
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MYO10-PFN2 (FusionGDB2 ID:56558) |
Fusion Gene Summary for MYO10-PFN2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MYO10-PFN2 | Fusion gene ID: 56558 | Hgene | Tgene | Gene symbol | MYO10 | PFN2 | Gene ID | 4651 | 5217 |
| Gene name | myosin X | profilin 2 | |
| Synonyms | - | D3S1319E|PFL | |
| Cytomap | 5p15.1 | 3q25.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | unconventional myosin-Xunconventional myosin-10unconventionnal myosin-X | profilin-2profilin II | |
| Modification date | 20200313 | 20200322 | |
| UniProtAcc | Q9HD67 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000507288, ENST00000513610, ENST00000274203, ENST00000427430, ENST00000505695, ENST00000512061, ENST00000515803, | ENST00000475518, ENST00000481275, ENST00000481767, ENST00000489155, ENST00000490975, ENST00000494827, ENST00000497148, ENST00000498307, ENST00000461868, ENST00000461930, ENST00000239940, ENST00000423691, ENST00000452853, | |
| Fusion gene scores | * DoF score | 20 X 14 X 10=2800 | 5 X 4 X 3=60 |
| # samples | 25 | 5 | |
| ** MAII score | log2(25/2800*10)=-3.48542682717024 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/60*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: MYO10 [Title/Abstract] AND PFN2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MYO10(16877717)-PFN2(149686337), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | PFN2 | GO:0030837 | negative regulation of actin filament polymerization | 7758455 |
| Tgene | PFN2 | GO:0032781 | positive regulation of ATPase activity | 7758455 |
| Tgene | PFN2 | GO:0050821 | protein stabilization | 18573880 |
 Fusion gene breakpoints across MYO10 (5'-gene) Fusion gene breakpoints across MYO10 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
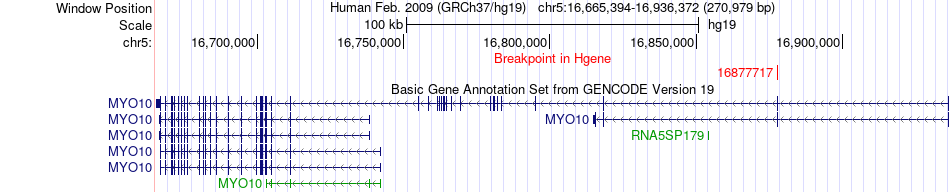 |
 Fusion gene breakpoints across PFN2 (3'-gene) Fusion gene breakpoints across PFN2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
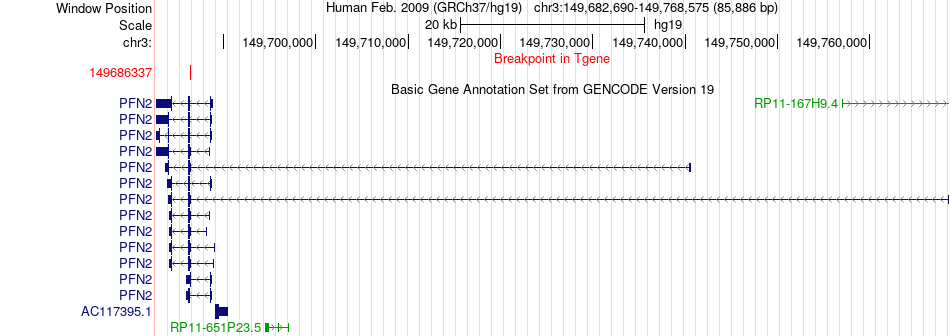 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ESCA | TCGA-R6-A6DQ | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
Top |
Fusion Gene ORF analysis for MYO10-PFN2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000507288 | ENST00000475518 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| 5CDS-5UTR | ENST00000507288 | ENST00000481275 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| 5CDS-5UTR | ENST00000507288 | ENST00000481767 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| 5CDS-5UTR | ENST00000507288 | ENST00000489155 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| 5CDS-5UTR | ENST00000507288 | ENST00000490975 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| 5CDS-5UTR | ENST00000507288 | ENST00000494827 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| 5CDS-5UTR | ENST00000507288 | ENST00000497148 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| 5CDS-5UTR | ENST00000507288 | ENST00000498307 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| 5CDS-5UTR | ENST00000513610 | ENST00000475518 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| 5CDS-5UTR | ENST00000513610 | ENST00000481275 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| 5CDS-5UTR | ENST00000513610 | ENST00000481767 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| 5CDS-5UTR | ENST00000513610 | ENST00000489155 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| 5CDS-5UTR | ENST00000513610 | ENST00000490975 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| 5CDS-5UTR | ENST00000513610 | ENST00000494827 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| 5CDS-5UTR | ENST00000513610 | ENST00000497148 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| 