
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:NAMPT-CEP290 (FusionGDB2 ID:57162) |
Fusion Gene Summary for NAMPT-CEP290 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: NAMPT-CEP290 | Fusion gene ID: 57162 | Hgene | Tgene | Gene symbol | NAMPT | CEP290 | Gene ID | 10135 | 80184 |
| Gene name | nicotinamide phosphoribosyltransferase | centrosomal protein 290 | |
| Synonyms | 1110035O14Rik|PBEF|PBEF1|VF|VISFATIN | 3H11Ag|BBS14|CT87|JBTS5|LCA10|MKS4|NPHP6|POC3|SLSN6|rd16 | |
| Cytomap | 7q22.3 | 12q21.32 | |
| Type of gene | protein-coding | protein-coding | |
| Description | nicotinamide phosphoribosyltransferaseNAmPRTasepre-B cell-enhancing factorpre-B-cell colony-enhancing factor 1 | centrosomal protein of 290 kDaBardet-Biedl syndrome 14 proteinCTCL tumor antigen se2-2Meckel syndrome, type 4POC3 centriolar protein homologcancer/testis antigen 87centrosomal protein 290kDamonoclonal antibody 3H11 antigennephrocytsin-6prostate c | |
| Modification date | 20200322 | 20200328 | |
| UniProtAcc | P43490 | O15078 | |
| Ensembl transtripts involved in fusion gene | ENST00000484527, ENST00000222553, ENST00000354289, | ENST00000309041, ENST00000397838, ENST00000547691, ENST00000552810, | |
| Fusion gene scores | * DoF score | 6 X 6 X 3=108 | 9 X 9 X 6=486 |
| # samples | 6 | 9 | |
| ** MAII score | log2(6/108*10)=-0.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/486*10)=-2.43295940727611 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: NAMPT [Title/Abstract] AND CEP290 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | NAMPT(105915391)-CEP290(88452797), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | NAMPT | GO:0008286 | insulin receptor signaling pathway | 17983582 |
| Hgene | NAMPT | GO:0045944 | positive regulation of transcription by RNA polymerase II | 23001182 |
| Hgene | NAMPT | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process | 19727662 |
| Hgene | NAMPT | GO:0060612 | adipose tissue development | 17983582 |
| Tgene | CEP290 | GO:0045893 | positive regulation of transcription, DNA-templated | 16682973 |
| Tgene | CEP290 | GO:0060271 | cilium assembly | 26386044 |
 Fusion gene breakpoints across NAMPT (5'-gene) Fusion gene breakpoints across NAMPT (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
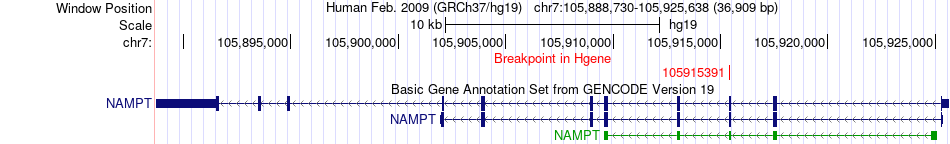 |
 Fusion gene breakpoints across CEP290 (3'-gene) Fusion gene breakpoints across CEP290 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
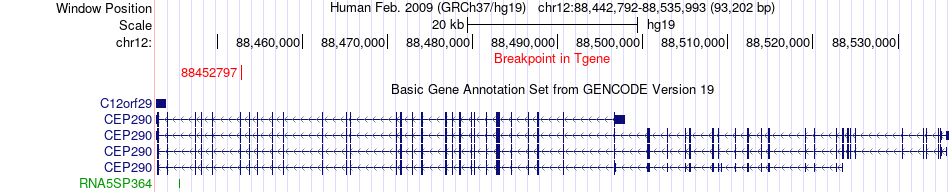 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ESCA | TCGA-VR-A8EW | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - |
Top |
Fusion Gene ORF analysis for NAMPT-CEP290 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5UTR-3CDS | ENST00000484527 | ENST00000309041 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - |
| 5UTR-3CDS | ENST00000484527 | ENST00000397838 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - |
| 5UTR-3CDS | ENST00000484527 | ENST00000547691 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - |
| 5UTR-3CDS | ENST00000484527 | ENST00000552810 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - |
| In-frame | ENST00000222553 | ENST00000309041 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - |
| In-frame | ENST00000222553 | ENST00000397838 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - |
| In-frame | ENST00000222553 | ENST00000547691 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - |
| In-frame | ENST00000222553 | ENST00000552810 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - |
| In-frame | ENST00000354289 | ENST00000309041 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - |
| In-frame | ENST00000354289 | ENST00000397838 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - |
| In-frame | ENST00000354289 | ENST00000547691 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - |
| In-frame | ENST00000354289 | ENST00000552810 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000222553 | NAMPT | chr7 | 105915391 | - | ENST00000547691 | CEP290 | chr12 | 88452797 | - | 1589 | 626 | 308 | 1420 | 370 |
| ENST00000222553 | NAMPT | chr7 | 105915391 | - | ENST00000552810 | CEP290 | chr12 | 88452797 | - | 1585 | 626 | 308 | 1420 | 370 |
| ENST00000222553 | NAMPT | chr7 | 105915391 | - | ENST00000397838 | CEP290 | chr12 | 88452797 | - | 1421 | 626 | 308 | 1420 | 371 |
| ENST00000222553 | NAMPT | chr7 | 105915391 | - | ENST00000309041 | CEP290 | chr12 | 88452797 | - | 1421 | 626 | 308 | 1420 | 371 |
| ENST00000354289 | NAMPT | chr7 | 105915391 | - | ENST00000547691 | CEP290 | chr12 | 88452797 | - | 1318 | 355 | 37 | 1149 | 370 |
| ENST00000354289 | NAMPT | chr7 | 105915391 | - | ENST00000552810 | CEP290 | chr12 | 88452797 | - | 1314 | 355 | 37 | 1149 | 370 |
| ENST00000354289 | NAMPT | chr7 | 105915391 | - | ENST00000397838 | CEP290 | chr12 | 88452797 | - | 1150 | 355 | 37 | 1149 | 371 |
| ENST00000354289 | NAMPT | chr7 | 105915391 | - | ENST00000309041 | CEP290 | chr12 | 88452797 | - | 1150 | 355 | 37 | 1149 | 371 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000222553 | ENST00000547691 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - | 0.000361118 | 0.9996389 |
| ENST00000222553 | ENST00000552810 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - | 0.000368276 | 0.99963176 |
| ENST00000222553 | ENST00000397838 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - | 0.00067925 | 0.99932075 |
| ENST00000222553 | ENST00000309041 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - | 0.00067925 | 0.99932075 |
| ENST00000354289 | ENST00000547691 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - | 0.000522511 | 0.99947757 |
| ENST00000354289 | ENST00000552810 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - | 0.000530193 | 0.9994698 |
| ENST00000354289 | ENST00000397838 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - | 0.001025128 | 0.9989749 |
| ENST00000354289 | ENST00000309041 | NAMPT | chr7 | 105915391 | - | CEP290 | chr12 | 88452797 | - | 0.001025128 | 0.9989749 |
Top |
Fusion Genomic Features for NAMPT-CEP290 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for NAMPT-CEP290 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:105915391/chr12:88452797) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NAMPT | CEP290 |
