
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:AAK1-FNTA (FusionGDB2 ID:57) |
Fusion Gene Summary for AAK1-FNTA |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: AAK1-FNTA | Fusion gene ID: 57 | Hgene | Tgene | Gene symbol | AAK1 | FNTA | Gene ID | 112268437 | 2339 |
| Gene name | uncharacterized protein FLJ45252 | farnesyltransferase, CAAX box, alpha | |
| Synonyms | AAK1 | FPTA|PGGT1A|PTAR2 | |
| Cytomap | - | 8p11.21 | |
| Type of gene | protein-coding | protein-coding | |
| Description | uncharacterized protein FLJ45252alternative protein AAK1 | protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alphaFTase-alphaGGTase-I-alphafarnesyl-protein transferase alpha-subunitprotein prenyltransferase alpha subunit repeat containing 2ras proteins prenyltransferase subunit alphatype | |
| Modification date | 20200303 | 20200313 | |
| UniProtAcc | Q2M2I8 | P49354 | |
| Ensembl transtripts involved in fusion gene | ENST00000406297, ENST00000409085, ENST00000409068, ENST00000470281, | ENST00000524546, ENST00000529687, ENST00000342116, ENST00000302279, | |
| Fusion gene scores | * DoF score | 19 X 13 X 11=2717 | 10 X 11 X 7=770 |
| # samples | 22 | 14 | |
| ** MAII score | log2(22/2717*10)=-3.62643913669732 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(14/770*10)=-2.4594316186373 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: AAK1 [Title/Abstract] AND FNTA [Title/Abstract] AND fusion [Title/Abstract] | ||
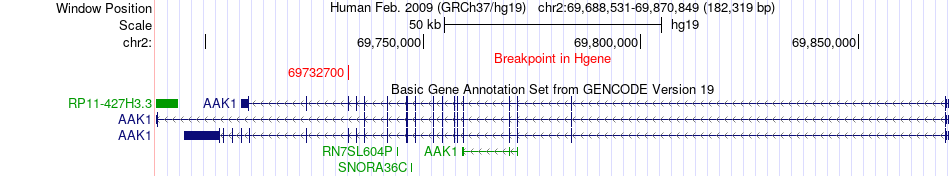
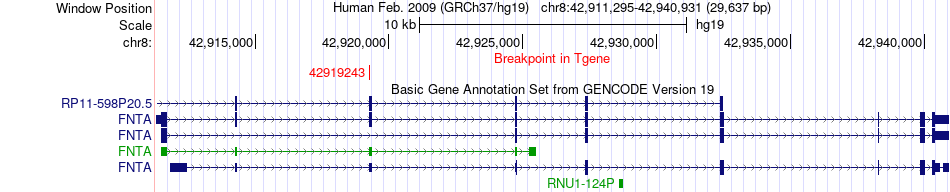
| Most frequent breakpoint | AAK1(69732700)-FNTA(42919243), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | FNTA | GO:0018343 | protein farnesylation | 16893176|19228685 |
| Tgene | FNTA | GO:0018344 | protein geranylgeranylation | 16893176 |
| Tgene | FNTA | GO:0090044 | positive regulation of tubulin deacetylation | 19228685 |
| Tgene | FNTA | GO:0090045 | positive regulation of deacetylase activity | 19228685 |
 Fusion gene breakpoints across AAK1 (5'-gene) Fusion gene breakpoints across AAK1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across FNTA (3'-gene) Fusion gene breakpoints across FNTA (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ESCA | TCGA-L5-A8NQ | AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + |
Top |
Fusion Gene ORF analysis for AAK1-FNTA |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000406297 | ENST00000524546 | AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + |
| 5CDS-3UTR | ENST00000409085 | ENST00000524546 | AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + |
| 5CDS-5UTR | ENST00000406297 | ENST00000529687 | AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + |
