
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:MYO1C-SMTNL2 (FusionGDB2 ID:56638) |
Fusion Gene Summary for MYO1C-SMTNL2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: MYO1C-SMTNL2 | Fusion gene ID: 56638 | Hgene | Tgene | Gene symbol | MYO1C | SMTNL2 | Gene ID | 4643 | 342527 |
| Gene name | myosin IE | smoothelin like 2 | |
| Synonyms | FSGS6|HuncM-IC|MYO1C | - | |
| Cytomap | 15q22.2 | 17p13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | unconventional myosin-IeMYO1E variant proteinmyosin-ICunconventional myosin 1E | smoothelin-like protein 2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000359786, ENST00000361007, ENST00000438665, ENST00000545534, ENST00000575158, ENST00000573198, | ENST00000338859, ENST00000389313, | |
| Fusion gene scores | * DoF score | 6 X 8 X 5=240 | 3 X 3 X 3=27 |
| # samples | 7 | 3 | |
| ** MAII score | log2(7/240*10)=-1.77760757866355 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(3/27*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: MYO1C [Title/Abstract] AND SMTNL2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | MYO1C(1381910)-SMTNL2(4510656), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | MYO1C-SMTNL2 seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across MYO1C (5'-gene) Fusion gene breakpoints across MYO1C (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
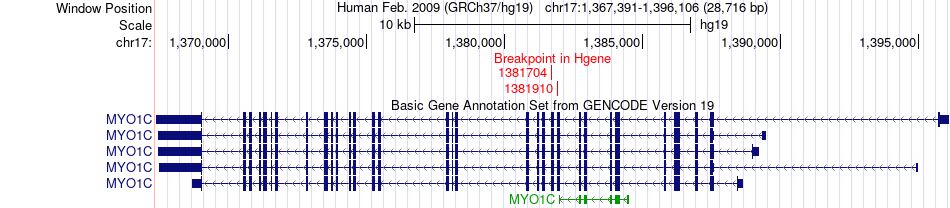 |
 Fusion gene breakpoints across SMTNL2 (3'-gene) Fusion gene breakpoints across SMTNL2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
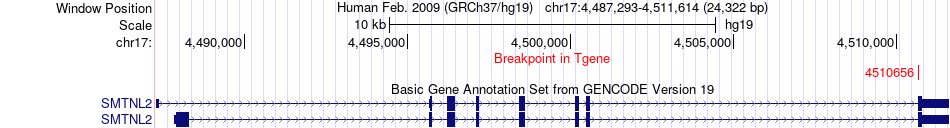 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | UCEC | TCGA-AJ-A3BD-01A | MYO1C | chr17 | 1381704 | - | SMTNL2 | chr17 | 4510656 | + |
| ChimerDB4 | UCEC | TCGA-AJ-A3BD-01A | MYO1C | chr17 | 1381910 | - | SMTNL2 | chr17 | 4510656 | + |
Top |
Fusion Gene ORF analysis for MYO1C-SMTNL2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000359786 | ENST00000338859 | MYO1C | chr17 | 1381910 | - | SMTNL2 | chr17 | 4510656 | + |
| Frame-shift | ENST00000359786 | ENST00000389313 | MYO1C | chr17 | 1381704 | - | SMTNL2 | chr17 | 4510656 | + |
| Frame-shift | ENST00000359786 | ENST00000389313 | MYO1C | chr17 | 1381910 | - | SMTNL2 | chr17 | 4510656 | + |
| Frame-shift | ENST00000361007 | ENST00000338859 | MYO1C | chr17 | 1381910 | - | SMTNL2 | chr17 | 4510656 | + |
| Frame-shift | ENST00000361007 | ENST00000389313 | MYO1C | chr17 | 1381704 | - | SMTNL2 | chr17 | 4510656 | + |
| Frame-shift | ENST00000361007 | ENST00000389313 | MYO1C | chr17 | 1381910 | - | SMTNL2 | chr17 | 4510656 | + |
| Frame-shift | ENST00000438665 | ENST00000338859 | MYO1C | chr17 | 1381910 | - | SMTNL2 | chr17 | 4510656 | + |
| Frame-shift | ENST00000438665 | ENST00000389313 | MYO1C | chr17 | 1381704 | - | SMTNL2 | chr17 | 4510656 | + |
| Frame-shift | ENST00000438665 | ENST00000389313 | MYO1C | chr17 | 1381910 | - | SMTNL2 | chr17 | 4510656 | + |
| Frame-shift | ENST00000545534 | ENST00000338859 | MYO1C | chr17 | 1381910 | - | SMTNL2 | chr17 | 4510656 | + |
| Frame-shift | ENST00000545534 | ENST00000389313 | MYO1C | chr17 | 1381704 | - | SMTNL2 | chr17 | 4510656 | + |
| Frame-shift | ENST00000545534 | ENST00000389313 | MYO1C | chr17 | 1381910 | - | SMTNL2 | chr17 | 4510656 | + |
| Frame-shift | ENST00000575158 | ENST00000338859 | MYO1C | chr17 | 1381704 | - | SMTNL2 | chr17 | 4510656 | + |
| Frame-shift | ENST00000575158 | ENST00000338859 | MYO1C | chr17 | 1381910 | - | SMTNL2 | chr17 | 4510656 | + |
| Frame-shift | ENST00000575158 | ENST00000389313 | MYO1C | chr17 | 1381704 | - | SMTNL2 | chr17 | 4510656 | + |
| In-frame | ENST00000359786 | ENST00000338859 | MYO1C | chr17 | 1381704 | - | SMTNL2 | chr17 | 4510656 | + |