5CDS-5UTR | ENST00000513610 | ENST00000498307 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| 5CDS-intron | ENST00000507288 | ENST00000461868 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| 5CDS-intron | ENST00000507288 | ENST00000461930 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| 5CDS-intron | ENST00000513610 | ENST00000461868 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| 5CDS-intron | ENST00000513610 | ENST00000461930 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| In-frame | ENST00000507288 | ENST00000239940 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| In-frame | ENST00000507288 | ENST00000423691 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| In-frame | ENST00000507288 | ENST00000452853 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| In-frame | ENST00000513610 | ENST00000239940 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| In-frame | ENST00000513610 | ENST00000423691 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| In-frame | ENST00000513610 | ENST00000452853 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-3CDS | ENST00000274203 | ENST00000239940 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-3CDS | ENST00000274203 | ENST00000423691 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-3CDS | ENST00000274203 | ENST00000452853 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-3CDS | ENST00000427430 | ENST00000239940 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-3CDS | ENST00000427430 | ENST00000423691 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-3CDS | ENST00000427430 | ENST00000452853 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-3CDS | ENST00000505695 | ENST00000239940 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-3CDS | ENST00000505695 | ENST00000423691 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-3CDS | ENST00000505695 | ENST00000452853 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-3CDS | ENST00000512061 | ENST00000239940 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-3CDS | ENST00000512061 | ENST00000423691 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-3CDS | ENST00000512061 | ENST00000452853 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-3CDS | ENST00000515803 | ENST00000239940 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-3CDS | ENST00000515803 | ENST00000423691 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-3CDS | ENST00000515803 | ENST00000452853 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000274203 | ENST00000475518 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000274203 | ENST00000481275 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000274203 | ENST00000481767 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000274203 | ENST00000489155 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000274203 | ENST00000490975 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000274203 | ENST00000494827 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000274203 | ENST00000497148 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000274203 | ENST00000498307 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000427430 | ENST00000475518 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000427430 | ENST00000481275 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000427430 | ENST00000481767 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000427430 | ENST00000489155 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000427430 | ENST00000490975 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000427430 | ENST00000494827 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000427430 | ENST00000497148 