| 5CDS-5UTR | ENST00000409085 | ENST00000529687 | AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + |
| 5CDS-intron | ENST00000406297 | ENST00000342116 | AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + |
| 5CDS-intron | ENST00000409085 | ENST00000342116 | AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + |
| In-frame | ENST00000406297 | ENST00000302279 | AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + |
| In-frame | ENST00000409085 | ENST00000302279 | AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + |
| intron-3CDS | ENST00000409068 | ENST00000302279 | AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + |
| intron-3CDS | ENST00000470281 | ENST00000302279 | AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + |
| intron-3UTR | ENST00000409068 | ENST00000524546 | AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + |
| intron-3UTR | ENST00000470281 | ENST00000524546 | AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + |
| intron-5UTR | ENST00000409068 | ENST00000529687 | AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + |
| intron-5UTR | ENST00000470281 | ENST00000529687 | AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + |
| intron-intron | ENST00000409068 | ENST00000342116 | AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + |
| intron-intron | ENST00000470281 | ENST00000342116 | AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000409085 | AAK1 | chr2 | 69732700 | - | ENST00000302279 | FNTA | chr8 | 42919243 | + | 4006 | 2646 | 278 | 3499 | 1073 |
| ENST00000406297 | AAK1 | chr2 | 69732700 | - | ENST00000302279 | FNTA | chr8 | 42919243 | + | 4006 | 2646 | 278 | 3499 | 1073 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000409085 | ENST00000302279 | AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + | 0.004193157 | 0.9958068 |
| ENST00000406297 | ENST00000302279 | AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + | 0.004193157 | 0.9958068 |
Top |
Fusion Genomic Features for AAK1-FNTA |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + | 1.75E-08 | 1 |
| AAK1 | chr2 | 69732700 | - | FNTA | chr8 | 42919243 | + | 1.75E-08 | 1 |
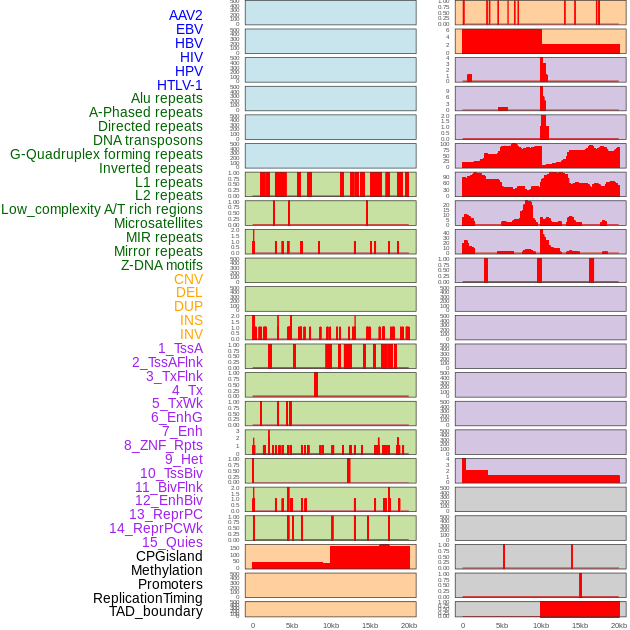
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
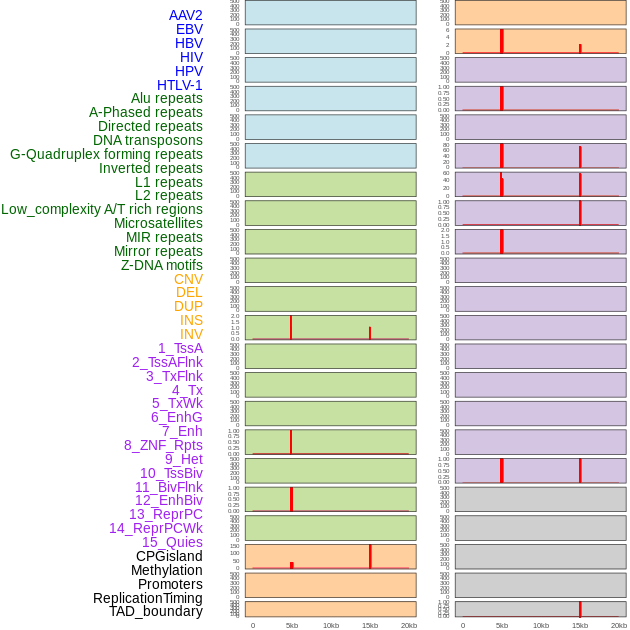
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for AAK1-FNTA |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:69732700/chr8:42919243) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| AAK1 | FNTA |
