
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MYLK-NXPH1 (FusionGDB2 ID:56525) |
Fusion Gene Summary for MYLK-NXPH1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MYLK-NXPH1 | Fusion gene ID: 56525 | Hgene | Tgene | Gene symbol | MYLK | NXPH1 | Gene ID | 4638 | 30010 |
| Gene name | myosin light chain kinase | neurexophilin 1 | |
| Synonyms | AAT7|KRP|MLCK|MLCK1|MLCK108|MLCK210|MMIHS|MSTP083|MYLK1|smMLCK | NPH1|Nbla00697 | |
| Cytomap | 3q21.1 | 7p21.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | myosin light chain kinase, smooth musclekinase-related proteinmyosin, light polypeptide kinasesmooth muscle myosin light chain kinasetelokin | neurexophilin-1 | |
| Modification date | 20200320 | 20200313 | |
| UniProtAcc | Q15746 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000346322, ENST00000354792, ENST00000359169, ENST00000360304, ENST00000360772, ENST00000475616, ENST00000418370, ENST00000510775, ENST00000578202, ENST00000583087, | ENST00000497400, ENST00000602349, ENST00000405863, | |
| Fusion gene scores | * DoF score | 17 X 15 X 7=1785 | 6 X 4 X 5=120 |
| # samples | 18 | 7 | |
| ** MAII score | log2(18/1785*10)=-3.30985526258679 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/120*10)=-0.777607578663552 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MYLK [Title/Abstract] AND NXPH1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MYLK(123367818)-NXPH1(8790638), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | MYLK-NXPH1 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. MYLK-NXPH1 seems lost the major protein functional domain in Hgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | MYLK | GO:0030335 | positive regulation of cell migration | 19826488 |
| Hgene | MYLK | GO:0051928 | positive regulation of calcium ion transport | 16284075 |
| Hgene | MYLK | GO:0071476 | cellular hypotonic response | 11976941 |
| Hgene | MYLK | GO:0090303 | positive regulation of wound healing | 15825080 |
 Fusion gene breakpoints across MYLK (5'-gene) Fusion gene breakpoints across MYLK (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
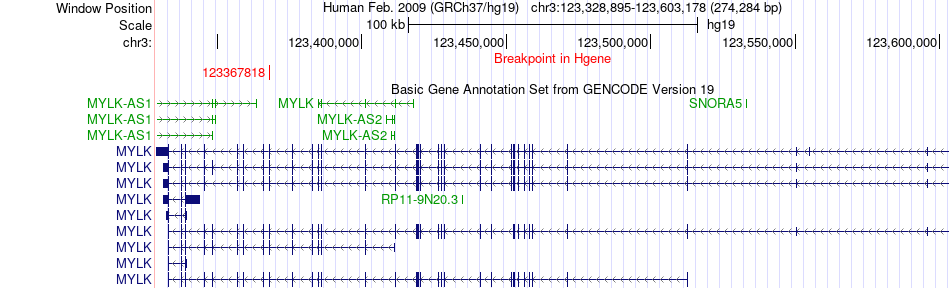 |
 Fusion gene breakpoints across NXPH1 (3'-gene) Fusion gene breakpoints across NXPH1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
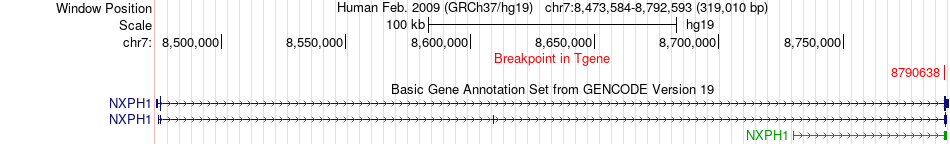 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | KIRC | TCGA-BP-4781-01A | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
Top |
Fusion Gene ORF analysis for MYLK-NXPH1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000346322 | ENST00000497400 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| 5CDS-3UTR | ENST00000354792 | ENST00000497400 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| 5CDS-3UTR | ENST00000359169 | ENST00000497400 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| 5CDS-3UTR | ENST00000360304 | ENST00000497400 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| 5CDS-3UTR | ENST00000360772 | ENST00000497400 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| 5CDS-3UTR | ENST00000475616 | ENST00000497400 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| 5CDS-5UTR | ENST00000346322 | ENST00000602349 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| 5CDS-5UTR | ENST00000354792 | ENST00000602349 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| 5CDS-5UTR | ENST00000359169 | ENST00000602349 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| 5CDS-5UTR | ENST00000360304 | ENST00000602349 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| 5CDS-5UTR | ENST00000360772 | ENST00000602349 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| 5CDS-5UTR | ENST00000475616 | ENST00000602349 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| Frame-shift | ENST00000354792 | ENST00000405863 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| Frame-shift | ENST00000360772 | ENST00000405863 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| Frame-shift | ENST00000475616 | ENST00000405863 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| In-frame | ENST00000346322 | ENST00000405863 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| In-frame | ENST00000359169 | ENST00000405863 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| In-frame | ENST00000360304 | ENST00000405863 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| intron-3CDS | ENST00000418370 | ENST00000405863 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| intron-3CDS | ENST00000510775 | ENST00000405863 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| intron-3CDS | ENST00000578202 | ENST00000405863 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| intron-3CDS | ENST00000583087 | ENST00000405863 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| intron-3UTR | ENST00000418370 | ENST00000497400 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| intron-3UTR | ENST00000510775 | ENST00000497400 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| intron-3UTR | ENST00000578202 | ENST00000497400 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| intron-3UTR | ENST00000583087 | ENST00000497400 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| intron-5UTR | ENST00000418370 | ENST00000602349 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| intron-5UTR | ENST00000510775 | ENST00000602349 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| intron-5UTR | ENST00000578202 | ENST00000602349 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
| intron-5UTR | ENST00000583087 | ENST00000602349 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000359169 | MYLK | chr3 | 123367818 | - | ENST00000405863 | NXPH1 | chr7 | 8790638 | + | 6653 | 4697 | 240 | 4715 | 1491 |
| ENST00000360304 | MYLK | chr3 | 123367818 | - | ENST00000405863 | NXPH1 | chr7 | 8790638 | + | 6653 | 4697 | 240 | 4715 | 1491 |
| ENST00000346322 | MYLK | chr3 | 123367818 | - | ENST00000405863 | NXPH1 | chr7 | 8790638 | + | 6458 | 4502 | 252 | 4520 | 1422 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000359169 | ENST00000405863 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + | 0.001699623 | 0.9983004 |
| ENST00000360304 | ENST00000405863 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + | 0.001699623 | 0.9983004 |
| ENST00000346322 | ENST00000405863 | MYLK | chr3 | 123367818 | - | NXPH1 | chr7 | 8790638 | + | 0.001592805 | 0.9984072 |
Top |
Fusion Genomic Features for MYLK-NXPH1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
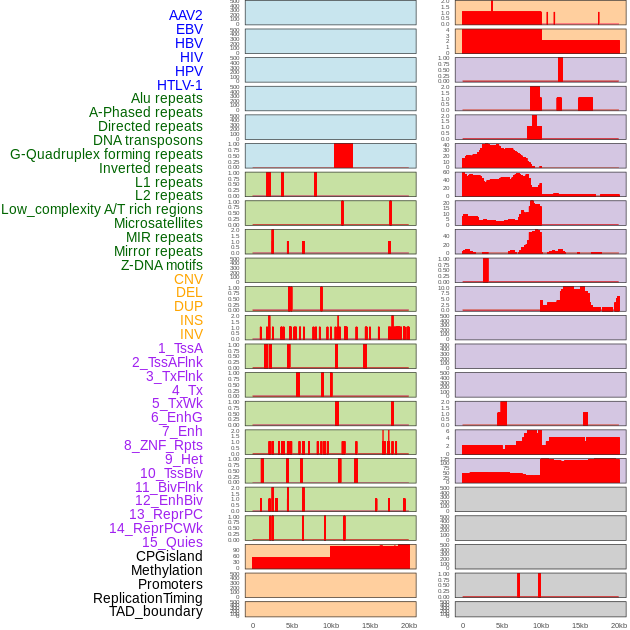 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
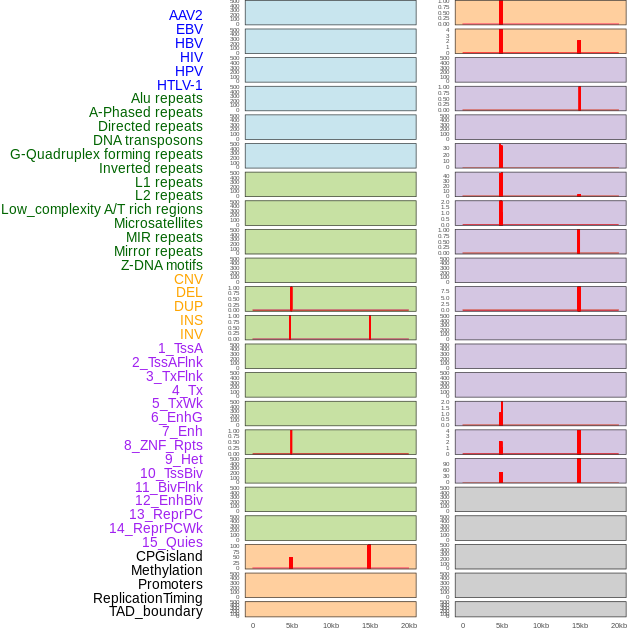 |
Top |
Fusion Protein Features for MYLK-NXPH1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:123367818/chr7:8790638) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MYLK | . |