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000427430 | ENST00000498307 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000505695 | ENST00000475518 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000505695 | ENST00000481275 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000505695 | ENST00000481767 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000505695 | ENST00000489155 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000505695 | ENST00000490975 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000505695 | ENST00000494827 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000505695 | ENST00000497148 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000505695 | ENST00000498307 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000512061 | ENST00000475518 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000512061 | ENST00000481275 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000512061 | ENST00000481767 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000512061 | ENST00000489155 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000512061 | ENST00000490975 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000512061 | ENST00000494827 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000512061 | ENST00000497148 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000512061 | ENST00000498307 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000515803 | ENST00000475518 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000515803 | ENST00000481275 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000515803 | ENST00000481767 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000515803 | ENST00000489155 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000515803 | ENST00000490975 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000515803 | ENST00000494827 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000515803 | ENST00000497148 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-5UTR | ENST00000515803 | ENST00000498307 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-intron | ENST00000274203 | ENST00000461868 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-intron | ENST00000274203 | ENST00000461930 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-intron | ENST00000427430 | ENST00000461868 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-intron | ENST00000427430 | ENST00000461930 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-intron | ENST00000505695 | ENST00000461868 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-intron | ENST00000505695 | ENST00000461930 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-intron | ENST00000512061 | ENST00000461868 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-intron | ENST00000512061 | ENST00000461930 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-intron | ENST00000515803 | ENST00000461868 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
| intron-intron | ENST00000515803 | ENST00000461930 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000513610 | MYO10 | chr5 | 16877717 | - | ENST00000452853 | PFN2 | chr3 | 149686337 | - | 2129 | 575 | 546 | 1 | 182 |
| ENST00000513610 | MYO10 | chr5 | 16877717 | - | ENST00000239940 | PFN2 | chr3 | 149686337 | - | 2451 | 575 | 401 | 865 | 154 |
| ENST00000513610 | MYO10 | chr5 | 16877717 | - | ENST00000423691 | PFN2 | chr3 | 149686337 | - | 1288 | 575 | 401 | 1009 | 202 |
| ENST00000507288 | MYO10 | chr5 | 16877717 | - | ENST00000452853 | PFN2 | chr3 | 149686337 | - | 2125 | 571 | 542 | 0 | 181 |
| ENST00000507288 | MYO10 | chr5 | 16877717 | - | ENST00000239940 | PFN2 | chr3 | 149686337 | - | 2447 | 571 | 397 | 861 | 154 |