| FUNCTION: Regulates clathrin-mediated endocytosis by phosphorylating the AP2M1/mu2 subunit of the adaptor protein complex 2 (AP-2) which ensures high affinity binding of AP-2 to cargo membrane proteins during the initial stages of endocytosis (PubMed:17494869, PubMed:11877457, PubMed:11877461, PubMed:12952931, PubMed:14617351, PubMed:25653444). Isoform 1 and isoform 2 display similar levels of kinase activity towards AP2M1 (PubMed:17494869). Preferentially, may phosphorylate substrates on threonine residues (PubMed:11877457, PubMed:18657069). Regulates phosphorylation of other AP-2 subunits as well as AP-2 localization and AP-2-mediated internalization of ligand complexes (PubMed:12952931). Phosphorylates NUMB and regulates its cellular localization, promoting NUMB localization to endosomes (PubMed:18657069). Binds to and stabilizes the activated form of NOTCH1, increases its localization in endosomes and regulates its transcriptional activity (PubMed:21464124). {ECO:0000269|PubMed:11877457, ECO:0000269|PubMed:11877461, ECO:0000269|PubMed:12952931, ECO:0000269|PubMed:14617351, ECO:0000269|PubMed:17494869, ECO:0000269|PubMed:18657069, ECO:0000269|PubMed:21464124, ECO:0000269|PubMed:25653444}.; FUNCTION: (Microbial infection) By regulating clathrin-mediated endocytosis, AAK1 plays a role in the entry of hepatitis C virus as well as for the lifecycle of other viruses such as Ebola and Dengue. {ECO:0000269|PubMed:25653444, ECO:0000305|PubMed:31136173}. | FUNCTION: Essential subunit of both the farnesyltransferase and the geranylgeranyltransferase complex. Contributes to the transfer of a farnesyl or geranylgeranyl moiety from farnesyl or geranylgeranyl diphosphate to a cysteine at the fourth position from the C-terminus of several proteins having the C-terminal sequence Cys-aliphatic-aliphatic-X. May positively regulate neuromuscular junction development downstream of MUSK via its function in RAC1 prenylation and activation. {ECO:0000269|PubMed:12036349, ECO:0000269|PubMed:12825937, ECO:0000269|PubMed:16893176, ECO:0000269|PubMed:19246009, ECO:0000269|PubMed:8419339, ECO:0000269|PubMed:8494894}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | AAK1 | chr2:69732700 | chr8:42919243 | ENST00000406297 | - | 16 | 18 | 12_42 | 756 | 864.0 | Compositional bias | Note=Gly-rich |
| Hgene | AAK1 | chr2:69732700 | chr8:42919243 | ENST00000406297 | - | 16 | 18 | 397_614 | 756 | 864.0 | Compositional bias | Note=Gln-rich |
| Hgene | AAK1 | chr2:69732700 | chr8:42919243 | ENST00000406297 | - | 16 | 18 | 658_663 | 756 | 864.0 | Compositional bias | Note=Poly-Ala |
| Hgene | AAK1 | chr2:69732700 | chr8:42919243 | ENST00000409085 | - | 16 | 22 | 12_42 | 756 | 962.0 | Compositional bias | Note=Gly-rich |
| Hgene | AAK1 | chr2:69732700 | chr8:42919243 | ENST00000409085 | - | 16 | 22 | 397_614 | 756 | 962.0 | Compositional bias | Note=Gln-rich |