| FUNCTION: Catalyzes the condensation of nicotinamide with 5-phosphoribosyl-1-pyrophosphate to yield nicotinamide mononucleotide, an intermediate in the biosynthesis of NAD. It is the rate limiting component in the mammalian NAD biosynthesis pathway. The secreted form behaves both as a cytokine with immunomodulating properties and an adipokine with anti-diabetic properties, it has no enzymatic activity, partly because of lack of activation by ATP, which has a low level in extracellular space and plasma. Plays a role in the modulation of circadian clock function. NAMPT-dependent oscillatory production of NAD regulates oscillation of clock target gene expression by releasing the core clock component: CLOCK-ARNTL/BMAL1 heterodimer from NAD-dependent SIRT1-mediated suppression (By similarity). {ECO:0000250|UniProtKB:Q99KQ4, ECO:0000269|PubMed:24130902}. | FUNCTION: Involved in early and late steps in cilia formation. Its association with CCP110 is required for inhibition of primary cilia formation by CCP110 (PubMed:18694559). May play a role in early ciliogenesis in the disappearance of centriolar satellites and in the transition of primary ciliar vesicles (PCVs) to capped ciliary vesicles (CCVs). Required for the centrosomal recruitment of RAB8A and for the targeting of centriole satellite proteins to centrosomes such as of PCM1 (PubMed:24421332). Required for the correct localization of ciliary and phototransduction proteins in retinal photoreceptor cells; may play a role in ciliary transport processes (By similarity). Required for efficient recruitment of RAB8A to primary cilium (PubMed:17705300). In the ciliary transition zone is part of the tectonic-like complex which is required for tissue-specific ciliogenesis and may regulate ciliary membrane composition (By similarity). Involved in regulation of the BBSome complex integrity, specifically for presence of BBS2, BBS5 and BBS8/TTC8 in the complex, and in ciliary targeting of selected BBSome cargos. May play a role in controlling entry of the BBSome complex to cilia possibly implicating IQCB1/NPHP5 (PubMed:25552655). Activates ATF4-mediated transcription (PubMed:16682973). {ECO:0000250|UniProtKB:Q6A078, ECO:0000269|PubMed:16682973, ECO:0000269|PubMed:17705300, ECO:0000269|PubMed:18694559, ECO:0000269|PubMed:24421332, ECO:0000269|PubMed:25552655}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000397838 | 39 | 46 | 1533_1584 | 1275 | 1540.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000397838 | 39 | 46 | 1635_2452 | 1275 | 1540.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000547691 | 22 | 29 | 1533_1584 | 1275 | 1540.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000547691 | 22 | 29 | 1635_2452 | 1275 | 1540.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000397838 | 39 | 46 | 1966_2479 | 1275 | 1540.0 | Region | Self-association (with itself or N-terminus) | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000547691 | 22 | 29 | 1966_2479 | 1275 | 1540.0 | Region | Self-association (with itself or N-terminus) |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NAMPT | chr7:105915391 | chr12:88452797 | ENST00000222553 | - | 3 | 11 | 311_313 | 106 | 492.0 | Region | Note=Nicotinamide ribonucleotide binding |