| ENST00000507288 | MYO10 | chr5 | 16877717 | - | ENST00000423691 | PFN2 | chr3 | 149686337 | - | 1284 | 571 | 397 | 1005 | 202 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000513610 | ENST00000452853 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - | 0.00758072 | 0.9924193 |
| ENST00000513610 | ENST00000239940 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - | 0.013655179 | 0.9863448 |
| ENST00000513610 | ENST00000423691 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - | 0.005476511 | 0.9945235 |
| ENST00000507288 | ENST00000452853 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - | 0.007839215 | 0.99216074 |
| ENST00000507288 | ENST00000239940 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - | 0.008250928 | 0.99174905 |
| ENST00000507288 | ENST00000423691 | MYO10 | chr5 | 16877717 | - | PFN2 | chr3 | 149686337 | - | 0.00524512 | 0.9947549 |
Top |
Fusion Genomic Features for MYO10-PFN2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
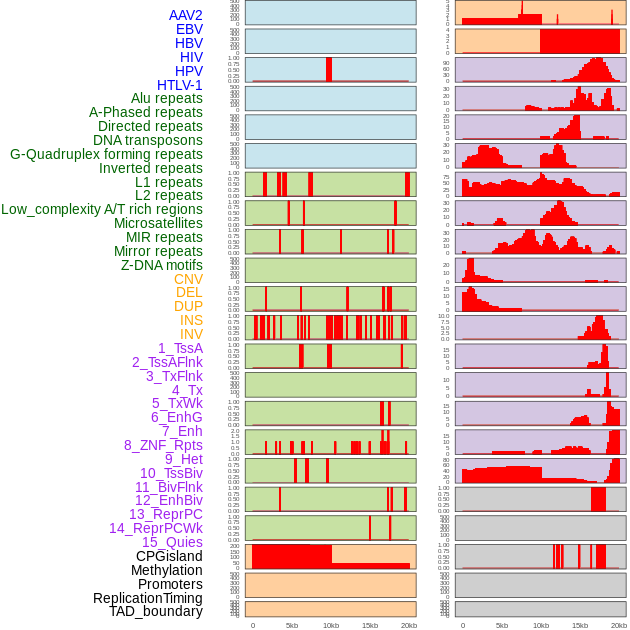 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for MYO10-PFN2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:16877717/chr3:149686337) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MYO10 | . |
| FUNCTION: Myosins are actin-based motor molecules with ATPase activity. Unconventional myosins serve in intracellular movements. MYO10 binds to actin filaments and actin bundles and functions as plus end-directed motor. The tail domain binds to membranous compartments containing phosphatidylinositol 3,4,5-trisphosphate or integrins, and mediates cargo transport along actin filaments. Regulates cell shape, cell spreading and cell adhesion. Stimulates the formation and elongation of filopodia. May play a role in neurite outgrowth and axon guidance. In hippocampal neurons it induces the formation of dendritic filopodia by trafficking the actin-remodeling protein VASP to the tips of filopodia, where it promotes actin elongation. Plays a role in formation of the podosome belt in osteoclasts. {ECO:0000269|PubMed:16894163, ECO:0000269|PubMed:18570893}.; FUNCTION: [Isoform Headless]: Functions as a dominant-negative regulator of isoform 1, suppressing its filopodia-inducing and axon outgrowth-promoting activities. In hippocampal neurons, it increases VASP retention in spine heads to induce spine formation and spine head expansion (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MYO10 | chr5:16877717 | chr3:149686337 | ENST00000513610 | - | 2 | 41 | 1212_1310 | 40 | 2059.0 | Domain | PH 1 |
| Hgene | MYO10 | chr5:16877717 | chr3:149686337 | ENST00000513610 | - | 2 | 41 | 1392_1497 | 40 | 2059.0 | Domain | PH 2 |
| Hgene | MYO10 | chr5:16877717 | chr3:149686337 | ENST00000513610 | - | 2 | 41 | 1547_1695 | 40 | 2059.0 | Domain | MyTH4 |
| Hgene | MYO10 | chr5:16877717 | chr3:149686337 | ENST00000513610 | - | 2 | 41 | 1700_2044 | 40 | 2059.0 | Domain | FERM |
| Hgene | MYO10 | chr5:16877717 | chr3:149686337 | ENST00000513610 | - | 2 | 41 | 63_739 | 40 | 2059.0 | Domain | Myosin motor |
| Hgene | MYO10 | chr5:16877717 | chr3:149686337 | ENST00000513610 | - | 2 | 41 | 742_763 | 40 | 2059.0 | Domain | IQ 1 |
| Hgene | MYO10 | chr5:16877717 | chr3:149686337 | ENST00000513610 | - | 2 | 41 | 764_787 | 40 | 2059.0 | Domain | IQ 2 |
| Hgene | MYO10 | chr5:16877717 | chr3:149686337 | ENST00000513610 | - | 2 | 41 | 788_817 | 40 | 2059.0 | Domain | IQ 3 |
| Hgene | MYO10 | chr5:16877717 | chr3:149686337 | ENST00000513610 | - | 2 | 41 | 157_164 | 40 | 2059.0 | Nucleotide binding | ATP |
| Hgene | MYO10 | chr5:16877717 | chr3:149686337 | ENST00000513610 | - | 2 | 41 | 619_641 | 40 | 2059.0 | Region | Actin-binding |
| Hgene | MYO10 | chr5:16877717 | chr3:149686337 | ENST00000513610 | - | 2 | 41 | 815_908 | 40 | 2059.0 | Region | SAH |