| Hgene | AAK1 | chr2:69732700 | chr8:42919243 | ENST00000409085 | - | 16 | 22 | 658_663 | 756 | 962.0 | Compositional bias | Note=Poly-Ala |
| Hgene | AAK1 | chr2:69732700 | chr8:42919243 | ENST00000406297 | - | 16 | 18 | 46_315 | 756 | 864.0 | Domain | Protein kinase |
| Hgene | AAK1 | chr2:69732700 | chr8:42919243 | ENST00000409085 | - | 16 | 22 | 46_315 | 756 | 962.0 | Domain | Protein kinase |
| Hgene | AAK1 | chr2:69732700 | chr8:42919243 | ENST00000406297 | - | 16 | 18 | 52_60 | 756 | 864.0 | Nucleotide binding | ATP |
| Hgene | AAK1 | chr2:69732700 | chr8:42919243 | ENST00000409085 | - | 16 | 22 | 52_60 | 756 | 962.0 | Nucleotide binding | ATP |
| Tgene | FNTA | chr2:69732700 | chr8:42919243 | ENST00000342116 | 0 | 7 | 22_31 | 0 | 313.0 | Compositional bias | Note=Pro-rich | |
| Tgene | FNTA | chr2:69732700 | chr8:42919243 | ENST00000302279 | 1 | 9 | 112_146 | 95 | 380.0 | Repeat | Note=PFTA 1 | |
| Tgene | FNTA | chr2:69732700 | chr8:42919243 | ENST00000302279 | 1 | 9 | 147_180 | 95 | 380.0 | Repeat | Note=PFTA 2 | |
| Tgene | FNTA | chr2:69732700 | chr8:42919243 | ENST00000302279 | 1 | 9 | 181_215 | 95 | 380.0 | Repeat | Note=PFTA 3 | |
| Tgene | FNTA | chr2:69732700 | chr8:42919243 | ENST00000302279 | 1 | 9 | 216_249 | 95 | 380.0 | Repeat | Note=PFTA 4 | |
| Tgene | FNTA | chr2:69732700 | chr8:42919243 | ENST00000302279 | 1 | 9 | 255_289 | 95 | 380.0 | Repeat | Note=PFTA 5 | |
| Tgene | FNTA | chr2:69732700 | chr8:42919243 | ENST00000342116 | 0 | 7 | 112_146 | 0 | 313.0 | Repeat | Note=PFTA 1 | |
| Tgene | FNTA | chr2:69732700 | chr8:42919243 | ENST00000342116 | 0 | 7 | 147_180 | 0 | 313.0 | Repeat | Note=PFTA 2 | |
| Tgene | FNTA | chr2:69732700 | chr8:42919243 | ENST00000342116 | 0 | 7 | 181_215 | 0 | 313.0 | Repeat | Note=PFTA 3 | |
| Tgene | FNTA | chr2:69732700 | chr8:42919243 | ENST00000342116 | 0 | 7 | 216_249 | 0 | 313.0 | Repeat | Note=PFTA 4 | |
| Tgene | FNTA | chr2:69732700 | chr8:42919243 | ENST00000342116 | 0 | 7 | 255_289 | 0 | 313.0 | Repeat | Note=PFTA 5 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | AAK1 | chr2:69732700 | chr8:42919243 | ENST00000406297 | - | 16 | 18 | 823_960 | 756 | 864.0 | Region | Clathrin-binding domain (CBD) |
| Hgene | AAK1 | chr2:69732700 | chr8:42919243 | ENST00000409085 | - | 16 | 22 | 823_960 | 756 | 962.0 | Region | Clathrin-binding domain (CBD) |
| Tgene | FNTA | chr2:69732700 | chr8:42919243 | ENST00000302279 | 1 | 9 | 22_31 | 95 | 380.0 | Compositional bias | Note=Pro-rich |
Top |