| In-frame | ENST00000361007 | ENST00000338859 | MYO1C | chr17 | 1381704 | - | SMTNL2 | chr17 | 4510656 | + |
| In-frame | ENST00000438665 | ENST00000338859 | MYO1C | chr17 | 1381704 | - | SMTNL2 | chr17 | 4510656 | + |
| In-frame | ENST00000545534 | ENST00000338859 | MYO1C | chr17 | 1381704 | - | SMTNL2 | chr17 | 4510656 | + |
| In-frame | ENST00000575158 | ENST00000389313 | MYO1C | chr17 | 1381910 | - | SMTNL2 | chr17 | 4510656 | + |
| intron-3CDS | ENST00000573198 | ENST00000338859 | MYO1C | chr17 | 1381704 | - | SMTNL2 | chr17 | 4510656 | + |
| intron-3CDS | ENST00000573198 | ENST00000338859 | MYO1C | chr17 | 1381910 | - | SMTNL2 | chr17 | 4510656 | + |
| intron-3CDS | ENST00000573198 | ENST00000389313 | MYO1C | chr17 | 1381704 | - | SMTNL2 | chr17 | 4510656 | + |
| intron-3CDS | ENST00000573198 | ENST00000389313 | MYO1C | chr17 | 1381910 | - | SMTNL2 | chr17 | 4510656 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000575158 | MYO1C | chr17 | 1381910 | - | ENST00000389313 | SMTNL2 | chr17 | 4510656 | + | 2238 | 1284 | 150 | 1301 | 383 |
| ENST00000359786 | MYO1C | chr17 | 1381704 | - | ENST00000338859 | SMTNL2 | chr17 | 4510656 | + | 2579 | 1620 | 277 | 1746 | 489 |
| ENST00000438665 | MYO1C | chr17 | 1381704 | - | ENST00000338859 | SMTNL2 | chr17 | 4510656 | + | 2420 | 1461 | 40 | 1587 | 515 |
| ENST00000361007 | MYO1C | chr17 | 1381704 | - | ENST00000338859 | SMTNL2 | chr17 | 4510656 | + | 2258 | 1299 | 82 | 1425 | 447 |
| ENST00000545534 | MYO1C | chr17 | 1381704 | - | ENST00000338859 | SMTNL2 | chr17 | 4510656 | + | 2425 | 1466 | 243 | 1592 | 449 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000575158 | ENST00000389313 | MYO1C | chr17 | 1381910 | - | SMTNL2 | chr17 | 4510656 | + | 0.003341897 | 0.9966581 |
| ENST00000359786 | ENST00000338859 | MYO1C | chr17 | 1381704 | - | SMTNL2 | chr17 | 4510656 | + | 0.005054842 | 0.9949451 |
| ENST00000438665 | ENST00000338859 | MYO1C | chr17 | 1381704 | - | SMTNL2 | chr17 | 4510656 | + | 0.003047015 | 0.996953 |
| ENST00000361007 | ENST00000338859 | MYO1C | chr17 | 1381704 | - | SMTNL2 | chr17 | 4510656 | + | 0.002499822 | 0.9975001 |
| ENST00000545534 | ENST00000338859 | MYO1C | chr17 | 1381704 | - | SMTNL2 | chr17 | 4510656 | + | 0.001708124 | 0.9982919 |
Top |
Fusion Genomic Features for MYO1C-SMTNL2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
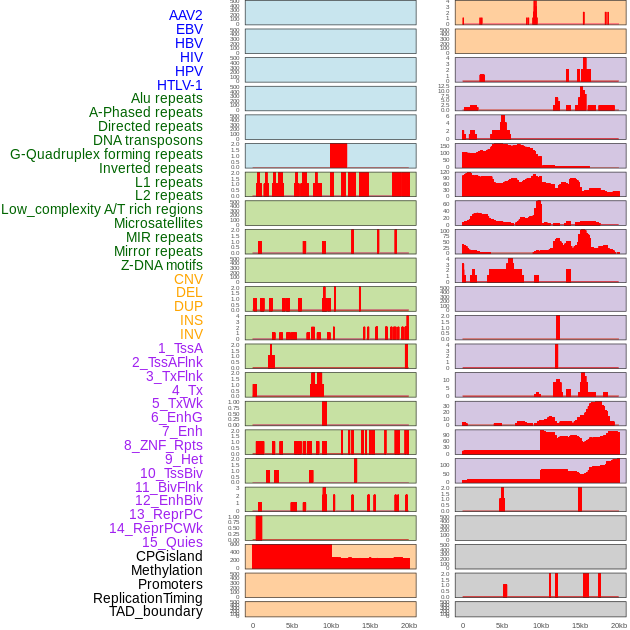 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
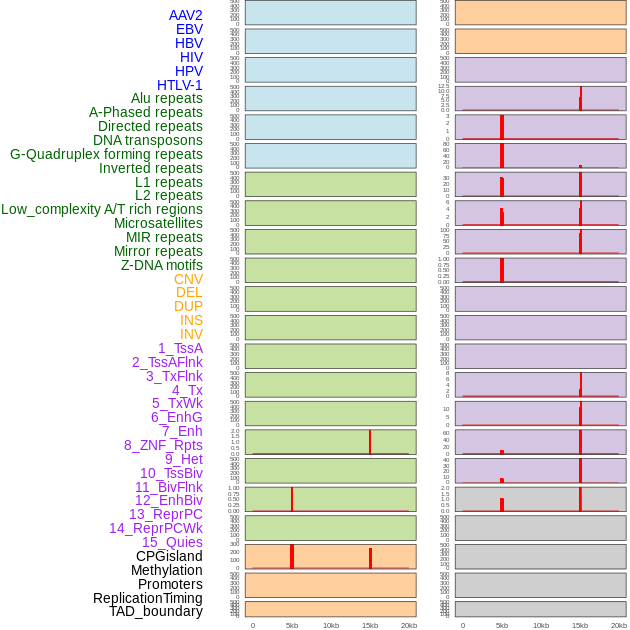 |
Top |
Fusion Protein Features for MYO1C-SMTNL2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:1381910/chr17:4510656) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000359786 | - | 11 | 32 | 139_148 | 431 | 1064.0 | Nucleotide binding | Note=ATP |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000359786 | - | 11 | 32 | 192_196 | 431 | 1064.0 | Nucleotide binding | Note=ATP |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000361007 | - | 11 | 32 | 139_148 | 396 | 1029.0 | Nucleotide binding | Note=ATP |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000361007 | - | 11 | 32 | 192_196 | 396 | 1029.0 | Nucleotide binding | Note=ATP |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000438665 | - | 11 | 32 | 139_148 | 412 | 1045.0 | Nucleotide binding | Note=ATP |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000438665 | - | 11 | 32 | 192_196 | 412 | 1045.0 | Nucleotide binding | Note=ATP |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000575158 | - | 11 | 32 | 139_148 | 396 | 1029.0 | Nucleotide binding | Note=ATP |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000575158 | - | 11 | 32 | 192_196 | 396 | 1029.0 | Nucleotide binding | Note=ATP |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000359786 | - | 10 | 32 | 139_148 | 404 | 1064.0 | Nucleotide binding | Note=ATP |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000359786 | - | 10 | 32 | 192_196 | 404 | 1064.0 | Nucleotide binding | Note=ATP |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000361007 | - | 10 | 32 | 139_148 | 369 | 1029.0 | Nucleotide binding | Note=ATP |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000361007 | - | 10 | 32 | 192_196 | 369 | 1029.0 | Nucleotide binding | Note=ATP |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000438665 | - | 10 | 32 | 139_148 | 385 | 1045.0 | Nucleotide binding | Note=ATP |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000438665 | - | 10 | 32 | 192_196 | 385 | 1045.0 | Nucleotide binding | Note=ATP |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000575158 | - | 10 | 32 | 139_148 | 369 | 1029.0 | Nucleotide binding | Note=ATP |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000575158 | - | 10 | 32 | 192_196 | 369 | 1029.0 | Nucleotide binding | Note=ATP |
| Tgene | SMTNL2 | chr17:1381704 | chr17:4510656 | ENST00000338859 | 6 | 8 | 351_458 | 275 | 318.0 | Domain | Calponin-homology (CH) | |
| Tgene | SMTNL2 | chr17:1381910 | chr17:4510656 | ENST00000338859 | 6 | 8 | 351_458 | 275 | 318.0 | Domain | Calponin-homology (CH) |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000359786 | - | 11 | 32 | 47_731 | 431 | 1064.0 | Domain | Myosin motor |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000359786 | - | 11 | 32 | 734_757 | 431 | 1064.0 | Domain | IQ 1 |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000359786 | - | 11 | 32 | 758_786 | 431 | 1064.0 | Domain | IQ 2 |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000359786 | - | 11 | 32 | 885_1059 | 431 | 1064.0 | Domain | TH1 |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000361007 | - | 11 | 32 | 47_731 | 396 | 1029.0 | Domain | Myosin motor |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000361007 | - | 11 | 32 | 734_757 | 396 | 1029.0 | Domain | IQ 1 |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000361007 | - | 11 | 32 | 758_786 | 396 | 1029.0 | Domain | IQ 2 |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000361007 | - | 11 | 32 | 885_1059 | 396 | 1029.0 | Domain | TH1 |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000438665 | - | 11 | 32 | 47_731 | 412 | 1045.0 | Domain | Myosin motor |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000438665 | - | 11 | 32 | 734_757 | 412 | 1045.0 | Domain | IQ 1 |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000438665 | - | 11 | 32 | 758_786 | 412 | 1045.0 | Domain | IQ 2 |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000438665 | - | 11 | 32 | 885_1059 | 412 | 1045.0 | Domain | TH1 |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000575158 | - | 11 | 32 | 47_731 | 396 | 1029.0 | Domain | Myosin motor |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000575158 | - | 11 | 32 | 734_757 | 396 | 1029.0 | Domain | IQ 1 |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000575158 | - | 11 | 32 | 758_786 | 396 | 1029.0 | Domain | IQ 2 |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000575158 | - | 11 | 32 | 885_1059 | 396 | 1029.0 | Domain | TH1 |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000359786 | - | 10 | 32 | 47_731 | 404 | 1064.0 | Domain | Myosin motor |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000359786 | - | 10 | 32 | 734_757 | 404 | 1064.0 | Domain | IQ 1 |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000359786 | - | 10 | 32 | 758_786 | 404 | 1064.0 | Domain | IQ 2 |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000359786 | - | 10 | 32 | 885_1059 | 404 | 1064.0 | Domain | TH1 |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000361007 | - | 10 | 32 | 47_731 | 369 | 1029.0 | Domain | Myosin motor |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000361007 | - | 10 | 32 | 734_757 | 369 | 1029.0 | Domain | IQ 1 |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000361007 | - | 10 | 32 | 758_786 | 369 | 1029.0 | Domain | IQ 2 |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000361007 | - | 10 | 32 | 885_1059 | 369 | 1029.0 | Domain | TH1 |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000438665 | - | 10 | 32 | 47_731 | 385 | 1045.0 | Domain | Myosin motor |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000438665 | - | 10 | 32 | 734_757 | 385 | 1045.0 | Domain | IQ 1 |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000438665 | - | 10 | 32 | 758_786 | 385 | 1045.0 | Domain | IQ 2 |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000438665 | - | 10 | 32 | 885_1059 | 385 | 1045.0 | Domain | TH1 |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000575158 | - | 10 | 32 | 47_731 | 369 | 1029.0 | Domain | Myosin motor |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000575158 | - | 10 | 32 | 734_757 | 369 | 1029.0 | Domain | IQ 1 |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000575158 | - | 10 | 32 | 758_786 | 369 | 1029.0 | Domain | IQ 2 |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000575158 | - | 10 | 32 | 885_1059 | 369 | 1029.0 | Domain | TH1 |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000359786 | - | 11 | 32 | 608_630 | 431 | 1064.0 | Region | Actin-binding |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000361007 | - | 11 | 32 | 608_630 | 396 | 1029.0 | Region | Actin-binding |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000438665 | - | 11 | 32 | 608_630 | 412 | 1045.0 | Region | Actin-binding |
| Hgene | MYO1C | chr17:1381704 | chr17:4510656 | ENST00000575158 | - | 11 | 32 | 608_630 | 396 | 1029.0 | Region | Actin-binding |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000359786 | - | 10 | 32 | 608_630 | 404 | 1064.0 | Region | Actin-binding |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000361007 | - | 10 | 32 | 608_630 | 369 | 1029.0 | Region | Actin-binding |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000438665 | - | 10 | 32 | 608_630 | 385 | 1045.0 | Region | Actin-binding |
| Hgene | MYO1C | chr17:1381910 | chr17:4510656 | ENST00000575158 | - | 10 | 32 | 608_630 | 369 | 1029.0 | Region | Actin-binding |
| Tgene | SMTNL2 | chr17:1381704 | chr17:4510656 | ENST00000338859 | 6 | 8 | 55_88 | 275 | 318.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | SMTNL2 | chr17:1381704 | chr17:4510656 | ENST00000389313 | 6 | 8 | 55_88 | 419 | 462.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | SMTNL2 | chr17:1381910 | chr17:4510656 | ENST00000338859 | 6 | 8 | 55_88 | 275 | 318.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | SMTNL2 | chr17:1381910 | chr17:4510656 | ENST00000389313 | 6 | 8 | 55_88 | 419 | 462.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | SMTNL2 | chr17:1381704 | chr17:4510656 | ENST00000389313 | 6 | 8 | 351_458 | 419 | 462.0 | Domain | Calponin-homology (CH) | |
| Tgene | SMTNL2 | chr17:1381910 | chr17:4510656 | ENST00000389313 | 6 | 8 | 351_458 | 419 | 462.0 | Domain | Calponin-homology (CH) |
Top |
Fusion Gene Sequence for MYO1C-SMTNL2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >56638_56638_1_MYO1C-SMTNL2_MYO1C_chr17_1381704_ENST00000359786_SMTNL2_chr17_4510656_ENST00000338859_length(transcript)=2579nt_BP=1620nt GGAGCCCAGGGTCCCCCATGGGTGGCTGCATCGGCTCCTCTGGTTACCTGGGACAACAGGCATTTCCTCCATCCCCAGCCCCTCCTCTTC CTGCTCCAACCATCCGGGCCAAGAGGCCTCCCAGTCAGACCTGGACGGCTCCAGCCGTGTCCTGAGGAGCTGGACCAGCCACATCCCCTG GGGCTGCAGTTGAAGCAGAACCAAGTGGCCATCCCGGCGTTAGACCGTAGGTTCCTGGTCCCGGAGTGGTCGGAGCCCGCCAGTGGGCAG GCAGCTCTTGCTCACAGGCCGCGGTGCCCAGGCCGCTGGCTCTCCGCAGGGCGGAATGGCGCTGCAAGTGGAGCTGGTACCCACCGGGGA