| FUNCTION: Calcium/calmodulin-dependent myosin light chain kinase implicated in smooth muscle contraction via phosphorylation of myosin light chains (MLC). Also regulates actin-myosin interaction through a non-kinase activity. Phosphorylates PTK2B/PYK2 and myosin light-chains. Involved in the inflammatory response (e.g. apoptosis, vascular permeability, leukocyte diapedesis), cell motility and morphology, airway hyperreactivity and other activities relevant to asthma. Required for tonic airway smooth muscle contraction that is necessary for physiological and asthmatic airway resistance. Necessary for gastrointestinal motility. Implicated in the regulation of endothelial as well as vascular permeability, probably via the regulation of cytoskeletal rearrangements. In the nervous system it has been shown to control the growth initiation of astrocytic processes in culture and to participate in transmitter release at synapses formed between cultured sympathetic ganglion cells. Critical participant in signaling sequences that result in fibroblast apoptosis. Plays a role in the regulation of epithelial cell survival. Required for epithelial wound healing, especially during actomyosin ring contraction during purse-string wound closure. Mediates RhoA-dependent membrane blebbing. Triggers TRPC5 channel activity in a calcium-dependent signaling, by inducing its subcellular localization at the plasma membrane. Promotes cell migration (including tumor cells) and tumor metastasis. PTK2B/PYK2 activation by phosphorylation mediates ITGB2 activation and is thus essential to trigger neutrophil transmigration during acute lung injury (ALI). May regulate optic nerve head astrocyte migration. Probably involved in mitotic cytoskeletal regulation. Regulates tight junction probably by modulating ZO-1 exchange in the perijunctional actomyosin ring. Mediates burn-induced microvascular barrier injury; triggers endothelial contraction in the development of microvascular hyperpermeability by phosphorylating MLC. Essential for intestinal barrier dysfunction. Mediates Giardia spp.-mediated reduced epithelial barrier function during giardiasis intestinal infection via reorganization of cytoskeletal F-actin and tight junctional ZO-1. Necessary for hypotonicity-induced Ca(2+) entry and subsequent activation of volume-sensitive organic osmolyte/anion channels (VSOAC) in cervical cancer cells. Responsible for high proliferative ability of breast cancer cells through anti-apoptosis. {ECO:0000269|PubMed:11113114, ECO:0000269|PubMed:11976941, ECO:0000269|PubMed:15020676, ECO:0000269|PubMed:15825080, ECO:0000269|PubMed:16284075, ECO:0000269|PubMed:16723733, ECO:0000269|PubMed:18587400, ECO:0000269|PubMed:18710790, ECO:0000269|PubMed:19826488, ECO:0000269|PubMed:20139351, ECO:0000269|PubMed:20181817, ECO:0000269|PubMed:20375339, ECO:0000269|PubMed:20453870}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 1098_1186 | 1402 | 1846.0 | Domain | Note=Ig-like C2-type 7 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 1238_1326 | 1402 | 1846.0 | Domain | Note=Ig-like C2-type 8 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 161_249 | 1402 | 1846.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 33_122 | 1402 | 1846.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 414_503 | 1402 | 1846.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 514_599 | 1402 | 1846.0 | Domain | Note=Ig-like C2-type 4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 620_711 | 1402 | 1846.0 | Domain | Note=Ig-like C2-type 5 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 721_821 | 1402 | 1846.0 | Domain | Note=Ig-like C2-type 6 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 1098_1186 | 1471 | 1864.0 | Domain | Note=Ig-like C2-type 7 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 1238_1326 | 1471 | 1864.0 | Domain | Note=Ig-like C2-type 8 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 1334_1426 | 1471 | 1864.0 | Domain | Fibronectin type-III |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 161_249 | 1471 | 1864.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 33_122 | 1471 | 1864.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 414_503 | 1471 | 1864.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 514_599 | 1471 | 1864.0 | Domain | Note=Ig-like C2-type 4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 620_711 | 1471 | 1864.0 | Domain | Note=Ig-like C2-type 5 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 721_821 | 1471 | 1864.0 | Domain | Note=Ig-like C2-type 6 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 1098_1186 | 1471 | 1915.0 | Domain | Note=Ig-like C2-type 7 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 1238_1326 | 1471 | 1915.0 | Domain | Note=Ig-like C2-type 8 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 1334_1426 | 1471 | 1915.0 | Domain | Fibronectin type-III |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 161_249 | 1471 | 1915.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 33_122 | 1471 | 1915.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 414_503 | 1471 | 1915.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 514_599 | 1471 | 1915.0 | Domain | Note=Ig-like C2-type 4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 620_711 | 1471 | 1915.0 | Domain | Note=Ig-like C2-type 5 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 721_821 | 1471 | 1915.0 | Domain | Note=Ig-like C2-type 6 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 1098_1186 | 1471 | 1864.0 | Domain | Note=Ig-like C2-type 7 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 1238_1326 | 1471 | 1864.0 | Domain | Note=Ig-like C2-type 8 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 1334_1426 | 1471 | 1864.0 | Domain | Fibronectin type-III |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 161_249 | 1471 | 1864.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 33_122 | 1471 | 1864.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 414_503 | 1471 | 1864.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 514_599 | 1471 | 1864.0 | Domain | Note=Ig-like C2-type 4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 620_711 | 1471 | 1864.0 | Domain | Note=Ig-like C2-type 5 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 721_821 | 1471 | 1864.0 | Domain | Note=Ig-like C2-type 6 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 1098_1186 | 1471 | 1915.0 | Domain | Note=Ig-like C2-type 7 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 1238_1326 | 1471 | 1915.0 | Domain | Note=Ig-like C2-type 8 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 1334_1426 | 1471 | 1915.0 | Domain | Fibronectin type-III |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 161_249 | 1471 | 1915.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 33_122 | 1471 | 1915.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 414_503 | 1471 | 1915.