Fusion Gene Sequence for AAK1-FNTA |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >57_57_1_AAK1-FNTA_AAK1_chr2_69732700_ENST00000406297_FNTA_chr8_42919243_ENST00000302279_length(transcript)=4006nt_BP=2646nt GATCAGCTGGCGGGCGGGCGGGAGCCGAGCGCGGCCCCGGCTCTCGCTGCAGCGCCGCCTCTTCTCTGCGTCGCAGGCCGGCCCGGCGGC CGTGACAATGTCGCGGGGCTGGTAGCAGGGCGCCGGCCGCCGAGCCGTCTCAAGTTTAAACTTACACGAATCGCTTTCTGGAGGAGGAGG GGACCCGCTGCGCGATTGACACGCATATTCCTATAGGCATCCTCCCTCAGCCCCCACCCCCACGGCCGGATTCGGGTGGCTCCTCTCCGA GGTGAAATCTGAGAAGAAATCCTTGGATCTCTTTTCTTAAAAAAAAAAAAAAAAAAAAAAAATCTAGAAACCATCGGTATTTTGCTTTGC TGCTCCCTATTCGCAAGATGAAGAAGTTTTTCGACTCCCGGCGAGAGCAGGGCGGCTCTGGCCTGGGCTCCGGCTCCAGCGGAGGAGGGG GCAGCACCTCGGGCCTGGGCAGTGGCTACATCGGAAGAGTCTTCGGCATCGGGCGACAGCAGGTCACAGTGGACGAGGTGTTGGCGGAAG GTGGATTTGCTATTGTATTTCTGGTGAGGACAAGCAATGGGATGAAATGTGCCTTGAAACGCATGTTTGTCAACAATGAGCATGATCTCC AGGTGTGCAAGAGAGAAATCCAGATAATGAGGGATCTTTCAGGGCACAAGAATATTGTGGGTTACATTGATTCTAGTATCAACAACGTGA GTAGCGGTGATGTATGGGAAGTGCTCATTCTGATGGACTTTTGTAGAGGTGGCCAGGTGGTAAACCTGATGAACCAGCGCCTGCAAACAG GCTTTACAGAGAATGAAGTGCTCCAGATATTTTGTGATACCTGTGAAGCTGTTGCCCGCCTGCATCAGTGCAAAACTCCTATTATCCACC GGGACCTGAAGGTTGAAAACATCCTCTTGCATGACCGAGGCCACTATGTCCTGTGTGACTTTGGAAGCGCCACCAACAAATTCCAGAATC CACAAACTGAGGGAGTCAATGCAGTAGAAGATGAGATTAAGAAATACACAACGCTGTCCTATCGAGCACCAGAAATGGTCAACCTGTACA GTGGCAAAATCATCACTACGAAGGCAGACATTTGGGCTCTTGGATGTTTGTTGTATAAATTATGCTACTTCACTTTGCCATTTGGGGAAA GTCAGGTGGCAATTTGTGATGGAAACTTCACAATTCCTGATAATTCTCGATATTCTCAAGACATGCACTGCCTAATTAGGTATATGTTGG AACCAGACCCTGACAAAAGGCCGGATATTTACCAGGTGTCCTACTTCTCATTTAAGCTACTCAAGAAAGAGTGCCCAATTCCAAATGTAC AGAACTCTCCCATTCCTGCAAAGCTTCCTGAACCAGTGAAAGCCAGTGAGGCAGCTGCAAAAAAGACCCAGCCAAAGGCCAGACTGACAG ATCCCATTCCCACCACAGAGACTTCAATTGCACCCCGCCAGAGGCCTAAAGCTGGGCAGACTCAGCCGAACCCAGGAATCCTTCCCATCC AGCCAGCGCTGACACCCCGGAAGAGGGCCACTGTTCAGCCCCCACCTCAGGCTGCAGGATCCAGCAATCAGCCTGGCCTTTTAGCCAGTG TTCCCCAACCAAAACCCCAAGCCCCACCCAGCCAGCCTCTGCCGCAAACTCAGGCCAAGCAGCCACAGGCTCCTCCCACTCCACAGCAGA CGCCTTCTACTCAGGCCCAGGGTCTGCCCGCTCAGGCCCAGGCCACACCCCAGCACCAGCAGCAACTCTTCCTCAAGCAGCAACAGCAGC AGCAACAGCCACCGCCAGCACAGCAGCAGCCGGCAGGCACGTTTTACCAGCAGCAGCAGGCCCAGACTCAGCAGTTTCAGGCAGTACATC CAGCAACCCAGAAACCAGCAATTGCTCAGTTCCCTGTGGTGTCCCAAGGAGGCTCTCAACAGCAGCTAATGCAGAATTTCTACCAGCAGC AGCAGCAGCAGCAACAACAACAGCAACAGCAACAGCTGGCCACAGCCCTGCATCAACAACAGCTGATGACTCAGCAGGCTGCCTTGCAGC AAAAGCCCACTATGGCAGCAGGACAGCAGCCCCAGCCACAGCCAGCTGCAGCCCCACAGCCAGCCCCTGCCCAGGAGCCAGCGATTCAAG CCCCAGTAAGACAACAGCCAAAGGTTCAGACAACCCCACCTCCTGCCGTCCAGGGGCAGAAAGTTGGATCTCTCACTCCACCCTCATCCC CCAAAACCCAACGTGCTGGGCACAGGCGTATTCTCAGTGACGTAACCCACAGTGCAGTCTTTGGGGTCCCTGCCAGCAAATCAACCCAGC TGCTCCAGGCAGCTGCAGCTGAGGCCAGTCTCAATAAGTCCAAGTCTGCAACCACCACTCCATCAGGCTCTCCTCGGACCTCTCAACAAA ACGTTTATAATCCTTCAGAAGGGTCTACGTGGAATCCCTTTGATGACGATAATTTCTCCAAACTCACAGCTGAAGAACTGCTAAACAAGG ACTTTGCCAAGCTTGGGGAAGGCAAACATCCCGAGAAGCTTGGAGGCTCAGCTGAGAGTTTGATCCCAGGCTTTCAATCAACCCAAGGTG