GATCATCCGCGTGGTTCATCCCCACAGGCCCTGCAAGCTTGCCCTGGGCAGTGACGGGGTTCGGGTGACCATGGAGAGTGCGCTCACCGC CCGTGACCGGGTGGGGGTGCAGGATTTCGTGCTGCTGGAGAACTTCACCAGCGAGGCCGCCTTCATCGAGAACCTGCGGCGGCGATTTCG GGAGAATCTCATCTACACCTACATTGGCCCCGTCCTGGTCTCTGTCAATCCCTACCGGGACCTGCAGATCTACAGCCGGCAGCATATGGA GCGTTACCGTGGCGTCAGCTTCTATGAAGTGCCCCCTCACCTGTTTGCCGTGGCGGACACTGTGTACCGAGCACTGCGCACGGAGCGTCG GGACCAGGCTGTGATGATCTCTGGGGAGAGCGGGGCAGGCAAGACCGAGGCCACCAAGAGGCTGCTGCAGTTCTATGCAGAGACCTGCCC AGCCCCCGAGCGCGGAGGTGCCGTGCGGGACCGGCTGCTACAGAGCAACCCGGTGCTGGAGGCCTTTGGAAATGCCAAGACCCTCCGGAA CGATAACTCCAGCAGGTTCGGGAAGTACATGGATGTGCAGTTTGACTTCAAGGGTGCCCCCGTGGGTGGCCACATCCTCAGTTACCTCCT GGAAAAGTCACGAGTGGTGCACCAGAATCATGGGGAGCGGAACTTCCACATCTTCTACCAGCTGCTGGAGGGGGGTGAGGAGGAGACTCT TCGCAGGCTGGGCTTGGAACGGAACCCCCAGAGCTACCTGTACCTGGTGAAGGGCCAGTGTGCCAAAGTCTCCTCCATCAACGACAAGAG TGACTGGAAGGTCGTCAGGAAGGCTCTGACAGTCATTGATTTCACCGAGGATGAAGTGGAGGACCTGCTGAGCATCGTGGCCAGCGTCCT TCATTTGGGCAACATCCACTTTGCTGCCAACGAGGAGAGCAATGCCCAGGTCACCACCGAGAACCAGCTCAAGTATCTGACCAGGCTCCT CAGCGTGGAAGGCTCGACGCTGCGAGAAGCCCTGACACACAGGAAGATCATCGCCAAGGGGGAGGAGCTCCTGAGCCCGCTGAACCTGGA GCAGGCCGCGTACGCACGAGACGCCCTCGCCAAGGCTGTGTACAGCCGCACTTTTACCTGGCTCGTCGGGAAGATCAACAGGTCGCTGGC CTCCAAGGACGTGGAGAGCCCCAGCTGGCGGAGCACCACGGTTCTCGGGCTCCTGGATATTTATGGCTTTGAAGTGTTTCAGCATAACAG GAATCTGGCCAACTGTGAGCGCCTCATCGAAGTGGAGGACATGATGGTGATGGGCCGCAAGCCGGACCCCATGTGTGTCTTCACCTACGT CCAGTCGCTGTACAACCACCTGCGTCGCTTCGAGTAAAGCCCCTGAGCCTGGATTGCCAAAGAGCAGCCCCAGGAAGAGGCCGGGGGTCC GCTTGCGATTCCCCAGCCAGGATGCCCCCAGGAGCCTTGCCGTTTGGTGTGAGCGCGCTGTTTGTTCTGTGGCATGTGACGGCACTCCCC TTCGAGCCCAGCTGTGTTACTGATTAAAAGTACTGCTGAGCTGTGGTCCGACAGCACTGATCACAGCCAAGGGCTTGGAGGAAAAGGAAA AATTATGAGAGAGAGAGAGACATTGGTGCTAAGTAATGATCTTCCTAAAGAAATGCTTGTGTTTATAGCTTCCAGAATGCTAATCTACAA TTTTCCCTCTGGTGAATTCGATACATCGGCTTTACAGGGTTACAGTGATTACCAAGTGTTTTTTTTTATCAAAATACCCAGAGTTTTTTA CTTCCTCACGCGATTGTAGGTTCCTCTCCTCCCTCCCTCTGGGCCACTGCCAGGAAACAGAGAGACCGCTTAATCAGCAGCTTGACAAAG AAGACCTCAAGTCTTGGGAAGAAACAGTTTAATCACTCCCAAGTCCTGGGCAACAGATGACCTTCAAGTCACCTCCGCTCTCCGGGGAGA TGGGAAGGCTCTCCTCTCGGTCCCAAAGTCCTCCTGTTCTTCCCAGGAGGCCCCACAAGTGTTTGGCTAAGCACAGGCTCTCGGGAATTT AACACTTTTGGGGAAGGAATAGGCCCTTTGTGCTGAGAGAGAGTTTTTATTCACATCTTTTTTAGGGGATTTGCTGCAGATATTTATAAA >56638_56638_1_MYO1C-SMTNL2_MYO1C_chr17_1381704_ENST00000359786_SMTNL2_chr17_4510656_ENST00000338859_length(amino acids)=489AA_BP=447 MLTGRGAQAAGSPQGGMALQVELVPTGEIIRVVHPHRPCKLALGSDGVRVTMESALTARDRVGVQDFVLLENFTSEAAFIENLRRRFREN LIYTYIGPVLVSVNPYRDLQIYSRQHMERYRGVSFYEVPPHLFAVADTVYRALRTERRDQAVMISGESGAGKTEATKRLLQFYAETCPAP ERGGAVRDRLLQSNPVLEAFGNAKTLRNDNSSRFGKYMDVQFDFKGAPVGGHILSYLLEKSRVVHQNHGERNFHIFYQLLEGGEEETLRR LGLERNPQSYLYLVKGQCAKVSSINDKSDWKVVRKALTVIDFTEDEVEDLLSIVASVLHLGNIHFAANEESNAQVTTENQLKYLTRLLSV EGSTLREALTHRKIIAKGEELLSPLNLEQAAYARDALAKAVYSRTFTWLVGKINRSLASKDVESPSWRSTTVLGLLDIYGFEVFQHNRNL -------------------------------------------------------------- >56638_56638_2_MYO1C-SMTNL2_MYO1C_chr17_1381704_ENST00000361007_SMTNL2_chr17_4510656_ENST00000338859_length(transcript)=2258nt_BP=1299nt AGAGTGCGCCCGGGCCGCGGATCCCCAAGCCCGCCACGCCGCCCAGCCCGGCCCCGCCCGCGCCGGAAGAGCCACGCCGGCCCTGGGCAG TGACGGGGTTCGGGTGACCATGGAGAGTGCGCTCACCGCCCGTGACCGGGTGGGGGTGCAGGATTTCGTGCTGCTGGAGAACTTCACCAG CGAGGCCGCCTTCATCGAGAACCTGCGGCGGCGATTTCGGGAGAATCTCATCTACACCTACATTGGCCCCGTCCTGGTCTCTGTCAATCC CTACCGGGACCTGCAGATCTACAGCCGGCAGCATATGGAGCGTTACCGTGGCGTCAGCTTCTATGAAGTGCCCCCTCACCTGTTTGCCGT GGCGGACACTGTGTACCGAGCACTGCGCACGGAGCGTCGGGACCAGGCTGTGATGATCTCTGGGGAGAGCGGGGCAGGCAAGACCGAGGC CACCAAGAGGCTGCTGCAGTTCTATGCAGAGACCTGCCCAGCCCCCGAGCGCGGAGGTGCCGTGCGGGACCGGCTGCTACAGAGCAACCC GGTGCTGGAGGCCTTTGGAAATGCCAAGACCCTCCGGAACGATAACTCCAGCAGGTTCGGGAAGTACATGGATGTGCAGTTTGACTTCAA GGGTGCCCCCGTGGGTGGCCACATCCTCAGTTACCTCCTGGAAAAGTCACGAGTGGTGCACCAGAATCATGGGGAGCGGAACTTCCACAT CTTCTACCAGCTGCTGGAGGGGGGTGAGGAGGAGACTCTTCGCAGGCTGGGCTTGGAACGGAACCCCCAGAGCTACCTGTACCTGGTGAA