| Hgene | NAMPT | chr7:105915391 | chr12:88452797 | ENST00000222553 | - | 3 | 11 | 353_354 | 106 | 492.0 | Region | Note=Nicotinamide ribonucleotide binding |
| Hgene | NAMPT | chr7:105915391 | chr12:88452797 | ENST00000354289 | - | 3 | 8 | 311_313 | 106 | 369.0 | Region | Note=Nicotinamide ribonucleotide binding |
| Hgene | NAMPT | chr7:105915391 | chr12:88452797 | ENST00000354289 | - | 3 | 8 | 353_354 | 106 | 369.0 | Region | Note=Nicotinamide ribonucleotide binding |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000397838 | 39 | 46 | 1071_1498 | 1275 | 1540.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000397838 | 39 | 46 | 598_664 | 1275 | 1540.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000397838 | 39 | 46 | 59_565 | 1275 | 1540.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000397838 | 39 | 46 | 697_931 | 1275 | 1540.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000397838 | 39 | 46 | 958_1027 | 1275 | 1540.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000547691 | 22 | 29 | 1071_1498 | 1275 | 1540.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000547691 | 22 | 29 | 598_664 | 1275 | 1540.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000547691 | 22 | 29 | 59_565 | 1275 | 1540.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000547691 | 22 | 29 | 697_931 | 1275 | 1540.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000547691 | 22 | 29 | 958_1027 | 1275 | 1540.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000552810 | 47 | 54 | 1071_1498 | 2215 | 2480.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000552810 | 47 | 54 | 1533_1584 | 2215 | 2480.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000552810 | 47 | 54 | 1635_2452 | 2215 | 2480.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000552810 | 47 | 54 | 598_664 | 2215 | 2480.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000552810 | 47 | 54 | 59_565 | 2215 | 2480.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000552810 | 47 | 54 | 697_931 | 2215 | 2480.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000552810 | 47 | 54 | 958_1027 | 2215 | 2480.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000397838 | 39 | 46 | 1_695 | 1275 | 1540.0 | Region | Self-association (with itself or C-terminus) | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000547691 | 22 | 29 | 1_695 | 1275 | 1540.0 | Region | Self-association (with itself or C-terminus) | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000552810 | 47 | 54 | 1966_2479 | 2215 | 2480.0 | Region | Self-association (with itself or N-terminus) | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000552810 | 47 | 54 | 1_695 | 2215 | 2480.0 | Region | Self-association (with itself or C-terminus) |
Top |