ATGCTTTTGCTACGACCTCATTTTCTGCTGGAACTGTTAGAGATGTTTATGATTACTTCCGAGCTGTCCTGCAGCGTGATGAAAGAAGTG AACGAGCTTTTAAGCTAACCCGGGATGCTATTGAGTTAAATGCAGCCAATTATACAGTGTGGCATTTCCGGAGAGTTCTTTTGAAGTCAC TTCAGAAGGATCTACATGAGGAAATGAACTACATCACTGCAATAATTGAGGAGCAGCCCAAAAACTATCAAGTTTGGCATCATAGGCGAG TATTAGTGGAATGGCTAAGAGATCCATCTCAGGAGCTTGAATTTATTGCTGATATTCTTAATCAGGATGCAAAGAATTATCATGCCTGGC AGCATCGACAATGGGTTATTCAGGAATTTAAACTTTGGGATAATGAGCTGCAGTATGTGGACCAACTTCTGAAAGAGGATGTGAGAAATA ACTCTGTCTGGAACCAAAGATACTTCGTTATTTCTAACACCACTGGCTACAATGATCGTGCTGTATTGGAGAGAGAAGTCCAATACACTC TGGAAATGATTAAACTAGTACCACATAATGAAAGTGCATGGAACTATTTGAAAGGGATTTTGCAGGATCGTGGTCTTTCCAAATATCCTA ATCTGTTAAATCAATTACTTGATTTACAACCAAGTCATAGTTCCCCCTACCTAATTGCCTTTCTTGTGGATATCTATGAAGACATGCTAG AAAATCAGTGTGACAATAAGGAAGACATTCTTAATAAAGCATTAGAGTTATGTGAAATCCTAGCTAAAGAAAAGGACACTATAAGAAAGG AATATTGGAGATACATTGGAAGATCCCTTCAAAGCAAACACAGCACAGAAAATGACTCACCAACAAATGTACAGCAATAACACCATCCAG AAGAACTTGATGGAATGCTTTTATTTTTTATTAAGGGACCCTGCAGGAGTTTCACACGAGAGTGGTCCTTCCCTTTGCCTGTGGTGTAAA AGTGCATCACACAGGTATTGCTTTTTAACAAGAACTGATGCTCCTTGGGTGCTGCTGCTACTCAGACTAGCTCTAAGTAATGTGATTCTT CTAAAGCAAAGTCATTGGATGGGAGGAGGAAGAAAAAGTCCCATAAAGGAACTTTTGTAGTCTTATCAACATATAATCTAATCCCTTAGC ATCAGCTCCTCCCTCAGTGGTACATGCGTCAAGATTTGTAGCAGTAATAACTGCAGGTCACTTGTATGTAATGGATGTGAGGTAGCCGAA GTTTGGTTCAGTAAGCAGGGAATACAGTCGTTCCATCAGAGCTGGTCTGCACACTCACATTATCTTGCTATCACTGTAACCAACTAATGC >57_57_1_AAK1-FNTA_AAK1_chr2_69732700_ENST00000406297_FNTA_chr8_42919243_ENST00000302279_length(amino acids)=1073AA_BP=789 MRRNPWISFLKKKKKKKKSRNHRYFALLLPIRKMKKFFDSRREQGGSGLGSGSSGGGGSTSGLGSGYIGRVFGIGRQQVTVDEVLAEGGF AIVFLVRTSNGMKCALKRMFVNNEHDLQVCKREIQIMRDLSGHKNIVGYIDSSINNVSSGDVWEVLILMDFCRGGQVVNLMNQRLQTGFT ENEVLQIFCDTCEAVARLHQCKTPIIHRDLKVENILLHDRGHYVLCDFGSATNKFQNPQTEGVNAVEDEIKKYTTLSYRAPEMVNLYSGK IITTKADIWALGCLLYKLCYFTLPFGESQVAICDGNFTIPDNSRYSQDMHCLIRYMLEPDPDKRPDIYQVSYFSFKLLKKECPIPNVQNS PIPAKLPEPVKASEAAAKKTQPKARLTDPIPTTETSIAPRQRPKAGQTQPNPGILPIQPALTPRKRATVQPPPQAAGSSNQPGLLASVPQ PKPQAPPSQPLPQTQAKQPQAPPTPQQTPSTQAQGLPAQAQATPQHQQQLFLKQQQQQQQPPPAQQQPAGTFYQQQQAQTQQFQAVHPAT QKPAIAQFPVVSQGGSQQQLMQNFYQQQQQQQQQQQQQQLATALHQQQLMTQQAALQQKPTMAAGQQPQPQPAAAPQPAPAQEPAIQAPV RQQPKVQTTPPPAVQGQKVGSLTPPSSPKTQRAGHRRILSDVTHSAVFGVPASKSTQLLQAAAAEASLNKSKSATTTPSGSPRTSQQNVY NPSEGSTWNPFDDDNFSKLTAEELLNKDFAKLGEGKHPEKLGGSAESLIPGFQSTQGDAFATTSFSAGTVRDVYDYFRAVLQRDERSERA FKLTRDAIELNAANYTVWHFRRVLLKSLQKDLHEEMNYITAIIEEQPKNYQVWHHRRVLVEWLRDPSQELEFIADILNQDAKNYHAWQHR QWVIQEFKLWDNELQYVDQLLKEDVRNNSVWNQRYFVISNTTGYNDRAVLEREVQYTLEMIKLVPHNESAWNYLKGILQDRGLSKYPNLL -------------------------------------------------------------- >57_57_2_AAK1-FNTA_AAK1_chr2_69732700_ENST00000409085_FNTA_chr8_42919243_ENST00000302279_length(transcript)=4006nt_BP=2646nt