTGCTC CGGGCCATTTCAAACGCCAGACCCGCCGTGGCTTCCTACCCGTTCACCACCCTGAAGCCCCACGTCGGGATCGTCCACTACGAAGGCCAC CTACAAATAGCAGCTGGAGCAGCTGCTGTTGCACCTGAAGGTGCTGTATGACGCCTACGCGGAGGCCGAGCTGGGCCAGGGCCGCCAGCC GCTCAGGTGGTAGCCACGCCAGAGCGGGGTCGCCTCTGGGCCTCTGTCTGAGCAAACCTGGGTGTGAATTCGGTGGTTTTGAATGCATAA AGTGCCTTGTGGACACGGGGGAGTTGTGGTGCTTCTGGGTCTCTGGGCCCCGCCTGCTGGCCTGAGATGCCCTCATGTTGGGAAGCATTC CGTGCCCCCTACCCCGCCTGCCCTCCGTATTTCCTGCACCTGTCAGCCTGCACCGACTGATGAGCCAGTTGCTCATTTGTGCTGATTAAC ACCCCTAATAAGGGGTTGGGGTGCCCATAACGGGGTGGCCCTGCCGCTGACTCAGGTCTCCGCCATGCACGCGTGGACTCTCGGATGAGC TCAGCAGAACCGCACAGCCAGAGCCCCAGGTCAGAAGTGCAGACCCATCTCTGACAGTTAGAGCCGATATCACTGGAAGATATTCAAATC GTCTCTATGCTTACGAACCTGCAGATACAGCTCTGTGTAAGTGTGACCTTCAATTCGGATGTTGCCCAAGAGTTGAACACCTCTATTGCT GTAGTGATGGGACTAGAGGCTGCATGGTGAGGTGAGAGAGAGCTAGGCTTAGGGACGCCTCGGACTCAGTTTGACCCTAACTAGCCTTGC ATCTTCAGGCAAATCACACTCTTTCCATCTCAGGTTTGTTTGTTTTTAATCTCAATTGAAGATGAATTATTTAGATTAGGTATCTTTTTC TCTAGGAGGATTCTATGACCACATTACCTATGAAACAACTGACAAACACCTTTCTCCTGATGGCCAGTATGTCCCCAGGATTATGTTTGT TGCTTTAAAGAAAGTGTTTGCTGAAAATAAAGAAATCCAGAAATTGGCAGAGCAGTTTGTCCTCCTCAATCTGGTTCAACAAACCCTTGA TGATTATTCATCACTTGGATGAGTGCCCACACAGTCAAGGTTGGGGTGACCAACTCATCTGGACTCAGACATATGAAGAAGCTCTATATA >55671_55671_4_MTG2-AGR2_MTG2_chr20_60776043_ENST00000536470_AGR2_chr7_16841341_ENST00000401412_length(amino acids)=172AA_BP= MRVDQQVKSLSSVLSRYQGFSGEDGGSKNCFGRSGAVLYIRVPVGTLVKEGGRVVADLSCVGDEYIAALGGAGGKGNRFFLANNNRAPVT -------------------------------------------------------------- >55671_55671_5_MTG2-AGR2_MTG2_chr20_60776043_ENST00000536470_AGR2_chr7_16841341_ENST00000419304_length(transcript)=2668nt_BP=1122nt GTGACGTGCTTTCCCGAGCCGAAAAGGTACTTTGTGGACTATCGGAGAGTGCTTGTCTGTGGAGGAAACGGAGGCGCTGGGGCAAGCTGC TTCCACAGTGAGCCCCGCAAGGAGTTTGGAGGCCCTGATGGAGGGGACGGAGGCAACGGTGGACACGTCATTCTGAGAGTTGACCAGCAA GTCAAGTCCCTGTCGTCGGTCCTGTCGCGGTACCAGGGTTTCAGTGGAGAAGATGGAGGGAGTAAAAACTGCTTCGGGCGCAGTGGCGCC GTCCTCTACATCCGGGTCCCCGTGGGCACGCTGGTGAAGGAGGGAGGCAGAGTTGTGGCCGACCTGTCTTGCGTGGGAGATGAGTACATT GCCGCGCTGGGCGGGGCAGGAGGGAAAGGCAACCGCTTCTTCCTGGCCAACAACAACCGTGCCCCTGTGACCTGTACCCCTGGACAGCCA GGACAGCAGCGAGTTCTCCACCTGGAGCTCAAGACGGTGGCCCACGCCGGAATGGTGGGATTCCCCAACGCCGGGAAGTCCTCACTGCTC CGGGCCATTTCAAACGCCAGACCCGCCGTGGCTTCCTACCCGTTCACCACCCTGAAGCCCCACGTCGGGATCGTCCACTACGAAGGCCAC CTACAAATAGCAGCTGGAGCAGCTGCTGTTGCACCTGAAGGTGCTGTATGACGCCTACGCGGAGGCCGAGCTGGGCCAGGGCCGCCAGCC