Fusion Gene Sequence for NAMPT-CEP290 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >57162_57162_1_NAMPT-CEP290_NAMPT_chr7_105915391_ENST00000222553_CEP290_chr12_88452797_ENST00000309041_length(transcript)=1421nt_BP=626nt GCTGCCGCGCCCCGCCCTTTCTCGGCCCCCGGAGGGTGACGGGGTGAAGGCGGGGGAACCGAGGTGGGGAGTCCGCCAGAGCTCCCAGAC TGCGAGCACGCGAGCCGCCGCAGCCGTCACCCGCGCCGCGTCACGGCTCCCGGGCCCGCCCTCCTCTGACCCCTCCCCTCTCTCCGTTTC CCCCTCTCCCCCTCCTCCGCCGACCGAGCAGTGACTTAAGCAACGGAGCGCGGTGAAGCTCATTTTTCTCCTTCCTCGCAGCCGCGCCAG GGAGCTCGCGGCGCGCGGCCCCTGTCCTCCGGCCCGAGATGAATCCTGCGGCAGAAGCCGAGTTCAACATCCTCCTGGCCACCGACTCCT ACAAGGTTACTCACTATAAACAATATCCACCCAACACAAGCAAAGTTTATTCCTACTTTGAATGCCGTGAAAAGAAGACAGAAAACTCCA AATTAAGGAAGGTGAAATATGAGGAAACAGTATTTTATGGGTTGCAGTACATTCTTAATAAGTACTTAAAAGGTAAAGTAGTAACCAAAG AGAAAATCCAGGAAGCCAAAGATGTCTACAAAGAACATTTCCAAGATGATGTCTTTAATGAAAAGGGATGGAACTACATTCTTGAGGAAA CTGATGCTGCAGAGAAATTACGGATAGCAAAGAATAATTTAGAGATATTAAATGAGAAGATGACAGTTCAACTAGAAGAGACTGGTAAGA GATTGCAGTTTGCAGAAAGCAGAGGTCCACAGCTTGAAGGTGCTGACAGTAAGAGCTGGAAATCCATTGTGGTTACAAGAATGTATGAAA CCAAGTTAAAAGAATTGGAAACTGATATTGCCAAAAAAAATCAAAGCATTACTGACCTTAAACAGCTTGTAAAAGAAGCAACAGAGAGAG AACAAAAAGTTAACAAATACAATGAAGACCTTGAACAACAGATTAAGATTCTTAAACATGTTCCTGAAGGTGCTGAGACAGAGCAAGGCC TTAAACGGGAGCTTCAAGTTCTTAGATTAGCTAATCATCAGCTGGATAAAGAGAAAGCAGAATTAATCCATCAGATAGAAGCTAACAAGG ACCAAAGTGGAGCTGAAAGCACCATACCTGATGCTGATCAACTAAAGGAAAAAATAAAAGATCTAGAGACACAGCTCAAAATGTCAGATC TAGAAAAGCAGCATTTGAAGGAGGAAATAAAGAAGCTGAAAAAAGAACTGGAAAATTTTGATCCTTCATTTTTTGAAGAAATTGAAGATC TTAAGTATAATTACAAGGAAGAAGTGAAGAAGAATATTCTCTTAGAAGAGAAGGTAAAAAAACTTTCAGAACAATTGGGAGTTGAATTAA >57162_57162_1_NAMPT-CEP290_NAMPT_chr7_105915391_ENST00000222553_CEP290_chr12_88452797_ENST00000309041_length(amino acids)=371AA_BP=106 MNPAAEAEFNILLATDSYKVTHYKQYPPNTSKVYSYFECREKKTENSKLRKVKYEETVFYGLQYILNKYLKGKVVTKEKIQEAKDVYKEH FQDDVFNEKGWNYILEETDAAEKLRIAKNNLEILNEKMTVQLEETGKRLQFAESRGPQLEGADSKSWKSIVVTRMYETKLKELETDIAKK NQSITDLKQLVKEATEREQKVNKYNEDLEQQIKILKHVPEGAETEQGLKRELQVLRLANHQLDKEKAELIHQIEANKDQSGAESTIPDAD QLKEKIKDLETQLKMSDLEKQHLKEEIKKLKKELENFDPSFFEEIEDLKYNYKEEVKKNILLEEKVKKLSEQLGVELTSPVAASEEFEDE -------------------------------------------------------------- >57162_57162_2_NAMPT-CEP290_NAMPT_chr7_105915391_ENST00000222553_CEP290_chr12_88452797_ENST00000397838_length(transcript)=1421nt_BP=626nt GCTGCCGCGCCCCGCCCTTTCTCGGCCCCCGGAGGGTGACGGGGTGAAGGCGGGGGAACCGAGGTGGGGAGTCCGCCAGAGCTCCCAGAC TGCGAGCACGCGAGCCGCCGCAGCCGTCACCCGCGCCGCGTCACGGCTCCCGGGCCCGCCCTCCTCTGACCCCTCCCCTCTCTCCGTTTC CCCCTCTCCCCCTCCTCCGCCGACCGAGCAGTGACTTAAGCAACGGAGCGCGGTGAAGCTCATTTTTCTCCTTCCTCGCAGCCGCGCCAG GGAGCTCGCGGCGCGCGGCCCCTGTCCTCCGGCCCGAGATGAATCCTGCGGCAGAAGCCGAGTTCAACATCCTCCTGGCCACCGACTCCT ACAAGGTTACTCACTATAAACAATATCCACCCAACACAAGCAAAGTTTATTCCTACTTTGAATGCCGTGAAAAGAAGACAGAAAACTCCA AATTAAGGAAGGTGAAATATGAGGAAACAGTATTTTATGGGTTGCAGTACATTCTTAATAAGTACTTAAAAGGTAAAGTAGTAACCAAAG AGAAAATCCAGGAAGCCAAAGATGTCTACAAAGAACATTTCCAAGATGATGTCTTTAATGAAAAGGGATGGAACTACATTCTTGAGGAAA CTGATGCTGCAGAGAAATTACGGATAGCAAAGAATAATTTAGAGATATTAAATGAGAAGATGACAGTTCAACTAGAAGAGACTGGTAAGA GATTGCAGTTTGCAGAAAGCAGAGGTCCACAGCTTGAAGGTGCTGACAGTAAGAGCTGGAAATCCATTGTGGTTACAAGAATGTATGAAA CCAAGTTAAAAGAATTGGAAACTGATATTGCCAAAAAAAATCAAAGCATTACTGACCTTAAACAGCTTGTAAAAGAAGCAACAGAGAGAG AACAAAAAGTTAACAAATACAATGAAGACCTTGAACAACAGATTAAGATTCTTAAACATGTTCCTGAAGGTGCTGAGACAGAGCAAGGCC TTAAACGGGAGCTTCAAGTTCTTAGATTAGCTAATCATCAGCTGGATAAAGAGAAAGCAGAATTAATCCATCAGATAGAAGCTAACAAGG