GGGCCAGTGTGCCAAAGTCTCCTCCATCAACGACAAGAGTGACTGGAAGGTCGTCAGGAAGGCTCTGACAGTCATTGATTTCACCGAGGA TGAAGTGGAGGACCTGCTGAGCATCGTGGCCAGCGTCCTTCATTTGGGCAACATCCACTTTGCTGCCAACGAGGAGAGCAATGCCCAGGT CACCACCGAGAACCAGCTCAAGTATCTGACCAGGCTCCTCAGCGTGGAAGGCTCGACGCTGCGAGAAGCCCTGACACACAGGAAGATCAT CGCCAAGGGGGAGGAGCTCCTGAGCCCGCTGAACCTGGAGCAGGCCGCGTACGCACGAGACGCCCTCGCCAAGGCTGTGTACAGCCGCAC TTTTACCTGGCTCGTCGGGAAGATCAACAGGTCGCTGGCCTCCAAGGACGTGGAGAGCCCCAGCTGGCGGAGCACCACGGTTCTCGGGCT CCTGGATATTTATGGCTTTGAAGTGTTTCAGCATAACAGGAATCTGGCCAACTGTGAGCGCCTCATCGAAGTGGAGGACATGATGGTGAT GGGCCGCAAGCCGGACCCCATGTGTGTCTTCACCTACGTCCAGTCGCTGTACAACCACCTGCGTCGCTTCGAGTAAAGCCCCTGAGCCTG GATTGCCAAAGAGCAGCCCCAGGAAGAGGCCGGGGGTCCGCTTGCGATTCCCCAGCCAGGATGCCCCCAGGAGCCTTGCCGTTTGGTGTG AGCGCGCTGTTTGTTCTGTGGCATGTGACGGCACTCCCCTTCGAGCCCAGCTGTGTTACTGATTAAAAGTACTGCTGAGCTGTGGTCCGA CAGCACTGATCACAGCCAAGGGCTTGGAGGAAAAGGAAAAATTATGAGAGAGAGAGAGACATTGGTGCTAAGTAATGATCTTCCTAAAGA AATGCTTGTGTTTATAGCTTCCAGAATGCTAATCTACAATTTTCCCTCTGGTGAATTCGATACATCGGCTTTACAGGGTTACAGTGATTA CCAAGTGTTTTTTTTTATCAAAATACCCAGAGTTTTTTACTTCCTCACGCGATTGTAGGTTCCTCTCCTCCCTCCCTCTGGGCCACTGCC AGGAAACAGAGAGACCGCTTAATCAGCAGCTTGACAAAGAAGACCTCAAGTCTTGGGAAGAAACAGTTTAATCACTCCCAAGTCCTGGGC AACAGATGACCTTCAAGTCACCTCCGCTCTCCGGGGAGATGGGAAGGCTCTCCTCTCGGTCCCAAAGTCCTCCTGTTCTTCCCAGGAGGC CCCACAAGTGTTTGGCTAAGCACAGGCTCTCGGGAATTTAACACTTTTGGGGAAGGAATAGGCCCTTTGTGCTGAGAGAGAGTTTTTATT CACATCTTTTTTAGGGGATTTGCTGCAGATATTTATAAAAAGTAACTCCCTCTGTACCACTGACCCATTTATACATAAAAAAGATGTGTT >56638_56638_2_MYO1C-SMTNL2_MYO1C_chr17_1381704_ENST00000361007_SMTNL2_chr17_4510656_ENST00000338859_length(amino acids)=447AA_BP=405 MGSDGVRVTMESALTARDRVGVQDFVLLENFTSEAAFIENLRRRFRENLIYTYIGPVLVSVNPYRDLQIYSRQHMERYRGVSFYEVPPHL FAVADTVYRALRTERRDQAVMISGESGAGKTEATKRLLQFYAETCPAPERGGAVRDRLLQSNPVLEAFGNAKTLRNDNSSRFGKYMDVQF DFKGAPVGGHILSYLLEKSRVVHQNHGERNFHIFYQLLEGGEEETLRRLGLERNPQSYLYLVKGQCAKVSSINDKSDWKVVRKALTVIDF TEDEVEDLLSIVASVLHLGNIHFAANEESNAQVTTENQLKYLTRLLSVEGSTLREALTHRKIIAKGEELLSPLNLEQAAYARDALAKAVY -------------------------------------------------------------- >56638_56638_3_MYO1C-SMTNL2_MYO1C_chr17_1381704_ENST00000438665_SMTNL2_chr17_4510656_ENST00000338859_length(transcript)=2420nt_BP=1461nt GGACCCCCCGCCCCGACACCCCCACCTCCACCAACCCCACCTGGAGGGGACCCCAGCCCGGCCGCGCGGGAGGCGGGGGAGGCCGGGGAA GCCCCGCGCTTCAGGGGCCCGGGTGGGGGCGGTGCGGGGGGCGGGCCCTCCCTCGCTGACGCCGCCGCCACCGCCGCCGAGCCCGGGACC CGGAGCGCGGGCAGCGGAGCGGGCGCCAGGCCGGCCGGGCAGGATGCGCTACCGGGCGTCGGCCCTGGGCAGTGACGGGGTTCGGGTGAC CATGGAGAGTGCGCTCACCGCCCGTGACCGGGTGGGGGTGCAGGATTTCGTGCTGCTGGAGAACTTCACCAGCGAGGCCGCCTTCATCGA GAACCTGCGGCGGCGATTTCGGGAGAATCTCATCTACACCTACATTGGCCCCGTCCTGGTCTCTGTCAATCCCTACCGGGACCTGCAGAT CTACAGCCGGCAGCATATGGAGCGTTACCGTGGCGTCAGCTTCTATGAAGTGCCCCCTCACCTGTTTGCCGTGGCGGACACTGTGTACCG AGCACTGCGCACGGAGCGTCGGGACCAGGCTGTGATGATCTCTGGGGAGAGCGGGGCAGGCAAGACCGAGGCCACCAAGAGGCTGCTGCA GTTCTATGCAGAGACCTGCCCAGCCCCCGAGCGCGGAGGTGCCGTGCGGGACCGGCTGCTACAGAGCAACCCGGTGCTGGAGGCCTTTGG AAATGCCAAGACCCTCCGGAACGATAACTCCAGCAGGTTCGGGAAGTACATGGATGTGCAGTTTGACTTCAAGGGTGCCCCCGTGGGTGG CCACATCCTCAGTTACCTCCTGGAAAAGTCACGAGTGGTGCACCAGAATCATGGGGAGCGGAACTTCCACATCTTCTACCAGCTGCTGGA GGGGGGTGAGGAGGAGACTCTTCGCAGGCTGGGCTTGGAACGGAACCCCCAGAGCTACCTGTACCTGGTGAAGGGCCAGTGTGCCAAAGT CTCCTCCATCAACGACAAGAGTGACTGGAAGGTCGTCAGGAAGGCTCTGACAGTCATTGATTTCACCGAGGATGAAGTGGAGGACCTGCT GAGCATCGTGGCCAGCGTCCTTCATTTGGGCAACATCCACTTTGCTGCCAACGAGGAGAGCAATGCCCAGGTCACCACCGAGAACCAGCT CAAGTATCTGACCAGGCTCCTCAGCGTGGAAGGCTCGACGCTGCGAGAAGCCCTGACACACAGGAAGATCATCGCCAAGGGGGAGGAGCT CCTGAGCCCGCTGAACCTGGAGCAGGCCGCGTACGCACGAGACGCCCTCGCCAAGGCTGTGTACAGCCGCACTTTTACCTGGCTCGTCGG GAAGATCAACAGGTCGCTGGCCTCCAAGGACGTGGAGAGCCCCAGCTGGCGGAGCACCACGGTTCTCGGGCTCCTGGATATTTATGGCTT TGAAGTGTTTCAGCATAACAGGAATCTGGCCAACTGTGAGCGCCTCATCGAAGTGGAGGACATGATGGTGATGGGCCGCAAGCCGGACCC CATGTGTGTCTTCACCTACGTCCAGTCGCTGTACAACCACCTGCGTCGCTTCGAGTAAAGCCCCTGAGCCTGGATTGCCAAAGAGCAGCC