GCTCAGGTGGTAGCCACGCCAGAGCGGGGTCGCCTCTGGGCCTCTGTCTGAGCAAACCTGGGTGTGAATTCGGTGGTTTTGAATGCATAA AGTGCCTTGTGGACACGGGGGAGTTGTGGTGCTTCTGGGTCTCTGGGCCCCGCCTGCTGGCCTGAGATGCCCTCATGTTGGGAAGCATTC CGTGCCCCCTACCCCGCCTGCCCTCCGTATTTCCTGCACCTGTCAGCCTGCACCGACTGATGAGCCAGTTGCTCATTTGTGCTGATTAAC ACCCCTAATAAGGGGTTGGGGTGCCCATAACGGGGTGGCCCTGCCGCTGACTCAGGTCTCCGCCATGCACGCGTGGACTCTCGGATGAGC TCAGCAGAACCGCACAGCCAGAGCCCCAGGTCAGAAGTGCAGTGCTTGACAACATGAAGAAAGCTCTCAAGTTGCTGAAGACTGAATTGT AAAGAAAAAAAATCTCCAAGCCCTTCTGTCTGTCAGGCCTTGAGACTTGAAACCAGAAGAAGTGTGAGAAGACTGGCTAGTGTGGAAGCA TAGTGAACACACTGATTAGGTTATGGTTTAATGTTACAACAACTATTTTTTAAGAAAAACAAGTTTTAGAAATTTGGTTTCAAGTGTACA TGTGTGAAAACAATATTGTATACTACCATAGTGAGCCATGATTTTCTAAAAAAAAAAATAAATGTTTTGGGGGTGTTCTGTTTTCTCCAA CTTGGTCTTTCACAGTGGTTCGTTTACCAAATAGGATTAAACACACACAAAATGCTCAAGGAAGGGACAAGACAAAACCAAAACTAGTTC AAATGATGAAGACCAAAGACCAAGTTATCATCTCACCACACCACAGGTTCTCACTAGATGACTGTAAGTAGACACGAGCTTAATCAACAG AAGTATCAAGCCATGTGCTTTAGCATAAAAGAATATTTAGAAAAACATCCCAAGAAAATCACATCACTACCTAGAGTCAACTCTGGCCAG GAACTCTAAGGTACACACTTTCATTTAGTAATTAAATTTTAGTCAGATTTTGCCCAACCTAATGCTCTCAGGGAAAGCCTCTGGCAAGTA GCTTTCTCCTTCAGAGGTCTAATTTAGTAGAAAGGTCATCCAAAGAACATCTGCACTCCTGAACACACCCTGAAGAAATCCTGGGAATTG ACCTTGTAATCGATTTGTCTGTCAAGGTCCTAAAGTACTGGAGTGAAATAAATTCAGCCAACATGTGACTAATTGGAAGAAGAGCAAAGG GTGGTGACGTGTTGATGAGGCAGATGGAGATCAGAGGTTACTAGGGTTTAGGAAACGTGAAAGGCTGTGGCATCAGGGTAGGGGAGCATT CTGCCTAACAGAAATTAGAATTGTGTGTTAATGTCTTCACTCTATACTTAATCTCACATTCATTAATATATGGAATTCCTCTACTGCCCA GCCCCTCCTGATTTCTTTGGCCCCTGGACTATGGTGCTGTATATAATGCTTTGCAGTATCTGTTGCTTGTCTTGATTAACTTTTTTGGAT AAAACCTTTTTTGAACAGAACCCATCTCTGACAGTTAGAGCCGATATCACTGGAAGATATTCAAATCGTCTCTATGCTTACGAACCTGCA GATACAGCTCTGTTATGAAACAACTGACAAACACCTTTCTCCTGATGGCCAGTATGTCCCCAGGATTATGTTTGTTGCTTTAAAGAAAGT GTTTGCTGAAAATAAAGAAATCCAGAAATTGGCAGAGCAGTTTGTCCTCCTCAATCTGGTTCAACAAACCCTTGATGATTATTCATCACT TGGATGAGTGCCCACACAGTCAAGGTTGGGGTGACCAACTCATCTGGACTCAGACATATGAAGAAGCTCTATATAAATCCAAGACAAGCT >55671_55671_5_MTG2-AGR2_MTG2_chr20_60776043_ENST00000536470_AGR2_chr7_16841341_ENST00000419304_length(amino acids)=172AA_BP= MRVDQQVKSLSSVLSRYQGFSGEDGGSKNCFGRSGAVLYIRVPVGTLVKEGGRVVADLSCVGDEYIAALGGAGGKGNRFFLANNNRAPVT -------------------------------------------------------------- >55671_55671_6_MTG2-AGR2_MTG2_chr20_60776043_ENST00000536470_AGR2_chr7_16841341_ENST00000419572_length(transcript)=1837nt_BP=1122nt