GATCAGCTGGCGGGCGGGCGGGAGCCGAGCGCGGCCCCGGCTCTCGCTGCAGCGCCGCCTCTTCTCTGCGTCGCAGGCCGGCCCGGCGGC CGTGACAATGTCGCGGGGCTGGTAGCAGGGCGCCGGCCGCCGAGCCGTCTCAAGTTTAAACTTACACGAATCGCTTTCTGGAGGAGGAGG GGACCCGCTGCGCGATTGACACGCATATTCCTATAGGCATCCTCCCTCAGCCCCCACCCCCACGGCCGGATTCGGGTGGCTCCTCTCCGA GGTGAAATCTGAGAAGAAATCCTTGGATCTCTTTTCTTAAAAAAAAAAAAAAAAAAAAAAAATCTAGAAACCATCGGTATTTTGCTTTGC TGCTCCCTATTCGCAAGATGAAGAAGTTTTTCGACTCCCGGCGAGAGCAGGGCGGCTCTGGCCTGGGCTCCGGCTCCAGCGGAGGAGGGG GCAGCACCTCGGGCCTGGGCAGTGGCTACATCGGAAGAGTCTTCGGCATCGGGCGACAGCAGGTCACAGTGGACGAGGTGTTGGCGGAAG GTGGATTTGCTATTGTATTTCTGGTGAGGACAAGCAATGGGATGAAATGTGCCTTGAAACGCATGTTTGTCAACAATGAGCATGATCTCC AGGTGTGCAAGAGAGAAATCCAGATAATGAGGGATCTTTCAGGGCACAAGAATATTGTGGGTTACATTGATTCTAGTATCAACAACGTGA GTAGCGGTGATGTATGGGAAGTGCTCATTCTGATGGACTTTTGTAGAGGTGGCCAGGTGGTAAACCTGATGAACCAGCGCCTGCAAACAG GCTTTACAGAGAATGAAGTGCTCCAGATATTTTGTGATACCTGTGAAGCTGTTGCCCGCCTGCATCAGTGCAAAACTCCTATTATCCACC GGGACCTGAAGGTTGAAAACATCCTCTTGCATGACCGAGGCCACTATGTCCTGTGTGACTTTGGAAGCGCCACCAACAAATTCCAGAATC CACAAACTGAGGGAGTCAATGCAGTAGAAGATGAGATTAAGAAATACACAACGCTGTCCTATCGAGCACCAGAAATGGTCAACCTGTACA GTGGCAAAATCATCACTACGAAGGCAGACATTTGGGCTCTTGGATGTTTGTTGTATAAATTATGCTACTTCACTTTGCCATTTGGGGAAA GTCAGGTGGCAATTTGTGATGGAAACTTCACAATTCCTGATAATTCTCGATATTCTCAAGACATGCACTGCCTAATTAGGTATATGTTGG AACCAGACCCTGACAAAAGGCCGGATATTTACCAGGTGTCCTACTTCTCATTTAAGCTACTCAAGAAAGAGTGCCCAATTCCAAATGTAC AGAACTCTCCCATTCCTGCAAAGCTTCCTGAACCAGTGAAAGCCAGTGAGGCAGCTGCAAAAAAGACCCAGCCAAAGGCCAGACTGACAG ATCCCATTCCCACCACAGAGACTTCAATTGCACCCCGCCAGAGGCCTAAAGCTGGGCAGACTCAGCCGAACCCAGGAATCCTTCCCATCC AGCCAGCGCTGACACCCCGGAAGAGGGCCACTGTTCAGCCCCCACCTCAGGCTGCAGGATCCAGCAATCAGCCTGGCCTTTTAGCCAGTG TTCCCCAACCAAAACCCCAAGCCCCACCCAGCCAGCCTCTGCCGCAAACTCAGGCCAAGCAGCCACAGGCTCCTCCCACTCCACAGCAGA CGCCTTCTACTCAGGCCCAGGGTCTGCCCGCTCAGGCCCAGGCCACACCCCAGCACCAGCAGCAACTCTTCCTCAAGCAGCAACAGCAGC AGCAACAGCCACCGCCAGCACAGCAGCAGCCGGCAGGCACGTTTTACCAGCAGCAGCAGGCCCAGACTCAGCAGTTTCAGGCAGTACATC CAGCAACCCAGAAACCAGCAATTGCTCAGTTCCCTGTGGTGTCCCAAGGAGGCTCTCAACAGCAGCTAATGCAGAATTTCTACCAGCAGC AGCAGCAGCAGCAACAACAACAGCAACAGCAACAGCTGGCCACAGCCCTGCATCAACAACAGCTGATGACTCAGCAGGCTGCCTTGCAGC AAAAGCCCACTATGGCAGCAGGACAGCAGCCCCAGCCACAGCCAGCTGCAGCCCCACAGCCAGCCCCTGCCCAGGAGCCAGCGATTCAAG CCCCAGTAAGACAACAGCCAAAGGTTCAGACAACCCCACCTCCTGCCGTCCAGGGGCAGAAAGTTGGATCTCTCACTCCACCCTCATCCC CCAAAACCCAACGTGCTGGGCACAGGCGTATTCTCAGTGACGTAACCCACAGTGCAGTCTTTGGGGTCCCTGCCAGCAAATCAACCCAGC TGCTCCAGGCAGCTGCAGCTGAGGCCAGTCTCAATAAGTCCAAGTCTGCAACCACCACTCCATCAGGCTCTCCTCGGACCTCTCAACAAA ACGTTTATAATCCTTCAGAAGGGTCTACGTGGAATCCCTTTGATGACGATAATTTCTCCAAACTCACAGCTGAAGAACTGCTAAACAAGG ACTTTGCCAAGCTTGGGGAAGGCAAACATCCCGAGAAGCTTGGAGGCTCAGCTGAGAGTTTGATCCCAGGCTTTCAATCAACCCAAGGTG ATGCTTTTGCTACGACCTCATTTTCTGCTGGAACTGTTAGAGATGTTTATGATTACTTCCGAGCTGTCCTGCAGCGTGATGAAAGAAGTG