| Hgene | MYO10 | chr5:16877717 | chr3:149686337 | ENST00000513610 | - | 2 | 41 | 883_933 | 40 | 2059.0 | Region | Mediates antiparallel dimerization |
Top |
Fusion Gene Sequence for MYO10-PFN2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >56558_56558_1_MYO10-PFN2_MYO10_chr5_16877717_ENST00000507288_PFN2_chr3_149686337_ENST00000239940_length(transcript)=2447nt_BP=571nt CGGCGCGGGACTGCTCACCTCCAAGCGCTCGCGCGGGGATCGCGGCTCCTGCTCACTTTGCGGCCCGCTGTCCTCCTGCCCGCCCCGAGG GCCCCCGGCCGGAGCGCAGAGGGAGGGGGCCGCGCTCGCCAGCACCCCGCCGCCTTCCCCCGCCTGGGGGAAGAATGTGCCACCAGCTGT TCTCCGCTTGCGAGCGCTGCGCCCAGTAGTGAGGAACTTGGAGGAAGAAGAGACAAAGGCTGCCGTCGGGACGGGCGAGTTAGGGACTTG GGTTTGGGCGAACAAAAGGTGAGAAGGACAAGAAGGGACCGGGCGATGGCAGCAGGGGAGCCCCGCGGGCGCGCGTCCTCGGGAGTGGCG CCGTGACACGCATGGTTTCCCCGGACCCGCGGCGGCGCTGACTTCCGCGAGTCGGAGCGGCACTCGGCGAGTCCGGGACTGCGCTGGAAC AATGGATAACTTCTTCACCGAGGGAACACGGGTCTGGCTGAGAGAAAATGGCCAGCATTTTCCAAGTACTGTAAATTCCTGTGCAGAAGG CATCGTCGTCTTCCGGACAGACTATGGTCAGCCAATAGAAATAGATATGATTGTAGGAAAAGACCGGGAAGGTTTCTTTACCAACGGTTT GACTCTTGGCGCGAAGAAATGCTCAGTGATCAGAGATAGTCTATACGTCGATGGTGACTGCACAATGGACATCCGGACAAAGAGTCAAGG TGGGGAGCCAACATACAATGTGGCTGTCGGCAGAGCTGGTAGAGTCTTGGTCTTTGTAATGGGAAAAGAAGGGGTCCATGGAGGCGGATT GAATAAGAAGGCATACTCAATGGCAAAATACTTGAGAGACTCTGGGTTCTAGCTGCTAGGCAGACTGTTAAGTATTAGGGGAAAATTGCT CTTAAACTTTCCTAGCTATAAGCTTAAGTCTTAATTCTGGAAATTTTATTAGCAATGCAGGGTGATGGGGTATGAACCTGTGTCTCCTTT GTATCCCTCTGTTGGTGGGGAAAGGTGTCTTTCTTTCTGCCCTCCCCCCCCAAAATAATTCTGTTCACTTTTGTTTTGTTTCCTTGTGTA CTCCAGCATTGGTTATAGTCATGGGAAAGGAAGGTGTCCACGGAGGCACACTTAACAAGAAAGCATATGAACTCGCTTTATACCTGAGGA GGTCTGATGTGTAAGCAGCCTCTCCCCATCTACCTAGCAACTGTCTTCATCAACAACCCTAATTATGGTCACAATGCTACCAAACTGTAG ATGGTAGCTAATTTTTCTTTACCTATTTTCTAATGTCATGATTCCTGTTTGCCCAATGGATCATTTGTATGTTAACCACTGTATGTAACC AACCCTTATCTGGCAACATAATTGCAGCACAATAATGATTTGCATGATACCTTGAAATTGGGGGGAGGGGGCATGCCAAGTTGGGCATCA CTTTGTCTTAGCAATTAATGGGATATTGATTACTAAAATAAGTTAATATTAAGCAAGGTGCCGGTTGTACAATCTCTGATCAGTGTCTTT TCAGCACTTTGAGCATTTACTTGGCTCATTTAGTCTTCCTTTTGTAGCGCATGGTTGGGAGGAAAAAGTGCATGCATCATTCCTTCACTC TTCTCTTTTTCCCGCCCCCCCTCCCTTCGCACATAGGCATTTGGTTTGCTTCCATCTTTTTTTATGCAGTGCCTGTTTTTTTTTAACCAA TTAAAATCCCTTTTGTTGATGAGCTATTGAGAGCTGCAGTAGTTTGCTTTTAGTATTGTTGTTGCACTTGAGCAGAGACAAACCTTTATT CATAGTGTCTACAGGACATATGAAGAGTGCAATGGCAAAACAAGAGCAAAAAGCACTTCCTCCCATGACCTTACAGTAACCATACTGATT GAATCCCCAGGGACATTCCATCATTGCAATAGCTCAGATTTTTCTTCCTTTTTCTTTGCACACCAGCTCTACTCTTTAGTAAAATTGTAA AAGGCTGCCATTATGGACATTAGGTATCCCAACATAACCATCTGGAGTGTGTCCAGTTTGTTCTTCATAGGACCAATTTTTATTTGCAGC TTGAGTTTTTATATGAAGTTGCATTATTGTGGACTTGGCTGTCTTGTGATGAATTTTTTTCATATGTATTCTGTGCCATACTATTGTTAA AATGAACTGTTGCTATTGTGAGATGGATTTTAACTGACCTATTAAGGGTTTCTTTCGAATGGCACTACTTTAGGGACATTCTAGTATTTG CTTCTATTGTTTGGGCCTTGTGGATAATGTACAGATTTAAAAACAAATCTTGTTGCTGATTTGTCCATTTCTTTCCCTGCACTTTGTTAC ATCTGGGATACAGTCTAACTCATCTGATTTAATATGCATTTAAAAAAATGCCATAACTATTAAACACCTTGTTTACAGACAGATGAAATA >56558_56558_1_MYO10-PFN2_MYO10_chr5_16877717_ENST00000507288_PFN2_chr3_149686337_ENST00000239940_length(amino acids)=154AA_BP=56 MTSASRSGTRRVRDCAGTMDNFFTEGTRVWLRENGQHFPSTVNSCAEGIVVFRTDYGQPIEIDMIVGKDREGFFTNGLTLGAKKCSVIRD -------------------------------------------------------------- >56558_56558_2_MYO10-PFN2_MYO10_chr5_16877717_ENST00000507288_PFN2_chr3_149686337_ENST00000423691_length(transcript)=1284nt_BP=571nt CGGCGCGGGACTGCTCACCTCCAAGCGCTCGCGCGGGGATCGCGGCTCCTGCTCACTTTGCGGCCCGCTGTCCTCCTGCCCGCCCCGAGG GCCCCCGGCCGGAGCGCAGAGGGAGGGGGCCGCGCTCGCCAGCACCCCGCCGCCTTCCCCCGCCTGGGGGAAGAATGTGCCACCAGCTGT TCTCCGCTTGCGAGCGCTGCGCCCAGTAGTGAGGAACTTGGAGGAAGAAGAGACAAAGGCTGCCGTCGGGACGGGCGAGTTAGGGACTTG GGTTTGGGCGAACAAAAGGTGAGAAGGACAAGAAGGGACCGGGCGATGGCAGCAGGGGAGCCCCGCGGGCGCGCGTCCTCGGGAGTGGCG CCGTGACACGCATGGTTTCCCCGGACCCGCGGCGGCGCTGACTTCCGCGAGTCGGAGCGGCACTCGGCGAGTCCGGGACTGCGCTGGAAC