GTGACGTGCTTTCCCGAGCCGAAAAGGTACTTTGTGGACTATCGGAGAGTGCTTGTCTGTGGAGGAAACGGAGGCGCTGGGGCAAGCTGC TTCCACAGTGAGCCCCGCAAGGAGTTTGGAGGCCCTGATGGAGGGGACGGAGGCAACGGTGGACACGTCATTCTGAGAGTTGACCAGCAA GTCAAGTCCCTGTCGTCGGTCCTGTCGCGGTACCAGGGTTTCAGTGGAGAAGATGGAGGGAGTAAAAACTGCTTCGGGCGCAGTGGCGCC GTCCTCTACATCCGGGTCCCCGTGGGCACGCTGGTGAAGGAGGGAGGCAGAGTTGTGGCCGACCTGTCTTGCGTGGGAGATGAGTACATT GCCGCGCTGGGCGGGGCAGGAGGGAAAGGCAACCGCTTCTTCCTGGCCAACAACAACCGTGCCCCTGTGACCTGTACCCCTGGACAGCCA GGACAGCAGCGAGTTCTCCACCTGGAGCTCAAGACGGTGGCCCACGCCGGAATGGTGGGATTCCCCAACGCCGGGAAGTCCTCACTGCTC CGGGCCATTTCAAACGCCAGACCCGCCGTGGCTTCCTACCCGTTCACCACCCTGAAGCCCCACGTCGGGATCGTCCACTACGAAGGCCAC CTACAAATAGCAGCTGGAGCAGCTGCTGTTGCACCTGAAGGTGCTGTATGACGCCTACGCGGAGGCCGAGCTGGGCCAGGGCCGCCAGCC GCTCAGGTGGTAGCCACGCCAGAGCGGGGTCGCCTCTGGGCCTCTGTCTGAGCAAACCTGGGTGTGAATTCGGTGGTTTTGAATGCATAA AGTGCCTTGTGGACACGGGGGAGTTGTGGTGCTTCTGGGTCTCTGGGCCCCGCCTGCTGGCCTGAGATGCCCTCATGTTGGGAAGCATTC CGTGCCCCCTACCCCGCCTGCCCTCCGTATTTCCTGCACCTGTCAGCCTGCACCGACTGATGAGCCAGTTGCTCATTTGTGCTGATTAAC ACCCCTAATAAGGGGTTGGGGTGCCCATAACGGGGTGGCCCTGCCGCTGACTCAGGTCTCCGCCATGCACGCGTGGACTCTCGGATGAGC TCAGCAGAACCGCACAGCCAGAGCCCCAGGTCAGAAGTGCAGTGCTTGACAACATGAAGAAAGCTCTCAAGTTGCTGAAGACTGAATTGT AAAGAAAAAAAATCTCCAAGCCCTTCTGTCTGTCAGGCCTTGAGACTTGAAACCAGAAGAAGTGTGAGAAGACTGGCTAGTGTGGAAGCA TAGTGAACACACTGATTAGGTTATGGTTTAATGTTACAACAACTATTTTTTAAGAAAAACAAGTTTTAGAAATTTGGTTTCAAGTGTACA TGTGTGAAAACAATATTGTATACTACCATAGTGAGCCATGATTTTCTAAAAAAAAAAATAAATGTTTTGGGGGTGTTCTGTTTTCTCCAC CCATCTCTGACAGTTAGAGCCGATATCACTGGAAGATATTCAAATCGTCTCTATGCTTACGAACCTGCAGATACAGCTCTGTTATGAAAC AACTGACAAACACCTTTCTCCTGATGGCCAGTATGTCCCCAGGATTATGTTTGTTGCTTTAAAGAAAGTGTTTGCTGAAAATAAAGAAAT CCAGAAATTGGCAGAGCAGTTTGTCCTCCTCAATCTGGTTCAACAAACCCTTGATGATTATTCATCACTTGGATGAGTGCCCACACAGTC AAGGTTGGGGTGACCAACTCATCTGGACTCAGACATATGAAGAAGCTCTATATAAATCCAAGACAAGCTGGAGCCAAAAAGGACACAAAG >55671_55671_6_MTG2-AGR2_MTG2_chr20_60776043_ENST00000536470_AGR2_chr7_16841341_ENST00000419572_length(amino acids)=172AA_BP= MRVDQQVKSLSSVLSRYQGFSGEDGGSKNCFGRSGAVLYIRVPVGTLVKEGGRVVADLSCVGDEYIAALGGAGGKGNRFFLANNNRAPVT -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MTG2-AGR2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MTG2-AGR2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MTG2-AGR2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