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 514_599 | 1471 | 1915.0 | Domain | Note=Ig-like C2-type 4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 620_711 | 1471 | 1915.0 | Domain | Note=Ig-like C2-type 5 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 721_821 | 1471 | 1915.0 | Domain | Note=Ig-like C2-type 6 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 868_998 | 1402 | 1846.0 | Region | Note=5 X 28 AA approximate tandem repeats |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 923_963 | 1402 | 1846.0 | Region | Actin-binding (calcium/calmodulin-sensitive) |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 948_963 | 1402 | 1846.0 | Region | Calmodulin-binding |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 999_1063 | 1402 | 1846.0 | Region | Note=6 X 12 AA approximate tandem repeats |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 1061_1460 | 1471 | 1864.0 | Region | Actin-binding (calcium/calmodulin-insensitive) |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 868_998 | 1471 | 1864.0 | Region | Note=5 X 28 AA approximate tandem repeats |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 923_963 | 1471 | 1864.0 | Region | Actin-binding (calcium/calmodulin-sensitive) |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 948_963 | 1471 | 1864.0 | Region | Calmodulin-binding |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 999_1063 | 1471 | 1864.0 | Region | Note=6 X 12 AA approximate tandem repeats |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 1061_1460 | 1471 | 1915.0 | Region | Actin-binding (calcium/calmodulin-insensitive) |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 868_998 | 1471 | 1915.0 | Region | Note=5 X 28 AA approximate tandem repeats |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 923_963 | 1471 | 1915.0 | Region | Actin-binding (calcium/calmodulin-sensitive) |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 948_963 | 1471 | 1915.0 | Region | Calmodulin-binding |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 999_1063 | 1471 | 1915.0 | Region | Note=6 X 12 AA approximate tandem repeats |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 1061_1460 | 1471 | 1864.0 | Region | Actin-binding (calcium/calmodulin-insensitive) |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 868_998 | 1471 | 1864.0 | Region | Note=5 X 28 AA approximate tandem repeats |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 923_963 | 1471 | 1864.0 | Region | Actin-binding (calcium/calmodulin-sensitive) |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 948_963 | 1471 | 1864.0 | Region | Calmodulin-binding |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 999_1063 | 1471 | 1864.0 | Region | Note=6 X 12 AA approximate tandem repeats |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 1061_1460 | 1471 | 1915.0 | Region | Actin-binding (calcium/calmodulin-insensitive) |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 868_998 | 1471 | 1915.0 | Region | Note=5 X 28 AA approximate tandem repeats |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 923_963 | 1471 | 1915.0 | Region | Actin-binding (calcium/calmodulin-sensitive) |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 948_963 | 1471 | 1915.0 | Region | Calmodulin-binding |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 999_1063 | 1471 | 1915.0 | Region | Note=6 X 12 AA approximate tandem repeats |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 1004_1015 | 1402 | 1846.0 | Repeat | Note=2-2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 1016_1027 | 1402 | 1846.0 | Repeat | Note=2-3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 1028_1039 | 1402 | 1846.0 | Repeat | Note=2-4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 1040_1051 | 1402 | 1846.0 | Repeat | Note=2-5 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 1052_1063 | 1402 | 1846.0 | Repeat | Note=2-6 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 868_895 | 1402 | 1846.0 | Repeat | Note=1-1 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 896_923 | 1402 | 1846.0 | Repeat | Note=1-2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 924_951 | 1402 | 1846.0 | Repeat | Note=1-3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 952_979 | 1402 | 1846.0 | Repeat | Note=1-4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 980_998 | 1402 | 1846.0 | Repeat | Note=1-5%3B truncated |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 999_1003 | 1402 | 1846.0 | Repeat | Note=2-1%3B truncated |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 1004_1015 | 1471 | 1864.0 | Repeat | Note=2-2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 1016_1027 | 1471 | 1864.0 | Repeat | Note=2-3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 1028_1039 | 1471 | 1864.0 | Repeat | Note=2-4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 1040_1051 | 1471 | 1864.0 | Repeat | Note=2-5 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 1052_1063 | 1471 | 1864.0 | Repeat | Note=2-6 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 868_895 | 1471 | 1864.0 | Repeat | Note=1-1 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 896_923 | 1471 | 1864.0 | Repeat | Note=1-2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 924_951 | 1471 | 1864.0 | Repeat | Note=1-3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 952_979 | 1471 | 1864.0 | Repeat | Note=1-4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 980_998 | 1471 | 1864.0 | Repeat | Note=1-5%3B truncated |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 999_1003 | 1471 | 1864.0 | Repeat | Note=2-1%3B truncated |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 1004_1015 | 1471 | 1915.0 | Repeat | Note=2-2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 1016_1027 | 1471 | 1915.0 | Repeat | Note=2-3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 1028_1039 | 1471 | 1915.0 | Repeat | Note=2-4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 1040_1051 | 1471 | 1915.0 | Repeat | Note=2-5 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 1052_1063 | 1471 | 1915.0 | Repeat | Note=2-6 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 868_895 | 1471 | 1915.0 | Repeat | Note=1-1 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 896_923 | 1471 | 1915.0 | Repeat | Note=1-2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 924_951 | 1471 | 1915.0 | Repeat | Note=1-3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 952_979 | 1471 | 1915.0 | Repeat | Note=1-4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 980_998 | 1471 | 1915.0 | Repeat | Note=1-5%3B truncated |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 999_1003 | 1471 | 1915.0 | Repeat | Note=2-1%3B truncated |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 1004_1015 | 1471 | 1864.0 | Repeat | Note=2-2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 1016_1027 | 1471 | 1864.0 | Repeat | Note=2-3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 1028_1039 | 1471 | 1864.0 | Repeat | Note=2-4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 1040_1051 | 1471 | 1864.0 | Repeat | Note=2-5 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 1052_1063 | 1471 | 1864.0 | Repeat | Note=2-6 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 868_895 | 1471 | 1864.0 | Repeat | Note=1-1 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 896_923 | 1471 | 1864.0 | Repeat | Note=1-2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 924_951 | 1471 | 1864.0 | Repeat | Note=1-3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 952_979 | 1471 | 1864.0 | Repeat | Note=1-4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 980_998 | 1471 | 1864.0 | Repeat | Note=1-5%3B truncated |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 999_1003 | 1471 | 1864.0 | Repeat | Note=2-1%3B truncated |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 1004_1015 | 1471 | 1915.0 | Repeat | Note=2-2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 1016_1027 | 1471 | 1915.0 | Repeat | Note=2-3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 1028_1039 | 1471 | 1915.0 | Repeat | Note=2-4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 1040_1051 | 1471 | 1915.0 | Repeat | Note=2-5 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 1052_1063 | 1471 | 1915.0 | Repeat | Note=2-6 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 868_895 | 1471 | 1915.0 | Repeat | Note=1-1 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 896_923 | 1471 | 1915.0 | Repeat | Note=1-2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 924_951 | 1471 | 1915.0 | Repeat | Note=1-3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 952_979 | 1471 | 1915.0 | Repeat | Note=1-4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 980_998 | 1471 | 1915.0 | Repeat | Note=1-5%3B truncated |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 999_1003 | 1471 | 1915.0 | Repeat | Note=2-1%3B truncated |
| Tgene | NXPH1 | chr3:123367818 | chr7:8790638 | ENST00000405863 | 1 | 3 | 177_185 | 18 | 272.0 | Region | Note=IV (linker domain) | |
| Tgene | NXPH1 | chr3:123367818 | chr7:8790638 | ENST00000405863 | 1 | 3 | 186_271 | 18 | 272.0 | Region | Note=V (Cys-rich) | |
| Tgene | NXPH1 | chr3:123367818 | chr7:8790638 | ENST00000405863 | 1 | 3 | 22_97 | 18 | 272.0 | Region | Note=II | |
| Tgene | NXPH1 | chr3:123367818 | chr7:8790638 | ENST00000405863 | 1 | 3 | 98_176 | 18 | 272.0 | Region | Note=III |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 1906_1914 | 1402 | 1846.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 1906_1914 | 1471 | 1864.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 1906_1914 | 1471 | 1915.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 1906_1914 | 1471 | 1864.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 1906_1914 | 0 | 155.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 1906_1914 | 1471 | 1915.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 1906_1914 | 0 | 155.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 1334_1426 | 1402 | 1846.0 | Domain | Fibronectin type-III |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 1464_1719 | 1402 | 1846.0 | Domain | Protein kinase |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 1809_1898 | 1402 | 1846.0 | Domain | Note=Ig-like C2-type 9 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 1464_1719 | 1471 | 1864.0 | Domain | Protein kinase |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 1809_1898 | 1471 | 1864.0 | Domain | Note=Ig-like C2-type 9 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 1464_1719 | 1471 | 1915.0 | Domain | Protein kinase |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 1809_1898 | 1471 | 1915.0 | Domain | Note=Ig-like C2-type 9 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 1464_1719 | 1471 | 1864.0 | Domain | Protein kinase |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 1809_1898 | 1471 | 1864.0 | Domain | Note=Ig-like C2-type 9 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 1098_1186 | 0 | 155.0 | Domain | Note=Ig-like C2-type 7 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 1238_1326 | 0 | 155.0 | Domain | Note=Ig-like C2-type 8 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 1334_1426 | 0 | 155.0 | Domain | Fibronectin type-III |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 1464_1719 | 0 | 155.0 | Domain | Protein kinase |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 161_249 | 0 | 155.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 1809_1898 | 0 | 155.0 | Domain | Note=Ig-like C2-type 9 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 33_122 | 0 | 155.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 414_503 | 0 | 155.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 514_599 | 0 | 155.0 | Domain | Note=Ig-like C2-type 4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 620_711 | 0 | 155.0 | Domain | Note=Ig-like C2-type 5 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 721_821 | 0 | 155.0 | Domain | Note=Ig-like C2-type 6 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 1464_1719 | 1471 | 1915.0 | Domain | Protein kinase |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 1809_1898 | 1471 | 1915.0 | Domain | Note=Ig-like C2-type 9 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 1098_1186 | 0 | 155.0 | Domain | Note=Ig-like C2-type 7 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 1238_1326 | 0 | 155.0 | Domain | Note=Ig-like C2-type 8 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 1334_1426 | 0 | 155.0 | Domain | Fibronectin type-III |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 1464_1719 | 0 | 155.0 | Domain | Protein kinase |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 161_249 | 0 | 155.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 1809_1898 | 0 | 155.0 | Domain | Note=Ig-like C2-type 9 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 33_122 | 0 | 155.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 414_503 | 0 | 155.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 514_599 | 0 | 155.0 | Domain | Note=Ig-like C2-type 4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 620_711 | 0 | 155.0 | Domain | Note=Ig-like C2-type 5 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 721_821 | 0 | 155.0 | Domain | Note=Ig-like C2-type 6 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 1470_1478 | 1402 | 1846.0 | Nucleotide binding | ATP |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 1470_1478 | 1471 | 1864.0 | Nucleotide binding | ATP |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 1470_1478 | 1471 | 1915.0 | Nucleotide binding | ATP |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 1470_1478 | 1471 | 1864.0 | Nucleotide binding | ATP |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 1470_1478 | 0 | 155.0 | Nucleotide binding | ATP |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 1470_1478 | 1471 | 1915.0 | Nucleotide binding | ATP |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 1470_1478 | 0 | 155.0 | Nucleotide binding | ATP |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 1061_1460 | 1402 | 1846.0 | Region | Actin-binding (calcium/calmodulin-insensitive) |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000346322 | - | 25 | 33 | 1711_1774 | 1402 | 1846.0 | Region | Note=Calmodulin-binding |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000359169 | - | 26 | 33 | 1711_1774 | 1471 | 1864.0 | Region | Note=Calmodulin-binding |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360304 | - | 26 | 34 | 1711_1774 | 1471 | 1915.0 | Region | Note=Calmodulin-binding |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000360772 | - | 27 | 34 | 1711_1774 | 1471 | 1864.0 | Region | Note=Calmodulin-binding |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 1061_1460 | 0 | 155.0 | Region | Actin-binding (calcium/calmodulin-insensitive) |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 1711_1774 | 0 | 155.0 | Region | Note=Calmodulin-binding |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 868_998 | 0 | 155.0 | Region | Note=5 X 28 AA approximate tandem repeats |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 923_963 | 0 | 155.0 | Region | Actin-binding (calcium/calmodulin-sensitive) |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 948_963 | 0 | 155.0 | Region | Calmodulin-binding |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 999_1063 | 0 | 155.0 | Region | Note=6 X 12 AA approximate tandem repeats |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000475616 | - | 23 | 31 | 1711_1774 | 1471 | 1915.0 | Region | Note=Calmodulin-binding |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 1061_1460 | 0 | 155.0 | Region | Actin-binding (calcium/calmodulin-insensitive) |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 1711_1774 | 0 | 155.0 | Region | Note=Calmodulin-binding |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 868_998 | 0 | 155.0 | Region | Note=5 X 28 AA approximate tandem repeats |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 923_963 | 0 | 155.0 | Region | Actin-binding (calcium/calmodulin-sensitive) |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 948_963 | 0 | 155.0 | Region | Calmodulin-binding |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 999_1063 | 0 | 155.0 | Region | Note=6 X 12 AA approximate tandem repeats |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 1004_1015 | 0 | 155.0 | Repeat | Note=2-2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 1016_1027 | 0 | 155.0 | Repeat | Note=2-3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 1028_1039 | 0 | 155.0 | Repeat | Note=2-4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 1040_1051 | 0 | 155.0 | Repeat | Note=2-5 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 1052_1063 | 0 | 155.0 | Repeat | Note=2-6 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 868_895 | 0 | 155.0 | Repeat | Note=1-1 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 896_923 | 0 | 155.0 | Repeat | Note=1-2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 924_951 | 0 | 155.0 | Repeat | Note=1-3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 952_979 | 0 | 155.0 | Repeat | Note=1-4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 980_998 | 0 | 155.0 | Repeat | Note=1-5%3B truncated |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000418370 | - | 1 | 3 | 999_1003 | 0 | 155.0 | Repeat | Note=2-1%3B truncated |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 1004_1015 | 0 | 155.0 | Repeat | Note=2-2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 1016_1027 | 0 | 155.0 | Repeat | Note=2-3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 1028_1039 | 0 | 155.0 | Repeat | Note=2-4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 1040_1051 | 0 | 155.0 | Repeat | Note=2-5 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 1052_1063 | 0 | 155.0 | Repeat | Note=2-6 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 868_895 | 0 | 155.0 | Repeat | Note=1-1 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 896_923 | 0 | 155.0 | Repeat | Note=1-2 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 924_951 | 0 | 155.0 | Repeat | Note=1-3 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 952_979 | 0 | 155.0 | Repeat | Note=1-4 |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 980_998 | 0 | 155.0 | Repeat | Note=1-5%3B truncated |
| Hgene | MYLK | chr3:123367818 | chr7:8790638 | ENST00000583087 | - | 1 | 4 | 999_1003 | 0 | 155.0 | Repeat | Note=2-1%3B truncated |
Top |
Fusion Gene Sequence for MYLK-NXPH1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >56525_56525_1_MYLK-NXPH1_MYLK_chr3_123367818_ENST00000346322_NXPH1_chr7_8790638_ENST00000405863_length(transcript)=6458nt_BP=4502nt GGTCGGCAGCAGGGCGCTGAGCGAGCTCGGAGCCCGCGCTGTGCGCCTGCGGCCGGGGCGCCCCGCCGAGCGCCGGTGCCCCGGCTCCCG GGCCGCCTTCGCCGCGCGGGAAGGATTCTTCAAAATTAACAGAAACCAATTCGGGCCAGCTGAAGAGAAAAAATAAAGGTGGCTCCCGGC TGCCTCTGCTGCAGTTCAGAGCAACTTCAGGAGCTTCCCAGCCGAGAGCTTCAGGACGCCTTTCCTGTCCCACTGGCCCAGTTGCCACAA CAAACAACAGAGAAGACGGTGACCATGGGGGATGTGAAGCTGGTTGCCTCGTCACACATTTCCAAAACCTCCCTCAGTGTGGATCCCTCA AGAGTTGACTCCATGCCCCTGACAGAGGCCCCTGCTTTCATTTTGCCCCCTCGGAACCTCTGCATCAAAGAAGGAGCCACCGCCAAGTTC GAAGGGCGGGTCCGGGGTTACCCAGAGCCCCAGGTGACATGGCACAGAAACGGGCAACCCATCACCAGCGGGGGCCGCTTCCTGCTGGAT TGCGGCATCCGGGGGACTTTCAGCCTTGTGATTCATGCTGTCCATGAGGAGGACAGGGGAAAGTATACCTGTGAAGCCACCAATGGCAGT GGTGCTCGCCAGGTGACAGTGGAGTTGACAGTAGAAGGAAGTTTTGCGAAGCAGCTTGGTCAGCCTGTTGTTTCCAAAACCTTAGGGGAT AGATTTTCAGCTCCAGCAGTGGAGACCCGTCCTAGCATCTGGGGGGAGTGCCCACCAAAGTTTGCTACCAAGCTGGGCCGAGTTGTGGTC AAAGAAGGACAGATGGGACGATTCTCCTGCAAGATCACTGGCCGGCCCCAACCGCAGGTCACCTGGCTCAAGGGAAATGTTCCACTGCAG CCGAGTGCCCGTGTGTCTGTGTCTGAGAAGAACGGCATGCAGGTTCTGGAAATCCATGGAGTCAACCAAGATGACGTGGGAGTGTACACG TGCCTGGTGGTGAACGGGTCGGGGAAGGCCTCGATGTCAGCTGAACTTTCCATCCAAGGTTTGGACAGTGCCAATAGGTCATTTGTGAGA GAAACAAAAGCCACCAATTCAGATGTCAGGAAAGAGGTGACCAATGTAATCTCAAAGGAGTCGAAGCTGGACAGTCTGGAGGCTGCAGCC AAAAGCAAGAACTGCTCCAGCCCCCAGAGAGGTGGCTCCCCACCCTGGGCTGCAAACAGCCAGCCTCAGCCCCCAAGGGAGTCCAAGCTG GAGTCATGCAAGGACTCGCCCAGAACGGCCCCGCAGACCCCGGTCCTTCAGAAGACTTCCAGCTCCATCACCCTGCAGGCCGCAAGAGTT CAGCCGGAACCAAGAGCACCAGGCCTGGGGGTCCTATCACCTTCTGGAGAAGAGAGGAAGAGGCCAGCTCCTCCCCGTCCAGCCACCTTC CCCACCAGGCAGCCTGGCCTGGGGAGCCAAGATGTTGTGAGCAAGGCTGCTAACAGGAGAATCCCCATGGAGGGCCAGAGGGATTCAGCA TTCCCCAAATTTGAGAGCAAGCCCCAAAGCCAGGAGGTCAAGGAAAATCAAACTGTCAAGTTCAGATGTGAAGGGCTTGCCGTGATGGAG GTGGCCCCCTCCTTCTCCAGTGTCCTGAAGGACTGCGCTGTTATTGAGGGCCAGGATTTTGTGCTGCAGTGCTCCGTACGGGGGACCCCA GTGCCCCGGATCACTTGGCTGCTGAATGGGCAGCCCATCCAGTACGCTCGCTCCACCTGCGAGGCCGGCGTGGCTGAGCTCCACATCCAG GATGCCCTGCCGGAGGACCATGGCACCTACACCTGCCTAGCTGAGAATGCCTTGGGGCAGGTGTCCTGCAGCGCCTGGGTCACCGTCCAT GAAAAGAAGAGTAGCAGGAAGAGTGAGTACCTTCTGCCTGTGGCTCCCAGCAAGCCCACTGCACCCATCTTCCTGCAGGGCCTCTCTGAT CTCAAAGTCATGGATGGAAGCCAGGTCACTATGACTGTCCAAGTGTCAGGGAATCCACCCCCTGAAGTCATCTGGCTGCACAATGGGAAT GAGATCCAAGAGTCAGAGGACTTCCACTTTGAACAGAGAGGAACTCAGCACAGCCTTTGTATCCAGGAAGTGTTCCCGGAGGACACGGGC ACGTACACCTGCGAGGCCTGGAACAGCGCTGGAGAGGTCCGCACCCAGGCCGTGCTCACGGTACAAGAGCCTCACGATGGCACCCAGCCC TGGTTCATCAGTAAGCCTCGCTCAGTGACAGCCTCCCTGGGCCAGAGTGTCCTCATCTCCTGCGCCATAGCTGGTGACCCCTTTCCTACC GTGCACTGGCTCAGAGATGGCAAAGCCCTCTGCAAAGACACTGGCCACTTCGAGGTGCTTCAGAATGAGGACGTGTTCACCCTGGTTCTA AAGAAGGTGCAGCCCTGGCATGCCGGCCAGTATGAGATCCTGCTCAAGAACCGGGTTGGCGAATGCAGTTGCCAGGTGTCACTGATGCTA CAGAACAGCTCTGCCAGAGCCCTTCCACGGGGGAGGGAGCCTGCCAGCTGCGAGGACCTCTGTGGTGGAGGAGTTGGTGCTGATGGTGGT GGTAGTGACCGCTATGGGTCCCTGAGGCCTGGCTGGCCAGCAAGAGGGCAGGGTTGGCTAGAGGAGGAAGACGGCGAGGACGTGCGAGGG GTGCTGAAGAGGCGCGTGGAGACGAGGCAGCACACTGAGGAGGCGATCCGCCAGCAGGAGGTGGAGCAGCTGGACTTCCGAGACCTCCTG GGGAAGAAGGTGAGTACAAAGACCCTATCGGAAGACGACCTGAAGGAGATCCCAGCCGAGCAGATGGATTTCCGTGCCAACCTGCAGCGG CAAGTGAAGCCAAAGACTGTGTCTGAGGAAGAGAGGAAGGTGCACAGCCCCCAGCAGGTCGATTTTCGCTCTGTCCTGGCCAAGAAGGGG ACTTCCAAGACCCCCGTGCCTGAGAAGGTGCCACCGCCAAAACCTGCCACCCCGGATTTTCGCTCAGTGCTGGGTGGCAAGAAGAAATTA CCAGCAGAGAATGGCAGCAGCAGTGCCGAGACCCTGAATGCCAAGGCAGTGGAGAGTTCCAAGCCCCTGAGCAATGCACAGCCTTCAGGG CCCTTGAAACCCGTGGGCAACGCCAAGCCTGCTGAGACCCTGAAGCCAATGGGCAACGCCAAGCCTGCCGAGACCCTGAAGCCCATGGGC AATGCCAAGCCTGATGAGAACCTGAAATCCGCTAGCAAAGAAGAACTCAAGAAAGACGTTAAGAATGATGTGAACTGCAAGAGAGGCCAT