AACGAGCTTTTAAGCTAACCCGGGATGCTATTGAGTTAAATGCAGCCAATTATACAGTGTGGCATTTCCGGAGAGTTCTTTTGAAGTCAC TTCAGAAGGATCTACATGAGGAAATGAACTACATCACTGCAATAATTGAGGAGCAGCCCAAAAACTATCAAGTTTGGCATCATAGGCGAG TATTAGTGGAATGGCTAAGAGATCCATCTCAGGAGCTTGAATTTATTGCTGATATTCTTAATCAGGATGCAAAGAATTATCATGCCTGGC AGCATCGACAATGGGTTATTCAGGAATTTAAACTTTGGGATAATGAGCTGCAGTATGTGGACCAACTTCTGAAAGAGGATGTGAGAAATA ACTCTGTCTGGAACCAAAGATACTTCGTTATTTCTAACACCACTGGCTACAATGATCGTGCTGTATTGGAGAGAGAAGTCCAATACACTC TGGAAATGATTAAACTAGTACCACATAATGAAAGTGCATGGAACTATTTGAAAGGGATTTTGCAGGATCGTGGTCTTTCCAAATATCCTA ATCTGTTAAATCAATTACTTGATTTACAACCAAGTCATAGTTCCCCCTACCTAATTGCCTTTCTTGTGGATATCTATGAAGACATGCTAG AAAATCAGTGTGACAATAAGGAAGACATTCTTAATAAAGCATTAGAGTTATGTGAAATCCTAGCTAAAGAAAAGGACACTATAAGAAAGG AATATTGGAGATACATTGGAAGATCCCTTCAAAGCAAACACAGCACAGAAAATGACTCACCAACAAATGTACAGCAATAACACCATCCAG AAGAACTTGATGGAATGCTTTTATTTTTTATTAAGGGACCCTGCAGGAGTTTCACACGAGAGTGGTCCTTCCCTTTGCCTGTGGTGTAAA AGTGCATCACACAGGTATTGCTTTTTAACAAGAACTGATGCTCCTTGGGTGCTGCTGCTACTCAGACTAGCTCTAAGTAATGTGATTCTT CTAAAGCAAAGTCATTGGATGGGAGGAGGAAGAAAAAGTCCCATAAAGGAACTTTTGTAGTCTTATCAACATATAATCTAATCCCTTAGC ATCAGCTCCTCCCTCAGTGGTACATGCGTCAAGATTTGTAGCAGTAATAACTGCAGGTCACTTGTATGTAATGGATGTGAGGTAGCCGAA GTTTGGTTCAGTAAGCAGGGAATACAGTCGTTCCATCAGAGCTGGTCTGCACACTCACATTATCTTGCTATCACTGTAACCAACTAATGC >57_57_2_AAK1-FNTA_AAK1_chr2_69732700_ENST00000409085_FNTA_chr8_42919243_ENST00000302279_length(amino acids)=1073AA_BP=789 MRRNPWISFLKKKKKKKKSRNHRYFALLLPIRKMKKFFDSRREQGGSGLGSGSSGGGGSTSGLGSGYIGRVFGIGRQQVTVDEVLAEGGF AIVFLVRTSNGMKCALKRMFVNNEHDLQVCKREIQIMRDLSGHKNIVGYIDSSINNVSSGDVWEVLILMDFCRGGQVVNLMNQRLQTGFT ENEVLQIFCDTCEAVARLHQCKTPIIHRDLKVENILLHDRGHYVLCDFGSATNKFQNPQTEGVNAVEDEIKKYTTLSYRAPEMVNLYSGK IITTKADIWALGCLLYKLCYFTLPFGESQVAICDGNFTIPDNSRYSQDMHCLIRYMLEPDPDKRPDIYQVSYFSFKLLKKECPIPNVQNS PIPAKLPEPVKASEAAAKKTQPKARLTDPIPTTETSIAPRQRPKAGQTQPNPGILPIQPALTPRKRATVQPPPQAAGSSNQPGLLASVPQ PKPQAPPSQPLPQTQAKQPQAPPTPQQTPSTQAQGLPAQAQATPQHQQQLFLKQQQQQQQPPPAQQQPAGTFYQQQQAQTQQFQAVHPAT QKPAIAQFPVVSQGGSQQQLMQNFYQQQQQQQQQQQQQQLATALHQQQLMTQQAALQQKPTMAAGQQPQPQPAAAPQPAPAQEPAIQAPV RQQPKVQTTPPPAVQGQKVGSLTPPSSPKTQRAGHRRILSDVTHSAVFGVPASKSTQLLQAAAAEASLNKSKSATTTPSGSPRTSQQNVY NPSEGSTWNPFDDDNFSKLTAEELLNKDFAKLGEGKHPEKLGGSAESLIPGFQSTQGDAFATTSFSAGTVRDVYDYFRAVLQRDERSERA FKLTRDAIELNAANYTVWHFRRVLLKSLQKDLHEEMNYITAIIEEQPKNYQVWHHRRVLVEWLRDPSQELEFIADILNQDAKNYHAWQHR QWVIQEFKLWDNELQYVDQLLKEDVRNNSVWNQRYFVISNTTGYNDRAVLEREVQYTLEMIKLVPHNESAWNYLKGILQDRGLSKYPNLL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for AAK1-FNTA |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for AAK1-FNTA |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for AAK1-FNTA |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