CCAGGAAGAGGCCGGGGGTCCGCTTGCGATTCCCCAGCCAGGATGCCCCCAGGAGCCTTGCCGTTTGGTGTGAGCGCGCTGTTTGTTCTG TGGCATGTGACGGCACTCCCCTTCGAGCCCAGCTGTGTTACTGATTAAAAGTACTGCTGAGCTGTGGTCCGACAGCACTGATCACAGCCA AGGGCTTGGAGGAAAAGGAAAAATTATGAGAGAGAGAGAGACATTGGTGCTAAGTAATGATCTTCCTAAAGAAATGCTTGTGTTTATAGC TTCCAGAATGCTAATCTACAATTTTCCCTCTGGTGAATTCGATACATCGGCTTTACAGGGTTACAGTGATTACCAAGTGTTTTTTTTTAT CAAAATACCCAGAGTTTTTTACTTCCTCACGCGATTGTAGGTTCCTCTCCTCCCTCCCTCTGGGCCACTGCCAGGAAACAGAGAGACCGC TTAATCAGCAGCTTGACAAAGAAGACCTCAAGTCTTGGGAAGAAACAGTTTAATCACTCCCAAGTCCTGGGCAACAGATGACCTTCAAGT CACCTCCGCTCTCCGGGGAGATGGGAAGGCTCTCCTCTCGGTCCCAAAGTCCTCCTGTTCTTCCCAGGAGGCCCCACAAGTGTTTGGCTA AGCACAGGCTCTCGGGAATTTAACACTTTTGGGGAAGGAATAGGCCCTTTGTGCTGAGAGAGAGTTTTTATTCACATCTTTTTTAGGGGA >56638_56638_3_MYO1C-SMTNL2_MYO1C_chr17_1381704_ENST00000438665_SMTNL2_chr17_4510656_ENST00000338859_length(amino acids)=515AA_BP=473 MEGTPARPRGRRGRPGKPRASGARVGAVRGAGPPSLTPPPPPPSPGPGARAAERAPGRPGRMRYRASALGSDGVRVTMESALTARDRVGV QDFVLLENFTSEAAFIENLRRRFRENLIYTYIGPVLVSVNPYRDLQIYSRQHMERYRGVSFYEVPPHLFAVADTVYRALRTERRDQAVMI SGESGAGKTEATKRLLQFYAETCPAPERGGAVRDRLLQSNPVLEAFGNAKTLRNDNSSRFGKYMDVQFDFKGAPVGGHILSYLLEKSRVV HQNHGERNFHIFYQLLEGGEEETLRRLGLERNPQSYLYLVKGQCAKVSSINDKSDWKVVRKALTVIDFTEDEVEDLLSIVASVLHLGNIH FAANEESNAQVTTENQLKYLTRLLSVEGSTLREALTHRKIIAKGEELLSPLNLEQAAYARDALAKAVYSRTFTWLVGKINRSLASKDVES -------------------------------------------------------------- >56638_56638_4_MYO1C-SMTNL2_MYO1C_chr17_1381704_ENST00000545534_SMTNL2_chr17_4510656_ENST00000338859_length(transcript)=2425nt_BP=1466nt GCCCCCGGCCAGAAATCTGGGCCTTTCGTGGTCAGAGGACACCGGCGGCCCCTTTGGGATCCACCCCCAATCACCACCACCTTGTTCTTC TCTGACCCTGGATACTCTGGGTGTCCATTTTTTGGAGCCCTGAGACCCTGTTTGAAGAGGGATCCATCTCAGCCCTGGTTGTGGGGCGAG GGTCTTGCTCAGCCTGCACCACCCTCCACCCTCCAGTAGGACCCAGAGGCAGCTGGGCTTTCCATGGCCCTGGGCAGTGACGGGGTTCGG GTGACCATGGAGAGTGCGCTCACCGCCCGTGACCGGGTGGGGGTGCAGGATTTCGTGCTGCTGGAGAACTTCACCAGCGAGGCCGCCTTC ATCGAGAACCTGCGGCGGCGATTTCGGGAGAATCTCATCTACACCTACATTGGCCCCGTCCTGGTCTCTGTCAATCCCTACCGGGACCTG CAGATCTACAGCCGGCAGCATATGGAGCGTTACCGTGGCGTCAGCTTCTATGAAGTGCCCCCTCACCTGTTTGCCGTGGCGGACACTGTG TACCGAGCACTGCGCACGGAGCGTCGGGACCAGGCTGTGATGATCTCTGGGGAGAGCGGGGCAGGCAAGACCGAGGCCACCAAGAGGCTG CTGCAGTTCTATGCAGAGACCTGCCCAGCCCCCGAGCGCGGAGGTGCCGTGCGGGACCGGCTGCTACAGAGCAACCCGGTGCTGGAGGCC TTTGGAAATGCCAAGACCCTCCGGAACGATAACTCCAGCAGGTTCGGGAAGTACATGGATGTGCAGTTTGACTTCAAGGGTGCCCCCGTG GGTGGCCACATCCTCAGTTACCTCCTGGAAAAGTCACGAGTGGTGCACCAGAATCATGGGGAGCGGAACTTCCACATCTTCTACCAGCTG CTGGAGGGGGGTGAGGAGGAGACTCTTCGCAGGCTGGGCTTGGAACGGAACCCCCAGAGCTACCTGTACCTGGTGAAGGGCCAGTGTGCC AAAGTCTCCTCCATCAACGACAAGAGTGACTGGAAGGTCGTCAGGAAGGCTCTGACAGTCATTGATTTCACCGAGGATGAAGTGGAGGAC CTGCTGAGCATCGTGGCCAGCGTCCTTCATTTGGGCAACATCCACTTTGCTGCCAACGAGGAGAGCAATGCCCAGGTCACCACCGAGAAC CAGCTCAAGTATCTGACCAGGCTCCTCAGCGTGGAAGGCTCGACGCTGCGAGAAGCCCTGACACACAGGAAGATCATCGCCAAGGGGGAG GAGCTCCTGAGCCCGCTGAACCTGGAGCAGGCCGCGTACGCACGAGACGCCCTCGCCAAGGCTGTGTACAGCCGCACTTTTACCTGGCTC GTCGGGAAGATCAACAGGTCGCTGGCCTCCAAGGACGTGGAGAGCCCCAGCTGGCGGAGCACCACGGTTCTCGGGCTCCTGGATATTTAT GGCTTTGAAGTGTTTCAGCATAACAGGAATCTGGCCAACTGTGAGCGCCTCATCGAAGTGGAGGACATGATGGTGATGGGCCGCAAGCCG GACCCCATGTGTGTCTTCACCTACGTCCAGTCGCTGTACAACCACCTGCGTCGCTTCGAGTAAAGCCCCTGAGCCTGGATTGCCAAAGAG CAGCCCCAGGAAGAGGCCGGGGGTCCGCTTGCGATTCCCCAGCCAGGATGCCCCCAGGAGCCTTGCCGTTTGGTGTGAGCGCGCTGTTTG TTCTGTGGCATGTGACGGCACTCCCCTTCGAGCCCAGCTGTGTTACTGATTAAAAGTACTGCTGAGCTGTGGTCCGACAGCACTGATCAC AGCCAAGGGCTTGGAGGAAAAGGAAAAATTATGAGAGAGAGAGAGACATTGGTGCTAAGTAATGATCTTCCTAAAGAAATGCTTGTGTTT ATAGCTTCCAGAATGCTAATCTACAATTTTCCCTCTGGTGAATTCGATACATCGGCTTTACAGGGTTACAGTGATTACCAAGTGTTTTTT TTTATCAAAATACCCAGAGTTTTTTACTTCCTCACGCGATTGTAGGTTCCTCTCCTCCCTCCCTCTGGGCCACTGCCAGGAAACAGAGAG ACCGCTTAATCAGCAGCTTGACAAAGAAGACCTCAAGTCTTGGGAAGAAACAGTTTAATCACTCCCAAGTCCTGGGCAACAGATGACCTT