GCAGGGACCACAGATAATGAAAAGAGATCAGAGAGCCAGGGGACAGCCCCAGCCTTCAAGCAGAAGCTGCAAGATGTTCATGTGGCAGAG GGCAAGAAGCTGCTGCTCCAGTGCCAGGTGTCTTCTGACCCCCCAGCCACCATCATCTGGACGCTGAACGGAAAGACCCTCAAGACCACC AAGTTCATCATCCTCTCCCAGGAAGGCTCACTCTGCTCCGTCTCCATCGAGAAGGCACTGCCTGAGGACAGAGGCTTATACAAGTGTGTA GCCAAGAATGACGCTGGCCAGGCGGAGTGCTCCTGCCAAGTCACCGTGGATGATGCTCCAGCCAGTGAGAACACCAAGGCCCCAGAGATG AAATCCCGGAGGCCCAAGAGCTCTCTTCCTCCCGTGCTAGGAACTGAGAGTGATGCGACTGTGAAAAAGAAACCTGCCCCCAAGACACCT CCGAAGGCAGCAATGCCCCCTCAGATCATCCAGTTCCCTGAGGACCAGAAGGTACGCGCAGGAGAGTCAGTGGAGCTGTTTGGCAAAGTG ACAGGCACTCAGCCCATCACCTGTACCTGGATGAAGTTCCGAAAGCAGATCCAGGAAAGCGAGCACATGAAGGTGGAGAACAGCGAGAAT GGCAGCAAGCTCACCATCCTGGCCGCGCGCCAGGAGCACTGCGGCTGCTACACACTGCTGGTGGAGAACAAGCTGGGCAGCAGGCAGGCC CAGGTCAACCTCACTGTCGTGGATAAGCCAGACCCCCCAGCTGGCACACCTTGTGCCTCTGACATTCGGAGCTCCTCACTGACCCTGTCC TGGTATGGCTCCTCATATGATGGGGGCAGTGCTGTACAGTCCTACAGCATCGAGATCTGGGACTCAGCCAACAAGACGTGGAAGGAACTA GCCACATGCCGCAGCACCTCTTTCAACGTCCAGGACCTGCTGCCTGACCACGAATATAAGTTCCGTGTACGTGCAATCAACGTGTATGGA ACCAGTGAGCCAAGCCAGGAGTCTGAACTCACAACGGTAGGAGAGAAACCTGAAGAGCCGAAGGATGAAGTGGAGGTGTCAGATGATGAT GAGAAGGAGCCCGAGGTTGATTACCGGACAGTGACAATCAATACTGAACAAAAAGTATCTGACTTCTACGACATTGAGGAGAGATTAGGA TCGTCACATGTGCCAATTTAACGAACGGTGGAAAGTCAGAACTTCTGAAATCAGGAAGCAGCAAATCCACACTAAAGCACATATGGACAG AAAGCAGCAAAGACTTGTCTATCAGCCGACTCCTGTCACAGACTTTTCGTGGCAAAGAGAATGATACAGATTTGGACCTGAGATATGACA CCCCAGAACCTTATTCTGAGCAAGACCTCTGGGACTGGCTGAGGAACTCCACAGACCTTCAAGAGCCTCGGCCCAGGGCCAAGAGAAGGC CCATTGTTAAAACGGGCAAGTTTAAGAAAATGTTTGGATGGGGCGATTTTCATTCCAACATCAAAACAGTGAAGCTGAACCTGTTGATAA CTGGGAAAATTGTAGATCATGGCAATGGGACATTTAGTGTTTATTTCAGGCATAATTCAACTGGTCAAGGGAATGTATCTGTCAGCTTGG TACCCCCTACAAAAATCGTGGAATTTGACTTGGCACAACAAACCGTGATTGATGCCAAAGATTCCAAGTCTTTTAATTGTCGCATTGAAT ATGAAAAGGTTGACAAGGCTACCAAGAACACACTCTGCAACTATGACCCTTCAAAAACCTGTTACCAGGAGCAAACCCAAAGTCATGTAT CCTGGCTCTGCTCCAAGCCCTTTAAGGTGATCTGTATTTACATTTCCTTTTATAGTACAGATTATAAACTGGTACAGAAAGTGTGCCCTG ACTACAACTACCACAGTGACACACCTTACTTTCCCTCGGGATGAAGGTGAACATGGGGGTGAGACTGAAGCCTGAGGAATTAAAGGTCAT ATGACAGGGCTGTTACCTCAAAGAAGAAGGTCACATCTGTTGCCTGGAATGTGTCTACACTGCTGCTCTTGTCAACTGGCTGCAAAATAC ACTAGTGGAAAACACTCTGATGTAATTTCTGCCCAGTCAGCTTCATCCCTCAGTATAATTGTAAATCATCACAGATTTTGAATTCACACC TGAAGACATGCTCTCACATATAGAGGTACACAAACACACCGTCATGCACATTTCAGCTTGCGTCTATCATGATTCCTGTTGAGAGGGCTT TCATTGTCTGACTCATAATGGTTCAGGATCAACTATCATCAAACGGAAGGATTAACTAGACAGAGAATGTTTCTAACAGTTGCTGTTATG GAAATCTCTTTTAAAGTCTTGAGTACATGCTAATCAATAATCTCCACTCATGCATTCCTACTGCTTGGAGTAGCTGTACTGGTAAATACT ACTGTAGGAGTATCTGCTTGTTAAAATGGAAAAATGTGTCTTTAGAGCTCAGTATTCTTTATTTTACAAACACAACAAAATGTAGTAACT TTTTTCCAGCATACAGTAGGCACATTCAAAGTGGTCCAAGATGGCTCTTTTTTCTTTGAAAGGGGCCTGTTCTCAGTAAAGATGAGCAAA CATTTGGAATTTACATGTGGGCAGACATTGGGATAACAACTTTCATCACCAATCATTGGACTTTTGTGAAGTTGACACCAGCTAAGGCTG CTTAAAATAAGTTCTGATCATTATATAAGAAGGGAAATGCCTGGCAGACACCATGTAAGTTATAAGTGTCTGTCTTATCTTTACTACACA TATTGTAACAAATTCAATATCCTAGTCTTCATTTGTATGAATGGTTTGTATTGTACATAGTTTAACCAAGTGTTATTTGAGCTGCTTATT AATATTAACTTGTACTTGTCTCTCTGCTTGTTATTGGTTAAGAAAAAAGGATATGAGGAATTCATTTTATCAATGTAGCTGTGAAGGCCA TTAAAAAGACAAACTTAATGTACAGAGCATTTATTCAGATCAAGTATTGTTGAAAGCTATACATATACAACATTACAGTCTGTCTGTATT >56525_56525_1_MYLK-NXPH1_MYLK_chr3_123367818_ENST00000346322_NXPH1_chr7_8790638_ENST00000405863_length(amino acids)=1422AA_BP= MAQLPQQTTEKTVTMGDVKLVASSHISKTSLSVDPSRVDSMPLTEAPAFILPPRNLCIKEGATAKFEGRVRGYPEPQVTWHRNGQPITSG GRFLLDCGIRGTFSLVIHAVHEEDRGKYTCEATNGSGARQVTVELTVEGSFAKQLGQPVVSKTLGDRFSAPAVETRPSIWGECPPKFATK LGRVVVKEGQMGRFSCKITGRPQPQVTWLKGNVPLQPSARVSVSEKNGMQVLEIHGVNQDDVGVYTCLVVNGSGKASMSAELSIQGLDSA NRSFVRETKATNSDVRKEVTNVISKESKLDSLEAAAKSKNCSSPQRGGSPPWAANSQPQPPRESKLESCKDSPRTAPQTPVLQKTSSSIT LQAARVQPEPRAPGLGVLSPSGEERKRPAPPRPATFPTRQPGLGSQDVVSKAANRRIPMEGQRDSAFPKFESKPQSQEVKENQTVKFRCE GLAVMEVAPSFSSVLKDCAVIEGQDFVLQCSVRGTPVPRITWLLNGQPIQYARSTCEAGVAELHIQDALPEDHGTYTCLAENALGQVSCS AWVTVHEKKSSRKSEYLLPVAPSKPTAPIFLQGLSDLKVMDGSQVTMTVQVSGNPPPEVIWLHNGNEIQESEDFHFEQRGTQHSLCIQEV FPEDTGTYTCEAWNSAGEVRTQAVLTVQEPHDGTQPWFISKPRSVTASLGQSVLISCAIAGDPFPTVHWLRDGKALCKDTGHFEVLQNED VFTLVLKKVQPWHAGQYEILLKNRVGECSCQVSLMLQNSSARALPRGREPASCEDLCGGGVGADGGGSDRYGSLRPGWPARGQGWLEEED GEDVRGVLKRRVETRQHTEEAIRQQEVEQLDFRDLLGKKVSTKTLSEDDLKEIPAEQMDFRANLQRQVKPKTVSEEERKVHSPQQVDFRS VLAKKGTSKTPVPEKVPPPKPATPDFRSVLGGKKKLPAENGSSSAETLNAKAVESSKPLSNAQPSGPLKPVGNAKPAETLKPMGNAKPAE TLKPMGNAKPDENLKSASKEELKKDVKNDVNCKRGHAGTTDNEKRSESQGTAPAFKQKLQDVHVAEGKKLLLQCQVSSDPPATIIWTLNG KTLKTTKFIILSQEGSLCSVSIEKALPEDRGLYKCVAKNDAGQAECSCQVTVDDAPASENTKAPEMKSRRPKSSLPPVLGTESDATVKKK PAPKTPPKAAMPPQIIQFPEDQKVRAGESVELFGKVTGTQPITCTWMKFRKQIQESEHMKVENSENGSKLTILAARQEHCGCYTLLVENK LGSRQAQVNLTVVDKPDPPAGTPCASDIRSSSLTLSWYGSSYDGGSAVQSYSIEIWDSANKTWKELATCRSTSFNVQDLLPDHEYKFRVR -------------------------------------------------------------- >56525_56525_2_MYLK-NXPH1_MYLK_chr3_123367818_ENST00000359169_NXPH1_chr7_8790638_ENST00000405863_length(transcript)=6653nt_BP=4697nt GGCGCTGAGCGAGCTCGGAGCCCGCGCTGTGCGCCTGCGGCCGGGGCGCCCCGCCGAGCGCCGGTGCCCCGGCTCCCGGGCCGCCTTCGC CGCGCGGGAAGGATTCTTCAAAATTAACAGAAACCAATTCGGGCCAGCTGAAGAGAAAAAATAAAGGTGGCTCCCGGCTGCCTCTGCTGC AGTTCAGAGCAACTTCAGGAGCTTCCCAGCCGAGAGCTTCAGGACGCCTTTCCTGTCCCACTGGCCCAGTTGCCACAACAAACAACAGAG AAGACGGTGACCATGGGGGATGTGAAGCTGGTTGCCTCGTCACACATTTCCAAAACCTCCCTCAGTGTGGATCCCTCAAGAGTTGACTCC ATGCCCCTGACAGAGGCCCCTGCTTTCATTTTGCCCCCTCGGAACCTCTGCATCAAAGAAGGAGCCACCGCCAAGTTCGAAGGGCGGGTC CGGGGTTACCCAGAGCCCCAGGTGACATGGCACAGAAACGGGCAACCCATCACCAGCGGGGGCCGCTTCCTGCTGGATTGCGGCATCCGG GGGACTTTCAGCCTTGTGATTCATGCTGTCCATGAGGAGGACAGGGGAAAGTATACCTGTGAAGCCACCAATGGCAGTGGTGCTCGCCAG GTGACAGTGGAGTTGACAGTAGAAGGAAGTTTTGCGAAGCAGCTTGGTCAGCCTGTTGTTTCCAAAACCTTAGGGGATAGATTTTCAGCT CCAGCAGTGGAGACCCGTCCTAGCATCTGGGGGGAGTGCCCACCAAAGTTTGCTACCAAGCTGGGCCGAGTTGTGGTCAAAGAAGGACAG ATGGGACGATTCTCCTGCAAGATCACTGGCCGGCCCCAACCGCAGGTCACCTGGCTCAAGGGAAATGTTCCACTGCAGCCGAGTGCCCGT GTGTCTGTGTCTGAGAAGAACGGCATGCAGGTTCTGGAAATCCATGGAGTCAACCAAGATGACGTGGGAGTGTACACGTGCCTGGTGGTG AACGGGTCGGGGAAGGCCTCGATGTCAGCTGAACTTTCCATCCAAGGTTTGGACAGTGCCAATAGGTCATTTGTGAGAGAAACAAAAGCC ACCAATTCAGATGTCAGGAAAGAGGTGACCAATGTAATCTCAAAGGAGTCGAAGCTGGACAGTCTGGAGGCTGCAGCCAAAAGCAAGAAC TGCTCCAGCCCCCAGAGAGGTGGCTCCCCACCCTGGGCTGCAAACAGCCAGCCTCAGCCCCCAAGGGAGTCCAAGCTGGAGTCATGCAAG GACTCGCCCAGAACGGCCCCGCAGACCCCGGTCCTTCAGAAGACTTCCAGCTCCATCACCCTGCAGGCCGCAAGAGTTCAGCCGGAACCA AGAGCACCAGGCCTGGGGGTCCTATCACCTTCTGGAGAAGAGAGGAAGAGGCCAGCTCCTCCCCGTCCAGCCACCTTCCCCACCAGGCAG CCTGGCCTGGGGAGCCAAGATGTTGTGAGCAAGGCTGCTAACAGGAGAATCCCCATGGAGGGCCAGAGGGATTCAGCATTCCCCAAATTT GAGAGCAAGCCCCAAAGCCAGGAGGTCAAGGAAAATCAAACTGTCAAGTTCAGATGTGAAGTTTCCGGGATTCCAAAGCCTGAAGTGGCC TGGTTCCTGGAAGGCACCCCCGTGAGGAGACAGGAAGGCAGCATTGAGGTTTATGAAGATGCTGGCTCCCATTACCTCTGCCTGCTGAAA GCCCGGACCAGGGACAGTGGGACATACAGCTGCACTGCTTCCAACGCCCAAGGCCAGCTGTCCTGTAGCTGGACCCTCCAAGTGGAAAGG CTTGCCGTGATGGAGGTGGCCCCCTCCTTCTCCAGTGTCCTGAAGGACTGCGCTGTTATTGAGGGCCAGGATTTTGTGCTGCAGTGCTCC GTACGGGGGACCCCAGTGCCCCGGATCACTTGGCTGCTGAATGGGCAGCCCATCCAGTACGCTCGCTCCACCTGCGAGGCCGGCGTGGCT GAGCTCCACATCCAGGATGCCCTGCCGGAGGACCATGGCACCTACACCTGCCTAGCTGAGAATGCCTTGGGGCAGGTGTCCTGCAGCGCC TGGGTCACCGTCCATGAAAAGAAGAGTAGCAGGAAGAGTGAGTACCTTCTGCCTGTGGCTCCCAGCAAGCCCACTGCACCCATCTTCCTG CAGGGCCTCTCTGATCTCAAAGTCATGGATGGAAGCCAGGTCACTATGACTGTCCAAGTGTCAGGGAATCCACCCCCTGAAGTCATCTGG CTGCACAATGGGAATGAGATCCAAGAGTCAGAGGACTTCCACTTTGAACAGAGAGGAACTCAGCACAGCCTTTGTATCCAGGAAGTGTTC CCGGAGGACACGGGCACGTACACCTGCGAGGCCTGGAACAGCGCTGGAGAGGTCCGCACCCAGGCCGTGCTCACGGTACAAGAGCCTCAC