AATGGATAACTTCTTCACCGAGGGAACACGGGTCTGGCTGAGAGAAAATGGCCAGCATTTTCCAAGTACTGTAAATTCCTGTGCAGAAGG CATCGTCGTCTTCCGGACAGACTATGGTCAGCCAATAGAAATAGATATGATTGTAGGAAAAGACCGGGAAGGTTTCTTTACCAACGGTTT GACTCTTGGCGCGAAGAAATGCTCAGTGATCAGAGATAGTCTATACGTCGATGGTGACTGCACAATGGACATCCGGACAAAGAGTCAAGG TGGGGAGCCAACATACAATGTGGCTGTCGGCAGAGCTGGTAGAGCATTGGTTATAGTCATGGGAAAGGAAGGTGTCCACGGAGGCACACT TAACAAGAAAGCATATGAACTCGCTTTATACCTGAGGAGGTATCCCAACATAACCATCTGGAGTGTGTCCAGTTTGTTCTTCATAGGACC AATTTTTATTTGCAGCTTGAGTTTTTATATGAAGTTGCATTATTGTGGACTTGGCTGTCTTGTGATGAATTTTTTTCATATGTATTCTGT GCCATACTATTGTTAAAATGAACTGTTGCTATTGTGAGATGGATTTTAACTGACCTATTAAGGGTTTCTTTCGAATGGCACTACTTTAGG GACATTCTAGTATTTGCTTCTATTGTTTGGGCCTTGTGGATAATGTACAGATTTAAAAACAAATCTTGTTGCTGATTTGTCCATTTCTTT CCCTGCACTTTGTTACATCTGGGATACAGTCTAACTCATCTGATTTAATATGCATTTAAAAAAATGCCATAACTATTAAACACCTTGTTT >56558_56558_2_MYO10-PFN2_MYO10_chr5_16877717_ENST00000507288_PFN2_chr3_149686337_ENST00000423691_length(amino acids)=202AA_BP=56 MTSASRSGTRRVRDCAGTMDNFFTEGTRVWLRENGQHFPSTVNSCAEGIVVFRTDYGQPIEIDMIVGKDREGFFTNGLTLGAKKCSVIRD SLYVDGDCTMDIRTKSQGGEPTYNVAVGRAGRALVIVMGKEGVHGGTLNKKAYELALYLRRYPNITIWSVSSLFFIGPIFICSLSFYMKL -------------------------------------------------------------- >56558_56558_3_MYO10-PFN2_MYO10_chr5_16877717_ENST00000507288_PFN2_chr3_149686337_ENST00000452853_length(transcript)=2125nt_BP=571nt CGGCGCGGGACTGCTCACCTCCAAGCGCTCGCGCGGGGATCGCGGCTCCTGCTCACTTTGCGGCCCGCTGTCCTCCTGCCCGCCCCGAGG GCCCCCGGCCGGAGCGCAGAGGGAGGGGGCCGCGCTCGCCAGCACCCCGCCGCCTTCCCCCGCCTGGGGGAAGAATGTGCCACCAGCTGT TCTCCGCTTGCGAGCGCTGCGCCCAGTAGTGAGGAACTTGGAGGAAGAAGAGACAAAGGCTGCCGTCGGGACGGGCGAGTTAGGGACTTG GGTTTGGGCGAACAAAAGGTGAGAAGGACAAGAAGGGACCGGGCGATGGCAGCAGGGGAGCCCCGCGGGCGCGCGTCCTCGGGAGTGGCG CCGTGACACGCATGGTTTCCCCGGACCCGCGGCGGCGCTGACTTCCGCGAGTCGGAGCGGCACTCGGCGAGTCCGGGACTGCGCTGGAAC AATGGATAACTTCTTCACCGAGGGAACACGGGTCTGGCTGAGAGAAAATGGCCAGCATTTTCCAAGTACTGTAAATTCCTGTGCAGAAGG CATCGTCGTCTTCCGGACAGACTATGGTCAGCCAATAGAAATAGATATGATTGTAGGAAAAGACCGGGAAGGTTTCTTTACCAACGGTTT GACTCTTGGCGCGAAGAAATGCTCAGTGATCAGAGATAGTCTATACGTCGATGGTGACTGCACAATGGACATCCGGACAAAGAGTCAAGG TGGGGAGCCAACATACAATGTGGCTGTCGGCAGAGCTGGTAGAGCATTGGTTATAGTCATGGGAAAGGAAGGTGTCCACGGAGGCACACT TAACAAGAAAGCATATGAACTCGCTTTATACCTGAGGAGGTCTGATGTGTAAGCAGCCTCTCCCCATCTACCTAGCAACTGTCTTCATCA ACAACCCTAATTATGGTCACAATGCTACCAAACTGTAGATGGTAGCTAATTTTTCTTTACCTATTTTCTAATGTCATGATTCCTGTTTGC CCAATGGATCATTTGTATGTTAACCACTGTATGTAACCAACCCTTATCTGGCAACATAATTGCAGCACAATAATGATTTGCATGATACCT TGAAATTGGGGGGAGGGGGCATGCCAAGTTGGGCATCACTTTGTCTTAGCAATTAATGGGATATTGATTACTAAAATAAGTTAATATTAA GCAAGGTGCCGGTTGTACAATCTCTGATCAGTGTCTTTTCAGCACTTTGAGCATTTACTTGGCTCATTTAGTCTTCCTTTTGTAGCGCAT GGTTGGGAGGAAAAAGTGCATGCATCATTCCTTCACTCTTCTCTTTTTCCCGCCCCCCCTCCCTTCGCACATAGGCATTTGGTTTGCTTC CATCTTTTTTTATGCAGTGCCTGTTTTTTTTTAACCAATTAAAATCCCTTTTGTTGATGAGCTATTGAGAGCTGCAGTAGTTTGCTTTTA GTATTGTTGTTGCACTTGAGCAGAGACAAACCTTTATTCATAGTGTCTACAGGACATATGAAGAGTGCAATGGCAAAACAAGAGCAAAAA GCACTTCCTCCCATGACCTTACAGTAACCATACTGATTGAATCCCCAGGGACATTCCATCATTGCAATAGCTCAGATTTTTCTTCCTTTT TCTTTGCACACCAGCTCTACTCTTTAGTAAAATTGTAAAAGGCTGCCATTATGGACATTAGGTATCCCAACATAACCATCTGGAGTGTGT CCAGTTTGTTCTTCATAGGACCAATTTTTATTTGCAGCTTGAGTTTTTATATGAAGTTGCATTATTGTGGACTTGGCTGTCTTGTGATGA ATTTTTTTCATATGTATTCTGTGCCATACTATTGTTAAAATGAACTGTTGCTATTGTGAGATGGATTTTAACTGACCTATTAAGGGTTTC TTTCGAATGGCACTACTTTAGGGACATTCTAGTATTTGCTTCTATTGTTTGGGCCTTGTGGATAATGTACAGATTTAAAAACAAATCTTG TTGCTGATTTGTCCATTTCTTTCCCTGCACTTTGTTACATCTGGGATACAGTCTAACTCATCTGATTTAATATGCATTTAAAAAAATGCC >56558_56558_3_MYO10-PFN2_MYO10_chr5_16877717_ENST00000507288_PFN2_chr3_149686337_ENST00000452853_length(amino acids)=181AA_BP= MPSAQEFTVLGKCWPFSLSQTRVPSVKKLSIVPAQSRTRRVPLRLAEVSAAAGPGKPCVSRRHSRGRAPAGLPCCHRPVPSCPSHLLFAQ TQVPNSPVPTAAFVSSSSKFLTTGRSARKRRTAGGTFFPQAGEGGGVLASAAPSLCAPAGGPRGGQEDSGPQSEQEPRSPRERLEVSSPA -------------------------------------------------------------- >56558_56558_4_MYO10-PFN2_MYO10_chr5_16877717_ENST00000513610_PFN2_chr3_149686337_ENST00000239940_length(transcript)=2451nt_BP=575nt GCCGCGGCGCGGGACTGCTCACCTCCAAGCGCTCGCGCGGGGATCGCGGCTCCTGCTCACTTTGCGGCCCGCTGTCCTCCTGCCCGCCCC GAGGGCCCCCGGCCGGAGCGCAGAGGGAGGGGGCCGCGCTCGCCAGCACCCCGCCGCCTTCCCCCGCCTGGGGGAAGAATGTGCCACCAG CTGTTCTCCGCTTGCGAGCGCTGCGCCCAGTAGTGAGGAACTTGGAGGAAGAAGAGACAAAGGCTGCCGTCGGGACGGGCGAGTTAGGGA CTTGGGTTTGGGCGAACAAAAGGTGAGAAGGACAAGAAGGGACCGGGCGATGGCAGCAGGGGAGCCCCGCGGGCGCGCGTCCTCGGGAGT GGCGCCGTGACACGCATGGTTTCCCCGGACCCGCGGCGGCGCTGACTTCCGCGAGTCGGAGCGGCACTCGGCGAGTCCGGGACTGCGCTG GAACAATGGATAACTTCTTCACCGAGGGAACACGGGTCTGGCTGAGAGAAAATGGCCAGCATTTTCCAAGTACTGTAAATTCCTGTGCAG AAGGCATCGTCGTCTTCCGGACAGACTATGGTCAGCCAATAGAAATAGATATGATTGTAGGAAAAGACCGGGAAGGTTTCTTTACCAACG GTTTGACTCTTGGCGCGAAGAAATGCTCAGTGATCAGAGATAGTCTATACGTCGATGGTGACTGCACAATGGACATCCGGACAAAGAGTC AAGGTGGGGAGCCAACATACAATGTGGCTGTCGGCAGAGCTGGTAGAGTCTTGGTCTTTGTAATGGGAAAAGAAGGGGTCCATGGAGGCG GATTGAATAAGAAGGCATACTCAATGGCAAAATACTTGAGAGACTCTGGGTTCTAGCTGCTAGGCAGACTGTTAAGTATTAGGGGAAAAT TGCTCTTAAACTTTCCTAGCTATAAGCTTAAGTCTTAATTCTGGAAATTTTATTAGCAATGCAGGGTGATGGGGTATGAACCTGTGTCTC CTTTGTATCCCTCTGTTGGTGGGGAAAGGTGTCTTTCTTTCTGCCCTCCCCCCCCAAAATAATTCTGTTCACTTTTGTTTTGTTTCCTTG TGTACTCCAGCATTGGTTATAGTCATGGGAAAGGAAGGTGTCCACGGAGGCACACTTAACAAGAAAGCATATGAACTCGCTTTATACCTG AGGAGGTCTGATGTGTAAGCAGCCTCTCCCCATCTACCTAGCAACTGTCTTCATCAACAACCCTAATTATGGTCACAATGCTACCAAACT GTAGATGGTAGCTAATTTTTCTTTACCTATTTTCTAATGTCATGATTCCTGTTTGCCCAATGGATCATTTGTATGTTAACCACTGTATGT AACCAACCCTTATCTGGCAACATAATTGCAGCACAATAATGATTTGCATGATACCTTGAAATTGGGGGGAGGGGGCATGCCAAGTTGGGC ATCACTTTGTCTTAGCAATTAATGGGATATTGATTACTAAAATAAGTTAATATTAAGCAAGGTGCCGGTTGTACAATCTCTGATCAGTGT CTTTTCAGCACTTTGAGCATTTACTTGGCTCATTTAGTCTTCCTTTTGTAGCGCATGGTTGGGAGGAAAAAGTGCATGCATCATTCCTTC ACTCTTCTCTTTTTCCCGCCCCCCCTCCCTTCGCACATAGGCATTTGGTTTGCTTCCATCTTTTTTTATGCAGTGCCTGTTTTTTTTTAA CCAATTAAAATCCCTTTTGTTGATGAGCTATTGAGAGCTGCAGTAGTTTGCTTTTAGTATTGTTGTTGCACTTGAGCAGAGACAAACCTT TATTCATAGTGTCTACAGGACATATGAAGAGTGCAATGGCAAAACAAGAGCAAAAAGCACTTCCTCCCATGACCTTACAGTAACCATACT GATTGAATCCCCAGGGACATTCCATCATTGCAATAGCTCAGATTTTTCTTCCTTTTTCTTTGCACACCAGCTCTACTCTTTAGTAAAATT GTAAAAGGCTGCCATTATGGACATTAGGTATCCCAACATAACCATCTGGAGTGTGTCCAGTTTGTTCTTCATAGGACCAATTTTTATTTG CAGCTTGAGTTTTTATATGAAGTTGCATTATTGTGGACTTGGCTGTCTTGTGATGAATTTTTTTCATATGTATTCTGTGCCATACTATTG TTAAAATGAACTGTTGCTATTGTGAGATGGATTTTAACTGACCTATTAAGGGTTTCTTTCGAATGGCACTACTTTAGGGACATTCTAGTA TTTGCTTCTATTGTTTGGGCCTTGTGGATAATGTACAGATTTAAAAACAAATCTTGTTGCTGATTTGTCCATTTCTTTCCCTGCACTTTG TTACATCTGGGATACAGTCTAACTCATCTGATTTAATATGCATTTAAAAAAATGCCATAACTATTAAACACCTTGTTTACAGACAGATGA >56558_56558_4_MYO10-PFN2_MYO10_chr5_16877717_ENST00000513610_PFN2_chr3_149686337_ENST00000239940_length(amino acids)=154AA_BP=56 MTSASRSGTRRVRDCAGTMDNFFTEGTRVWLRENGQHFPSTVNSCAEGIVVFRTDYGQPIEIDMIVGKDREGFFTNGLTLGAKKCSVIRD -------------------------------------------------------------- >56558_56558_5_MYO10-PFN2_MYO10_chr5_16877717_ENST00000513610_PFN2_chr3_149686337_ENST00000423691_length(transcript)=1288nt_BP=575nt GCCGCGGCGCGGGACTGCTCACCTCCAAGCGCTCGCGCGGGGATCGCGGCTCCTGCTCACTTTGCGGCCCGCTGTCCTCCTGCCCGCCCC GAGGGCCCCCGGCCGGAGCGCAGAGGGAGGGGGCCGCGCTCGCCAGCACCCCGCCGCCTTCCCCCGCCTGGGGGAAGAATGTGCCACCAG CTGTTCTCCGCTTGCGAGCGCTGCGCCCAGTAGTGAGGAACTTGGAGGAAGAAGAGACAAAGGCTGCCGTCGGGACGGGCGAGTTAGGGA CTTGGGTTTGGGCGAACAAAAGGTGAGAAGGACAAGAAGGGACCGGGCGATGGCAGCAGGGGAGCCCCGCGGGCGCGCGTCCTCGGGAGT GGCGCCGTGACACGCATGGTTTCCCCGGACCCGCGGCGGCGCTGACTTCCGCGAGTCGGAGCGGCACTCGGCGAGTCCGGGACTGCGCTG GAACAATGGATAACTTCTTCACCGAGGGAACACGGGTCTGGCTGAGAGAAAATGGCCAGCATTTTCCAAGTACTGTAAATTCCTGTGCAG AAGGCATCGTCGTCTTCCGGACAGACTATGGTCAGCCAATAGAAATAGATATGATTGTAGGAAAAGACCGGGAAGGTTTCTTTACCAACG GTTTGACTCTTGGCGCGAAGAAATGCTCAGTGATCAGAGATAGTCTATACGTCGATGGTGACTGCACAATGGACATCCGGACAAAGAGTC