CAAGTCACCTCCGCTCTCCGGGGAGATGGGAAGGCTCTCCTCTCGGTCCCAAAGTCCTCCTGTTCTTCCCAGGAGGCCCCACAAGTGTTT GGCTAAGCACAGGCTCTCGGGAATTTAACACTTTTGGGGAAGGAATAGGCCCTTTGTGCTGAGAGAGAGTTTTTATTCACATCTTTTTTA >56638_56638_4_MYO1C-SMTNL2_MYO1C_chr17_1381704_ENST00000545534_SMTNL2_chr17_4510656_ENST00000338859_length(amino acids)=449AA_BP=407 MALGSDGVRVTMESALTARDRVGVQDFVLLENFTSEAAFIENLRRRFRENLIYTYIGPVLVSVNPYRDLQIYSRQHMERYRGVSFYEVPP HLFAVADTVYRALRTERRDQAVMISGESGAGKTEATKRLLQFYAETCPAPERGGAVRDRLLQSNPVLEAFGNAKTLRNDNSSRFGKYMDV QFDFKGAPVGGHILSYLLEKSRVVHQNHGERNFHIFYQLLEGGEEETLRRLGLERNPQSYLYLVKGQCAKVSSINDKSDWKVVRKALTVI DFTEDEVEDLLSIVASVLHLGNIHFAANEESNAQVTTENQLKYLTRLLSVEGSTLREALTHRKIIAKGEELLSPLNLEQAAYARDALAKA -------------------------------------------------------------- >56638_56638_5_MYO1C-SMTNL2_MYO1C_chr17_1381910_ENST00000575158_SMTNL2_chr17_4510656_ENST00000389313_length(transcript)=2238nt_BP=1284nt GTATTTGGACATTGGTTCTATTTTTAACTTTCCCGGGACTCGAGTTCGGGGGTTGGGGGAGGAAAATCCCGCTTCCTCTGGGACCCGCAG GCCTCGGTGCCCGCCCAGCGCCCAGGCCCGCCGCGGAGTGTAGTCCCAGGCCCGGAAGCCCTGGGCAGTGACGGGGTTCGGGTGACCATG GAGAGTGCGCTCACCGCCCGTGACCGGGTGGGGGTGCAGGATTTCGTGCTGCTGGAGAACTTCACCAGCGAGGCCGCCTTCATCGAGAAC CTGCGGCGGCGATTTCGGGAGAATCTCATCTACACCTACATTGGCCCCGTCCTGGTCTCTGTCAATCCCTACCGGGACCTGCAGATCTAC AGCCGGCAGCATATGGAGCGTTACCGTGGCGTCAGCTTCTATGAAGTGCCCCCTCACCTGTTTGCCGTGGCGGACACTGTGTACCGAGCA CTGCGCACGGAGCGTCGGGACCAGGCTGTGATGATCTCTGGGGAGAGCGGGGCAGGCAAGACCGAGGCCACCAAGAGGCTGCTGCAGTTC TATGCAGAGACCTGCCCAGCCCCCGAGCGCGGAGGTGCCGTGCGGGACCGGCTGCTACAGAGCAACCCGGTGCTGGAGGCCTTTGGAAAT GCCAAGACCCTCCGGAACGATAACTCCAGCAGGTTCGGGAAGTACATGGATGTGCAGTTTGACTTCAAGGGTGCCCCCGTGGGTGGCCAC ATCCTCAGTTACCTCCTGGAAAAGTCACGAGTGGTGCACCAGAATCATGGGGAGCGGAACTTCCACATCTTCTACCAGCTGCTGGAGGGG GGTGAGGAGGAGACTCTTCGCAGGCTGGGCTTGGAACGGAACCCCCAGAGCTACCTGTACCTGGTGAAGGGCCAGTGTGCCAAAGTCTCC TCCATCAACGACAAGAGTGACTGGAAGGTCGTCAGGAAGGCTCTGACAGTCATTGATTTCACCGAGGATGAAGTGGAGGACCTGCTGAGC ATCGTGGCCAGCGTCCTTCATTTGGGCAACATCCACTTTGCTGCCAACGAGGAGAGCAATGCCCAGGTCACCACCGAGAACCAGCTCAAG TATCTGACCAGGCTCCTCAGCGTGGAAGGCTCGACGCTGCGAGAAGCCCTGACACACAGGAAGATCATCGCCAAGGGGGAGGAGCTCCTG AGCCCGCTGAACCTGGAGCAGGCCGCGTACGCACGAGACGCCCTCGCCAAGGCTGTGTACAGCCGCACTTTTACCTGGCTCGTCGGGAAG ATCAACAGGTCGCTGGCCTCCAAGGAATCTGGCCAACTGTGAGCGCCTCATCGAAGTGGAGGACATGATGGTGATGGGCCGCAAGCCGGA CCCCATGTGTGTCTTCACCTACGTCCAGTCGCTGTACAACCACCTGCGTCGCTTCGAGTAAAGCCCCTGAGCCTGGATTGCCAAAGAGCA GCCCCAGGAAGAGGCCGGGGGTCCGCTTGCGATTCCCCAGCCAGGATGCCCCCAGGAGCCTTGCCGTTTGGTGTGAGCGCGCTGTTTGTT CTGTGGCATGTGACGGCACTCCCCTTCGAGCCCAGCTGTGTTACTGATTAAAAGTACTGCTGAGCTGTGGTCCGACAGCACTGATCACAG CCAAGGGCTTGGAGGAAAAGGAAAAATTATGAGAGAGAGAGAGACATTGGTGCTAAGTAATGATCTTCCTAAAGAAATGCTTGTGTTTAT AGCTTCCAGAATGCTAATCTACAATTTTCCCTCTGGTGAATTCGATACATCGGCTTTACAGGGTTACAGTGATTACCAAGTGTTTTTTTT TATCAAAATACCCAGAGTTTTTTACTTCCTCACGCGATTGTAGGTTCCTCTCCTCCCTCCCTCTGGGCCACTGCCAGGAAACAGAGAGAC CGCTTAATCAGCAGCTTGACAAAGAAGACCTCAAGTCTTGGGAAGAAACAGTTTAATCACTCCCAAGTCCTGGGCAACAGATGACCTTCA AGTCACCTCCGCTCTCCGGGGAGATGGGAAGGCTCTCCTCTCGGTCCCAAAGTCCTCCTGTTCTTCCCAGGAGGCCCCACAAGTGTTTGG CTAAGCACAGGCTCTCGGGAATTTAACACTTTTGGGGAAGGAATAGGCCCTTTGTGCTGAGAGAGAGTTTTTATTCACATCTTTTTTAGG >56638_56638_5_MYO1C-SMTNL2_MYO1C_chr17_1381910_ENST00000575158_SMTNL2_chr17_4510656_ENST00000389313_length(amino acids)=383AA_BP= MGSDGVRVTMESALTARDRVGVQDFVLLENFTSEAAFIENLRRRFRENLIYTYIGPVLVSVNPYRDLQIYSRQHMERYRGVSFYEVPPHL FAVADTVYRALRTERRDQAVMISGESGAGKTEATKRLLQFYAETCPAPERGGAVRDRLLQSNPVLEAFGNAKTLRNDNSSRFGKYMDVQF DFKGAPVGGHILSYLLEKSRVVHQNHGERNFHIFYQLLEGGEEETLRRLGLERNPQSYLYLVKGQCAKVSSINDKSDWKVVRKALTVIDF TEDEVEDLLSIVASVLHLGNIHFAANEESNAQVTTENQLKYLTRLLSVEGSTLREALTHRKIIAKGEELLSPLNLEQAAYARDALAKAVY -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for MYO1C-SMTNL2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for MYO1C-SMTNL2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for MYO1C-SMTNL2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