GATGGCACCCAGCCCTGGTTCATCAGTAAGCCTCGCTCAGTGACAGCCTCCCTGGGCCAGAGTGTCCTCATCTCCTGCGCCATAGCTGGT GACCCCTTTCCTACCGTGCACTGGCTCAGAGATGGCAAAGCCCTCTGCAAAGACACTGGCCACTTCGAGGTGCTTCAGAATGAGGACGTG TTCACCCTGGTTCTAAAGAAGGTGCAGCCCTGGCATGCCGGCCAGTATGAGATCCTGCTCAAGAACCGGGTTGGCGAATGCAGTTGCCAG GTGTCACTGATGCTACAGAACAGCTCTGCCAGAGCCCTTCCACGGGGGAGGGAGCCTGCCAGCTGCGAGGACCTCTGTGGTGGAGGAGTT GGTGCTGATGGTGGTGGTAGTGACCGCTATGGGTCCCTGAGGCCTGGCTGGCCAGCAAGAGGGCAGGGTTGGCTAGAGGAGGAAGACGGC GAGGACGTGCGAGGGGTGCTGAAGAGGCGCGTGGAGACGAGGCAGCACACTGAGGAGGCGATCCGCCAGCAGGAGGTGGAGCAGCTGGAC TTCCGAGACCTCCTGGGGAAGAAGGTGAGTACAAAGACCCTATCGGAAGACGACCTGAAGGAGATCCCAGCCGAGCAGATGGATTTCCGT GCCAACCTGCAGCGGCAAGTGAAGCCAAAGACTGTGTCTGAGGAAGAGAGGAAGGTGCACAGCCCCCAGCAGGTCGATTTTCGCTCTGTC CTGGCCAAGAAGGGGACTTCCAAGACCCCCGTGCCTGAGAAGGTGCCACCGCCAAAACCTGCCACCCCGGATTTTCGCTCAGTGCTGGGT GGCAAGAAGAAATTACCAGCAGAGAATGGCAGCAGCAGTGCCGAGACCCTGAATGCCAAGGCAGTGGAGAGTTCCAAGCCCCTGAGCAAT GCACAGCCTTCAGGGCCCTTGAAACCCGTGGGCAACGCCAAGCCTGCTGAGACCCTGAAGCCAATGGGCAACGCCAAGCCTGCCGAGACC CTGAAGCCCATGGGCAATGCCAAGCCTGATGAGAACCTGAAATCCGCTAGCAAAGAAGAACTCAAGAAAGACGTTAAGAATGATGTGAAC TGCAAGAGAGGCCATGCAGGGACCACAGATAATGAAAAGAGATCAGAGAGCCAGGGGACAGCCCCAGCCTTCAAGCAGAAGCTGCAAGAT GTTCATGTGGCAGAGGGCAAGAAGCTGCTGCTCCAGTGCCAGGTGTCTTCTGACCCCCCAGCCACCATCATCTGGACGCTGAACGGAAAG ACCCTCAAGACCACCAAGTTCATCATCCTCTCCCAGGAAGGCTCACTCTGCTCCGTCTCCATCGAGAAGGCACTGCCTGAGGACAGAGGC TTATACAAGTGTGTAGCCAAGAATGACGCTGGCCAGGCGGAGTGCTCCTGCCAAGTCACCGTGGATGATGCTCCAGCCAGTGAGAACACC AAGGCCCCAGAGATGAAATCCCGGAGGCCCAAGAGCTCTCTTCCTCCCGTGCTAGGAACTGAGAGTGATGCGACTGTGAAAAAGAAACCT GCCCCCAAGACACCTCCGAAGGCAGCAATGCCCCCTCAGATCATCCAGTTCCCTGAGGACCAGAAGGTACGCGCAGGAGAGTCAGTGGAG CTGTTTGGCAAAGTGACAGGCACTCAGCCCATCACCTGTACCTGGATGAAGTTCCGAAAGCAGATCCAGGAAAGCGAGCACATGAAGGTG GAGAACAGCGAGAATGGCAGCAAGCTCACCATCCTGGCCGCGCGCCAGGAGCACTGCGGCTGCTACACACTGCTGGTGGAGAACAAGCTG GGCAGCAGGCAGGCCCAGGTCAACCTCACTGTCGTGGATAAGCCAGACCCCCCAGCTGGCACACCTTGTGCCTCTGACATTCGGAGCTCC TCACTGACCCTGTCCTGGTATGGCTCCTCATATGATGGGGGCAGTGCTGTACAGTCCTACAGCATCGAGATCTGGGACTCAGCCAACAAG ACGTGGAAGGAACTAGCCACATGCCGCAGCACCTCTTTCAACGTCCAGGACCTGCTGCCTGACCACGAATATAAGTTCCGTGTACGTGCA ATCAACGTGTATGGAACCAGTGAGCCAAGCCAGGAGTCTGAACTCACAACGGTAGGAGAGAAACCTGAAGAGCCGAAGGATGAAGTGGAG GTGTCAGATGATGATGAGAAGGAGCCCGAGGTTGATTACCGGACAGTGACAATCAATACTGAACAAAAAGTATCTGACTTCTACGACATT GAGGAGAGATTAGGATCGTCACATGTGCCAATTTAACGAACGGTGGAAAGTCAGAACTTCTGAAATCAGGAAGCAGCAAATCCACACTAA AGCACATATGGACAGAAAGCAGCAAAGACTTGTCTATCAGCCGACTCCTGTCACAGACTTTTCGTGGCAAAGAGAATGATACAGATTTGG ACCTGAGATATGACACCCCAGAACCTTATTCTGAGCAAGACCTCTGGGACTGGCTGAGGAACTCCACAGACCTTCAAGAGCCTCGGCCCA GGGCCAAGAGAAGGCCCATTGTTAAAACGGGCAAGTTTAAGAAAATGTTTGGATGGGGCGATTTTCATTCCAACATCAAAACAGTGAAGC TGAACCTGTTGATAACTGGGAAAATTGTAGATCATGGCAATGGGACATTTAGTGTTTATTTCAGGCATAATTCAACTGGTCAAGGGAATG TATCTGTCAGCTTGGTACCCCCTACAAAAATCGTGGAATTTGACTTGGCACAACAAACCGTGATTGATGCCAAAGATTCCAAGTCTTTTA ATTGTCGCATTGAATATGAAAAGGTTGACAAGGCTACCAAGAACACACTCTGCAACTATGACCCTTCAAAAACCTGTTACCAGGAGCAAA CCCAAAGTCATGTATCCTGGCTCTGCTCCAAGCCCTTTAAGGTGATCTGTATTTACATTTCCTTTTATAGTACAGATTATAAACTGGTAC AGAAAGTGTGCCCTGACTACAACTACCACAGTGACACACCTTACTTTCCCTCGGGATGAAGGTGAACATGGGGGTGAGACTGAAGCCTGA GGAATTAAAGGTCATATGACAGGGCTGTTACCTCAAAGAAGAAGGTCACATCTGTTGCCTGGAATGTGTCTACACTGCTGCTCTTGTCAA CTGGCTGCAAAATACACTAGTGGAAAACACTCTGATGTAATTTCTGCCCAGTCAGCTTCATCCCTCAGTATAATTGTAAATCATCACAGA TTTTGAATTCACACCTGAAGACATGCTCTCACATATAGAGGTACACAAACACACCGTCATGCACATTTCAGCTTGCGTCTATCATGATTC CTGTTGAGAGGGCTTTCATTGTCTGACTCATAATGGTTCAGGATCAACTATCATCAAACGGAAGGATTAACTAGACAGAGAATGTTTCTA ACAGTTGCTGTTATGGAAATCTCTTTTAAAGTCTTGAGTACATGCTAATCAATAATCTCCACTCATGCATTCCTACTGCTTGGAGTAGCT GTACTGGTAAATACTACTGTAGGAGTATCTGCTTGTTAAAATGGAAAAATGTGTCTTTAGAGCTCAGTATTCTTTATTTTACAAACACAA CAAAATGTAGTAACTTTTTTCCAGCATACAGTAGGCACATTCAAAGTGGTCCAAGATGGCTCTTTTTTCTTTGAAAGGGGCCTGTTCTCA GTAAAGATGAGCAAACATTTGGAATTTACATGTGGGCAGACATTGGGATAACAACTTTCATCACCAATCATTGGACTTTTGTGAAGTTGA CACCAGCTAAGGCTGCTTAAAATAAGTTCTGATCATTATATAAGAAGGGAAATGCCTGGCAGACACCATGTAAGTTATAAGTGTCTGTCT TATCTTTACTACACATATTGTAACAAATTCAATATCCTAGTCTTCATTTGTATGAATGGTTTGTATTGTACATAGTTTAACCAAGTGTTA TTTGAGCTGCTTATTAATATTAACTTGTACTTGTCTCTCTGCTTGTTATTGGTTAAGAAAAAAGGATATGAGGAATTCATTTTATCAATG TAGCTGTGAAGGCCATTAAAAAGACAAACTTAATGTACAGAGCATTTATTCAGATCAAGTATTGTTGAAAGCTATACATATACAACATTA >56525_56525_2_MYLK-NXPH1_MYLK_chr3_123367818_ENST00000359169_NXPH1_chr7_8790638_ENST00000405863_length(amino acids)=1491AA_BP= MAQLPQQTTEKTVTMGDVKLVASSHISKTSLSVDPSRVDSMPLTEAPAFILPPRNLCIKEGATAKFEGRVRGYPEPQVTWHRNGQPITSG GRFLLDCGIRGTFSLVIHAVHEEDRGKYTCEATNGSGARQVTVELTVEGSFAKQLGQPVVSKTLGDRFSAPAVETRPSIWGECPPKFATK LGRVVVKEGQMGRFSCKITGRPQPQVTWLKGNVPLQPSARVSVSEKNGMQVLEIHGVNQDDVGVYTCLVVNGSGKASMSAELSIQGLDSA NRSFVRETKATNSDVRKEVTNVISKESKLDSLEAAAKSKNCSSPQRGGSPPWAANSQPQPPRESKLESCKDSPRTAPQTPVLQKTSSSIT LQAARVQPEPRAPGLGVLSPSGEERKRPAPPRPATFPTRQPGLGSQDVVSKAANRRIPMEGQRDSAFPKFESKPQSQEVKENQTVKFRCE VSGIPKPEVAWFLEGTPVRRQEGSIEVYEDAGSHYLCLLKARTRDSGTYSCTASNAQGQLSCSWTLQVERLAVMEVAPSFSSVLKDCAVI EGQDFVLQCSVRGTPVPRITWLLNGQPIQYARSTCEAGVAELHIQDALPEDHGTYTCLAENALGQVSCSAWVTVHEKKSSRKSEYLLPVA PSKPTAPIFLQGLSDLKVMDGSQVTMTVQVSGNPPPEVIWLHNGNEIQESEDFHFEQRGTQHSLCIQEVFPEDTGTYTCEAWNSAGEVRT QAVLTVQEPHDGTQPWFISKPRSVTASLGQSVLISCAIAGDPFPTVHWLRDGKALCKDTGHFEVLQNEDVFTLVLKKVQPWHAGQYEILL KNRVGECSCQVSLMLQNSSARALPRGREPASCEDLCGGGVGADGGGSDRYGSLRPGWPARGQGWLEEEDGEDVRGVLKRRVETRQHTEEA IRQQEVEQLDFRDLLGKKVSTKTLSEDDLKEIPAEQMDFRANLQRQVKPKTVSEEERKVHSPQQVDFRSVLAKKGTSKTPVPEKVPPPKP ATPDFRSVLGGKKKLPAENGSSSAETLNAKAVESSKPLSNAQPSGPLKPVGNAKPAETLKPMGNAKPAETLKPMGNAKPDENLKSASKEE LKKDVKNDVNCKRGHAGTTDNEKRSESQGTAPAFKQKLQDVHVAEGKKLLLQCQVSSDPPATIIWTLNGKTLKTTKFIILSQEGSLCSVS IEKALPEDRGLYKCVAKNDAGQAECSCQVTVDDAPASENTKAPEMKSRRPKSSLPPVLGTESDATVKKKPAPKTPPKAAMPPQIIQFPED QKVRAGESVELFGKVTGTQPITCTWMKFRKQIQESEHMKVENSENGSKLTILAARQEHCGCYTLLVENKLGSRQAQVNLTVVDKPDPPAG TPCASDIRSSSLTLSWYGSSYDGGSAVQSYSIEIWDSANKTWKELATCRSTSFNVQDLLPDHEYKFRVRAINVYGTSEPSQESELTTVGE -------------------------------------------------------------- >56525_56525_3_MYLK-NXPH1_MYLK_chr3_123367818_ENST00000360304_NXPH1_chr7_8790638_ENST00000405863_length(transcript)=6653nt_BP=4697nt GGCGCTGAGCGAGCTCGGAGCCCGCGCTGTGCGCCTGCGGCCGGGGCGCCCCGCCGAGCGCCGGTGCCCCGGCTCCCGGGCCGCCTTCGC CGCGCGGGAAGGATTCTTCAAAATTAACAGAAACCAATTCGGGCCAGCTGAAGAGAAAAAATAAAGGTGGCTCCCGGCTGCCTCTGCTGC AGTTCAGAGCAACTTCAGGAGCTTCCCAGCCGAGAGCTTCAGGACGCCTTTCCTGTCCCACTGGCCCAGTTGCCACAACAAACAACAGAG AAGACGGTGACCATGGGGGATGTGAAGCTGGTTGCCTCGTCACACATTTCCAAAACCTCCCTCAGTGTGGATCCCTCAAGAGTTGACTCC ATGCCCCTGACAGAGGCCCCTGCTTTCATTTTGCCCCCTCGGAACCTCTGCATCAAAGAAGGAGCCACCGCCAAGTTCGAAGGGCGGGTC CGGGGTTACCCAGAGCCCCAGGTGACATGGCACAGAAACGGGCAACCCATCACCAGCGGGGGCCGCTTCCTGCTGGATTGCGGCATCCGG GGGACTTTCAGCCTTGTGATTCATGCTGTCCATGAGGAGGACAGGGGAAAGTATACCTGTGAAGCCACCAATGGCAGTGGTGCTCGCCAG GTGACAGTGGAGTTGACAGTAGAAGGAAGTTTTGCGAAGCAGCTTGGTCAGCCTGTTGTTTCCAAAACCTTAGGGGATAGATTTTCAGCT CCAGCAGTGGAGACCCGTCCTAGCATCTGGGGGGAGTGCCCACCAAAGTTTGCTACCAAGCTGGGCCGAGTTGTGGTCAAAGAAGGACAG ATGGGACGATTCTCCTGCAAGATCACTGGCCGGCCCCAACCGCAGGTCACCTGGCTCAAGGGAAATGTTCCACTGCAGCCGAGTGCCCGT GTGTCTGTGTCTGAGAAGAACGGCATGCAGGTTCTGGAAATCCATGGAGTCAACCAAGATGACGTGGGAGTGTACACGTGCCTGGTGGTG AACGGGTCGGGGAAGGCCTCGATGTCAGCTGAACTTTCCATCCAAGGTTTGGACAGTGCCAATAGGTCATTTGTGAGAGAAACAAAAGCC ACCAATTCAGATGTCAGGAAAGAGGTGACCAATGTAATCTCAAAGGAGTCGAAGCTGGACAGTCTGGAGGCTGCAGCCAAAAGCAAGAAC TGCTCCAGCCCCCAGAGAGGTGGCTCCCCACCCTGGGCTGCAAACAGCCAGCCTCAGCCCCCAAGGGAGTCCAAGCTGGAGTCATGCAAG