AAGGTGGGGAGCCAACATACAATGTGGCTGTCGGCAGAGCTGGTAGAGCATTGGTTATAGTCATGGGAAAGGAAGGTGTCCACGGAGGCA CACTTAACAAGAAAGCATATGAACTCGCTTTATACCTGAGGAGGTATCCCAACATAACCATCTGGAGTGTGTCCAGTTTGTTCTTCATAG GACCAATTTTTATTTGCAGCTTGAGTTTTTATATGAAGTTGCATTATTGTGGACTTGGCTGTCTTGTGATGAATTTTTTTCATATGTATT CTGTGCCATACTATTGTTAAAATGAACTGTTGCTATTGTGAGATGGATTTTAACTGACCTATTAAGGGTTTCTTTCGAATGGCACTACTT TAGGGACATTCTAGTATTTGCTTCTATTGTTTGGGCCTTGTGGATAATGTACAGATTTAAAAACAAATCTTGTTGCTGATTTGTCCATTT CTTTCCCTGCACTTTGTTACATCTGGGATACAGTCTAACTCATCTGATTTAATATGCATTTAAAAAAATGCCATAACTATTAAACACCTT >56558_56558_5_MYO10-PFN2_MYO10_chr5_16877717_ENST00000513610_PFN2_chr3_149686337_ENST00000423691_length(amino acids)=202AA_BP=56 MTSASRSGTRRVRDCAGTMDNFFTEGTRVWLRENGQHFPSTVNSCAEGIVVFRTDYGQPIEIDMIVGKDREGFFTNGLTLGAKKCSVIRD SLYVDGDCTMDIRTKSQGGEPTYNVAVGRAGRALVIVMGKEGVHGGTLNKKAYELALYLRRYPNITIWSVSSLFFIGPIFICSLSFYMKL -------------------------------------------------------------- >56558_56558_6_MYO10-PFN2_MYO10_chr5_16877717_ENST00000513610_PFN2_chr3_149686337_ENST00000452853_length(transcript)=2129nt_BP=575nt GCCGCGGCGCGGGACTGCTCACCTCCAAGCGCTCGCGCGGGGATCGCGGCTCCTGCTCACTTTGCGGCCCGCTGTCCTCCTGCCCGCCCC GAGGGCCCCCGGCCGGAGCGCAGAGGGAGGGGGCCGCGCTCGCCAGCACCCCGCCGCCTTCCCCCGCCTGGGGGAAGAATGTGCCACCAG CTGTTCTCCGCTTGCGAGCGCTGCGCCCAGTAGTGAGGAACTTGGAGGAAGAAGAGACAAAGGCTGCCGTCGGGACGGGCGAGTTAGGGA CTTGGGTTTGGGCGAACAAAAGGTGAGAAGGACAAGAAGGGACCGGGCGATGGCAGCAGGGGAGCCCCGCGGGCGCGCGTCCTCGGGAGT GGCGCCGTGACACGCATGGTTTCCCCGGACCCGCGGCGGCGCTGACTTCCGCGAGTCGGAGCGGCACTCGGCGAGTCCGGGACTGCGCTG GAACAATGGATAACTTCTTCACCGAGGGAACACGGGTCTGGCTGAGAGAAAATGGCCAGCATTTTCCAAGTACTGTAAATTCCTGTGCAG AAGGCATCGTCGTCTTCCGGACAGACTATGGTCAGCCAATAGAAATAGATATGATTGTAGGAAAAGACCGGGAAGGTTTCTTTACCAACG GTTTGACTCTTGGCGCGAAGAAATGCTCAGTGATCAGAGATAGTCTATACGTCGATGGTGACTGCACAATGGACATCCGGACAAAGAGTC AAGGTGGGGAGCCAACATACAATGTGGCTGTCGGCAGAGCTGGTAGAGCATTGGTTATAGTCATGGGAAAGGAAGGTGTCCACGGAGGCA CACTTAACAAGAAAGCATATGAACTCGCTTTATACCTGAGGAGGTCTGATGTGTAAGCAGCCTCTCCCCATCTACCTAGCAACTGTCTTC ATCAACAACCCTAATTATGGTCACAATGCTACCAAACTGTAGATGGTAGCTAATTTTTCTTTACCTATTTTCTAATGTCATGATTCCTGT TTGCCCAATGGATCATTTGTATGTTAACCACTGTATGTAACCAACCCTTATCTGGCAACATAATTGCAGCACAATAATGATTTGCATGAT ACCTTGAAATTGGGGGGAGGGGGCATGCCAAGTTGGGCATCACTTTGTCTTAGCAATTAATGGGATATTGATTACTAAAATAAGTTAATA TTAAGCAAGGTGCCGGTTGTACAATCTCTGATCAGTGTCTTTTCAGCACTTTGAGCATTTACTTGGCTCATTTAGTCTTCCTTTTGTAGC GCATGGTTGGGAGGAAAAAGTGCATGCATCATTCCTTCACTCTTCTCTTTTTCCCGCCCCCCCTCCCTTCGCACATAGGCATTTGGTTTG CTTCCATCTTTTTTTATGCAGTGCCTGTTTTTTTTTAACCAATTAAAATCCCTTTTGTTGATGAGCTATTGAGAGCTGCAGTAGTTTGCT TTTAGTATTGTTGTTGCACTTGAGCAGAGACAAACCTTTATTCATAGTGTCTACAGGACATATGAAGAGTGCAATGGCAAAACAAGAGCA AAAAGCACTTCCTCCCATGACCTTACAGTAACCATACTGATTGAATCCCCAGGGACATTCCATCATTGCAATAGCTCAGATTTTTCTTCC TTTTTCTTTGCACACCAGCTCTACTCTTTAGTAAAATTGTAAAAGGCTGCCATTATGGACATTAGGTATCCCAACATAACCATCTGGAGT GTGTCCAGTTTGTTCTTCATAGGACCAATTTTTATTTGCAGCTTGAGTTTTTATATGAAGTTGCATTATTGTGGACTTGGCTGTCTTGTG ATGAATTTTTTTCATATGTATTCTGTGCCATACTATTGTTAAAATGAACTGTTGCTATTGTGAGATGGATTTTAACTGACCTATTAAGGG TTTCTTTCGAATGGCACTACTTTAGGGACATTCTAGTATTTGCTTCTATTGTTTGGGCCTTGTGGATAATGTACAGATTTAAAAACAAAT CTTGTTGCTGATTTGTCCATTTCTTTCCCTGCACTTTGTTACATCTGGGATACAGTCTAACTCATCTGATTTAATATGCATTTAAAAAAA >56558_56558_6_MYO10-PFN2_MYO10_chr5_16877717_ENST00000513610_PFN2_chr3_149686337_ENST00000452853_length(amino acids)=182AA_BP= MPSAQEFTVLGKCWPFSLSQTRVPSVKKLSIVPAQSRTRRVPLRLAEVSAAAGPGKPCVSRRHSRGRAPAGLPCCHRPVPSCPSHLLFAQ TQVPNSPVPTAAFVSSSSKFLTTGRSARKRRTAGGTFFPQAGEGGGVLASAAPSLCAPAGGPRGGQEDSGPQSEQEPRSPRERLEVSSPA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MYO10-PFN2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MYO10-PFN2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MYO10-PFN2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