GACTCGCCCAGAACGGCCCCGCAGACCCCGGTCCTTCAGAAGACTTCCAGCTCCATCACCCTGCAGGCCGCAAGAGTTCAGCCGGAACCA AGAGCACCAGGCCTGGGGGTCCTATCACCTTCTGGAGAAGAGAGGAAGAGGCCAGCTCCTCCCCGTCCAGCCACCTTCCCCACCAGGCAG CCTGGCCTGGGGAGCCAAGATGTTGTGAGCAAGGCTGCTAACAGGAGAATCCCCATGGAGGGCCAGAGGGATTCAGCATTCCCCAAATTT GAGAGCAAGCCCCAAAGCCAGGAGGTCAAGGAAAATCAAACTGTCAAGTTCAGATGTGAAGTTTCCGGGATTCCAAAGCCTGAAGTGGCC TGGTTCCTGGAAGGCACCCCCGTGAGGAGACAGGAAGGCAGCATTGAGGTTTATGAAGATGCTGGCTCCCATTACCTCTGCCTGCTGAAA GCCCGGACCAGGGACAGTGGGACATACAGCTGCACTGCTTCCAACGCCCAAGGCCAGCTGTCCTGTAGCTGGACCCTCCAAGTGGAAAGG CTTGCCGTGATGGAGGTGGCCCCCTCCTTCTCCAGTGTCCTGAAGGACTGCGCTGTTATTGAGGGCCAGGATTTTGTGCTGCAGTGCTCC GTACGGGGGACCCCAGTGCCCCGGATCACTTGGCTGCTGAATGGGCAGCCCATCCAGTACGCTCGCTCCACCTGCGAGGCCGGCGTGGCT GAGCTCCACATCCAGGATGCCCTGCCGGAGGACCATGGCACCTACACCTGCCTAGCTGAGAATGCCTTGGGGCAGGTGTCCTGCAGCGCC TGGGTCACCGTCCATGAAAAGAAGAGTAGCAGGAAGAGTGAGTACCTTCTGCCTGTGGCTCCCAGCAAGCCCACTGCACCCATCTTCCTG CAGGGCCTCTCTGATCTCAAAGTCATGGATGGAAGCCAGGTCACTATGACTGTCCAAGTGTCAGGGAATCCACCCCCTGAAGTCATCTGG CTGCACAATGGGAATGAGATCCAAGAGTCAGAGGACTTCCACTTTGAACAGAGAGGAACTCAGCACAGCCTTTGTATCCAGGAAGTGTTC CCGGAGGACACGGGCACGTACACCTGCGAGGCCTGGAACAGCGCTGGAGAGGTCCGCACCCAGGCCGTGCTCACGGTACAAGAGCCTCAC GATGGCACCCAGCCCTGGTTCATCAGTAAGCCTCGCTCAGTGACAGCCTCCCTGGGCCAGAGTGTCCTCATCTCCTGCGCCATAGCTGGT GACCCCTTTCCTACCGTGCACTGGCTCAGAGATGGCAAAGCCCTCTGCAAAGACACTGGCCACTTCGAGGTGCTTCAGAATGAGGACGTG TTCACCCTGGTTCTAAAGAAGGTGCAGCCCTGGCATGCCGGCCAGTATGAGATCCTGCTCAAGAACCGGGTTGGCGAATGCAGTTGCCAG GTGTCACTGATGCTACAGAACAGCTCTGCCAGAGCCCTTCCACGGGGGAGGGAGCCTGCCAGCTGCGAGGACCTCTGTGGTGGAGGAGTT GGTGCTGATGGTGGTGGTAGTGACCGCTATGGGTCCCTGAGGCCTGGCTGGCCAGCAAGAGGGCAGGGTTGGCTAGAGGAGGAAGACGGC GAGGACGTGCGAGGGGTGCTGAAGAGGCGCGTGGAGACGAGGCAGCACACTGAGGAGGCGATCCGCCAGCAGGAGGTGGAGCAGCTGGAC TTCCGAGACCTCCTGGGGAAGAAGGTGAGTACAAAGACCCTATCGGAAGACGACCTGAAGGAGATCCCAGCCGAGCAGATGGATTTCCGT GCCAACCTGCAGCGGCAAGTGAAGCCAAAGACTGTGTCTGAGGAAGAGAGGAAGGTGCACAGCCCCCAGCAGGTCGATTTTCGCTCTGTC CTGGCCAAGAAGGGGACTTCCAAGACCCCCGTGCCTGAGAAGGTGCCACCGCCAAAACCTGCCACCCCGGATTTTCGCTCAGTGCTGGGT GGCAAGAAGAAATTACCAGCAGAGAATGGCAGCAGCAGTGCCGAGACCCTGAATGCCAAGGCAGTGGAGAGTTCCAAGCCCCTGAGCAAT GCACAGCCTTCAGGGCCCTTGAAACCCGTGGGCAACGCCAAGCCTGCTGAGACCCTGAAGCCAATGGGCAACGCCAAGCCTGCCGAGACC CTGAAGCCCATGGGCAATGCCAAGCCTGATGAGAACCTGAAATCCGCTAGCAAAGAAGAACTCAAGAAAGACGTTAAGAATGATGTGAAC TGCAAGAGAGGCCATGCAGGGACCACAGATAATGAAAAGAGATCAGAGAGCCAGGGGACAGCCCCAGCCTTCAAGCAGAAGCTGCAAGAT GTTCATGTGGCAGAGGGCAAGAAGCTGCTGCTCCAGTGCCAGGTGTCTTCTGACCCCCCAGCCACCATCATCTGGACGCTGAACGGAAAG ACCCTCAAGACCACCAAGTTCATCATCCTCTCCCAGGAAGGCTCACTCTGCTCCGTCTCCATCGAGAAGGCACTGCCTGAGGACAGAGGC TTATACAAGTGTGTAGCCAAGAATGACGCTGGCCAGGCGGAGTGCTCCTGCCAAGTCACCGTGGATGATGCTCCAGCCAGTGAGAACACC AAGGCCCCAGAGATGAAATCCCGGAGGCCCAAGAGCTCTCTTCCTCCCGTGCTAGGAACTGAGAGTGATGCGACTGTGAAAAAGAAACCT GCCCCCAAGACACCTCCGAAGGCAGCAATGCCCCCTCAGATCATCCAGTTCCCTGAGGACCAGAAGGTACGCGCAGGAGAGTCAGTGGAG CTGTTTGGCAAAGTGACAGGCACTCAGCCCATCACCTGTACCTGGATGAAGTTCCGAAAGCAGATCCAGGAAAGCGAGCACATGAAGGTG GAGAACAGCGAGAATGGCAGCAAGCTCACCATCCTGGCCGCGCGCCAGGAGCACTGCGGCTGCTACACACTGCTGGTGGAGAACAAGCTG GGCAGCAGGCAGGCCCAGGTCAACCTCACTGTCGTGGATAAGCCAGACCCCCCAGCTGGCACACCTTGTGCCTCTGACATTCGGAGCTCC TCACTGACCCTGTCCTGGTATGGCTCCTCATATGATGGGGGCAGTGCTGTACAGTCCTACAGCATCGAGATCTGGGACTCAGCCAACAAG ACGTGGAAGGAACTAGCCACATGCCGCAGCACCTCTTTCAACGTCCAGGACCTGCTGCCTGACCACGAATATAAGTTCCGTGTACGTGCA ATCAACGTGTATGGAACCAGTGAGCCAAGCCAGGAGTCTGAACTCACAACGGTAGGAGAGAAACCTGAAGAGCCGAAGGATGAAGTGGAG GTGTCAGATGATGATGAGAAGGAGCCCGAGGTTGATTACCGGACAGTGACAATCAATACTGAACAAAAAGTATCTGACTTCTACGACATT GAGGAGAGATTAGGATCGTCACATGTGCCAATTTAACGAACGGTGGAAAGTCAGAACTTCTGAAATCAGGAAGCAGCAAATCCACACTAA AGCACATATGGACAGAAAGCAGCAAAGACTTGTCTATCAGCCGACTCCTGTCACAGACTTTTCGTGGCAAAGAGAATGATACAGATTTGG ACCTGAGATATGACACCCCAGAACCTTATTCTGAGCAAGACCTCTGGGACTGGCTGAGGAACTCCACAGACCTTCAAGAGCCTCGGCCCA GGGCCAAGAGAAGGCCCATTGTTAAAACGGGCAAGTTTAAGAAAATGTTTGGATGGGGCGATTTTCATTCCAACATCAAAACAGTGAAGC TGAACCTGTTGATAACTGGGAAAATTGTAGATCATGGCAATGGGACATTTAGTGTTTATTTCAGGCATAATTCAACTGGTCAAGGGAATG TATCTGTCAGCTTGGTACCCCCTACAAAAATCGTGGAATTTGACTTGGCACAACAAACCGTGATTGATGCCAAAGATTCCAAGTCTTTTA ATTGTCGCATTGAATATGAAAAGGTTGACAAGGCTACCAAGAACACACTCTGCAACTATGACCCTTCAAAAACCTGTTACCAGGAGCAAA CCCAAAGTCATGTATCCTGGCTCTGCTCCAAGCCCTTTAAGGTGATCTGTATTTACATTTCCTTTTATAGTACAGATTATAAACTGGTAC AGAAAGTGTGCCCTGACTACAACTACCACAGTGACACACCTTACTTTCCCTCGGGATGAAGGTGAACATGGGGGTGAGACTGAAGCCTGA GGAATTAAAGGTCATATGACAGGGCTGTTACCTCAAAGAAGAAGGTCACATCTGTTGCCTGGAATGTGTCTACACTGCTGCTCTTGTCAA CTGGCTGCAAAATACACTAGTGGAAAACACTCTGATGTAATTTCTGCCCAGTCAGCTTCATCCCTCAGTATAATTGTAAATCATCACAGA TTTTGAATTCACACCTGAAGACATGCTCTCACATATAGAGGTACACAAACACACCGTCATGCACATTTCAGCTTGCGTCTATCATGATTC CTGTTGAGAGGGCTTTCATTGTCTGACTCATAATGGTTCAGGATCAACTATCATCAAACGGAAGGATTAACTAGACAGAGAATGTTTCTA ACAGTTGCTGTTATGGAAATCTCTTTTAAAGTCTTGAGTACATGCTAATCAATAATCTCCACTCATGCATTCCTACTGCTTGGAGTAGCT GTACTGGTAAATACTACTGTAGGAGTATCTGCTTGTTAAAATGGAAAAATGTGTCTTTAGAGCTCAGTATTCTTTATTTTACAAACACAA CAAAATGTAGTAACTTTTTTCCAGCATACAGTAGGCACATTCAAAGTGGTCCAAGATGGCTCTTTTTTCTTTGAAAGGGGCCTGTTCTCA GTAAAGATGAGCAAACATTTGGAATTTACATGTGGGCAGACATTGGGATAACAACTTTCATCACCAATCATTGGACTTTTGTGAAGTTGA CACCAGCTAAGGCTGCTTAAAATAAGTTCTGATCATTATATAAGAAGGGAAATGCCTGGCAGACACCATGTAAGTTATAAGTGTCTGTCT TATCTTTACTACACATATTGTAACAAATTCAATATCCTAGTCTTCATTTGTATGAATGGTTTGTATTGTACATAGTTTAACCAAGTGTTA TTTGAGCTGCTTATTAATATTAACTTGTACTTGTCTCTCTGCTTGTTATTGGTTAAGAAAAAAGGATATGAGGAATTCATTTTATCAATG TAGCTGTGAAGGCCATTAAAAAGACAAACTTAATGTACAGAGCATTTATTCAGATCAAGTATTGTTGAAAGCTATACATATACAACATTA >56525_56525_3_MYLK-NXPH1_MYLK_chr3_123367818_ENST00000360304_NXPH1_chr7_8790638_ENST00000405863_length(amino acids)=1491AA_BP= MAQLPQQTTEKTVTMGDVKLVASSHISKTSLSVDPSRVDSMPLTEAPAFILPPRNLCIKEGATAKFEGRVRGYPEPQVTWHRNGQPITSG GRFLLDCGIRGTFSLVIHAVHEEDRGKYTCEATNGSGARQVTVELTVEGSFAKQLGQPVVSKTLGDRFSAPAVETRPSIWGECPPKFATK LGRVVVKEGQMGRFSCKITGRPQPQVTWLKGNVPLQPSARVSVSEKNGMQVLEIHGVNQDDVGVYTCLVVNGSGKASMSAELSIQGLDSA NRSFVRETKATNSDVRKEVTNVISKESKLDSLEAAAKSKNCSSPQRGGSPPWAANSQPQPPRESKLESCKDSPRTAPQTPVLQKTSSSIT LQAARVQPEPRAPGLGVLSPSGEERKRPAPPRPATFPTRQPGLGSQDVVSKAANRRIPMEGQRDSAFPKFESKPQSQEVKENQTVKFRCE VSGIPKPEVAWFLEGTPVRRQEGSIEVYEDAGSHYLCLLKARTRDSGTYSCTASNAQGQLSCSWTLQVERLAVMEVAPSFSSVLKDCAVI EGQDFVLQCSVRGTPVPRITWLLNGQPIQYARSTCEAGVAELHIQDALPEDHGTYTCLAENALGQVSCSAWVTVHEKKSSRKSEYLLPVA PSKPTAPIFLQGLSDLKVMDGSQVTMTVQVSGNPPPEVIWLHNGNEIQESEDFHFEQRGTQHSLCIQEVFPEDTGTYTCEAWNSAGEVRT QAVLTVQEPHDGTQPWFISKPRSVTASLGQSVLISCAIAGDPFPTVHWLRDGKALCKDTGHFEVLQNEDVFTLVLKKVQPWHAGQYEILL KNRVGECSCQVSLMLQNSSARALPRGREPASCEDLCGGGVGADGGGSDRYGSLRPGWPARGQGWLEEEDGEDVRGVLKRRVETRQHTEEA IRQQEVEQLDFRDLLGKKVSTKTLSEDDLKEIPAEQMDFRANLQRQVKPKTVSEEERKVHSPQQVDFRSVLAKKGTSKTPVPEKVPPPKP ATPDFRSVLGGKKKLPAENGSSSAETLNAKAVESSKPLSNAQPSGPLKPVGNAKPAETLKPMGNAKPAETLKPMGNAKPDENLKSASKEE LKKDVKNDVNCKRGHAGTTDNEKRSESQGTAPAFKQKLQDVHVAEGKKLLLQCQVSSDPPATIIWTLNGKTLKTTKFIILSQEGSLCSVS IEKALPEDRGLYKCVAKNDAGQAECSCQVTVDDAPASENTKAPEMKSRRPKSSLPPVLGTESDATVKKKPAPKTPPKAAMPPQIIQFPED QKVRAGESVELFGKVTGTQPITCTWMKFRKQIQESEHMKVENSENGSKLTILAARQEHCGCYTLLVENKLGSRQAQVNLTVVDKPDPPAG TPCASDIRSSSLTLSWYGSSYDGGSAVQSYSIEIWDSANKTWKELATCRSTSFNVQDLLPDHEYKFRVRAINVYGTSEPSQESELTTVGE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MYLK-NXPH1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MYLK-NXPH1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MYLK-NXPH1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