ACCAAAGTGGAGCTGAAAGCACCATACCTGATGCTGATCAACTAAAGGAAAAAATAAAAGATCTAGAGACACAGCTCAAAATGTCAGATC TAGAAAAGCAGCATTTGAAGGAGGAAATAAAGAAGCTGAAAAAAGAACTGGAAAATTTTGATCCTTCATTTTTTGAAGAAATTGAAGATC TTAAGTATAATTACAAGGAAGAAGTGAAGAAGAATATTCTCTTAGAAGAGAAGGTAAAAAAACTTTCAGAACAATTGGGAGTTGAATTAA >57162_57162_2_NAMPT-CEP290_NAMPT_chr7_105915391_ENST00000222553_CEP290_chr12_88452797_ENST00000397838_length(amino acids)=371AA_BP=106 MNPAAEAEFNILLATDSYKVTHYKQYPPNTSKVYSYFECREKKTENSKLRKVKYEETVFYGLQYILNKYLKGKVVTKEKIQEAKDVYKEH FQDDVFNEKGWNYILEETDAAEKLRIAKNNLEILNEKMTVQLEETGKRLQFAESRGPQLEGADSKSWKSIVVTRMYETKLKELETDIAKK NQSITDLKQLVKEATEREQKVNKYNEDLEQQIKILKHVPEGAETEQGLKRELQVLRLANHQLDKEKAELIHQIEANKDQSGAESTIPDAD QLKEKIKDLETQLKMSDLEKQHLKEEIKKLKKELENFDPSFFEEIEDLKYNYKEEVKKNILLEEKVKKLSEQLGVELTSPVAASEEFEDE -------------------------------------------------------------- >57162_57162_3_NAMPT-CEP290_NAMPT_chr7_105915391_ENST00000222553_CEP290_chr12_88452797_ENST00000547691_length(transcript)=1589nt_BP=626nt GCTGCCGCGCCCCGCCCTTTCTCGGCCCCCGGAGGGTGACGGGGTGAAGGCGGGGGAACCGAGGTGGGGAGTCCGCCAGAGCTCCCAGAC TGCGAGCACGCGAGCCGCCGCAGCCGTCACCCGCGCCGCGTCACGGCTCCCGGGCCCGCCCTCCTCTGACCCCTCCCCTCTCTCCGTTTC CCCCTCTCCCCCTCCTCCGCCGACCGAGCAGTGACTTAAGCAACGGAGCGCGGTGAAGCTCATTTTTCTCCTTCCTCGCAGCCGCGCCAG GGAGCTCGCGGCGCGCGGCCCCTGTCCTCCGGCCCGAGATGAATCCTGCGGCAGAAGCCGAGTTCAACATCCTCCTGGCCACCGACTCCT ACAAGGTTACTCACTATAAACAATATCCACCCAACACAAGCAAAGTTTATTCCTACTTTGAATGCCGTGAAAAGAAGACAGAAAACTCCA AATTAAGGAAGGTGAAATATGAGGAAACAGTATTTTATGGGTTGCAGTACATTCTTAATAAGTACTTAAAAGGTAAAGTAGTAACCAAAG AGAAAATCCAGGAAGCCAAAGATGTCTACAAAGAACATTTCCAAGATGATGTCTTTAATGAAAAGGGATGGAACTACATTCTTGAGGAAA CTGATGCTGCAGAGAAATTACGGATAGCAAAGAATAATTTAGAGATATTAAATGAGAAGATGACAGTTCAACTAGAAGAGACTGGTAAGA GATTGCAGTTTGCAGAAAGCAGAGGTCCACAGCTTGAAGGTGCTGACAGTAAGAGCTGGAAATCCATTGTGGTTACAAGAATGTATGAAA CCAAGTTAAAAGAATTGGAAACTGATATTGCCAAAAAAAATCAAAGCATTACTGACCTTAAACAGCTTGTAAAAGAAGCAACAGAGAGAG AACAAAAAGTTAACAAATACAATGAAGACCTTGAACAACAGATTAAGATTCTTAAACATGTTCCTGAAGGTGCTGAGACAGAGCAAGGCC TTAAACGGGAGCTTCAAGTTCTTAGATTAGCTAATCATCAGCTGGATAAAGAGAAAGCAGAATTAATCCATCAGATAGAAGCTAACAAGG ACCAAAGTGGAGCTGAAAGCACCATACCTGATGCTGATCAACTAAAGGAAAAAATAAAAGATCTAGAGACACAGCTCAAAATGTCAGATC TAGAAAAGCAGCATTTGAAGGAGGAAATAAAGAAGCTGAAAAAAGAACTGGAAAATTTTGATCCTTCATTTTTTGAAGAAATTGAAGATC TTAAGTATAATTACAAGGAAGAAGTGAAGAAGAATATTCTCTTAGAAGAGAAGGTAAAAAAACTTTCAGAACAATTGGGAGTTGAATTAA CTAGCCCTGTTGCTGCTTCTGAAGAGTTTGAAGATGAAGAAGAAAGTCCTGTTAATTTCCCCATTTACTAAAGGTCACCTATAAACTTTG TTTCATTTAACTATTTATTAACTTTATAAGTTAAATATACTTGGAAATAAGCAGTTCTCCGAACTGTAGTATTTCCTTCTCACTACCTTG >57162_57162_3_NAMPT-CEP290_NAMPT_chr7_105915391_ENST00000222553_CEP290_chr12_88452797_ENST00000547691_length(amino acids)=370AA_BP=106 MNPAAEAEFNILLATDSYKVTHYKQYPPNTSKVYSYFECREKKTENSKLRKVKYEETVFYGLQYILNKYLKGKVVTKEKIQEAKDVYKEH FQDDVFNEKGWNYILEETDAAEKLRIAKNNLEILNEKMTVQLEETGKRLQFAESRGPQLEGADSKSWKSIVVTRMYETKLKELETDIAKK NQSITDLKQLVKEATEREQKVNKYNEDLEQQIKILKHVPEGAETEQGLKRELQVLRLANHQLDKEKAELIHQIEANKDQSGAESTIPDAD QLKEKIKDLETQLKMSDLEKQHLKEEIKKLKKELENFDPSFFEEIEDLKYNYKEEVKKNILLEEKVKKLSEQLGVELTSPVAASEEFEDE -------------------------------------------------------------- >57162_57162_4_NAMPT-CEP290_NAMPT_chr7_105915391_ENST00000222553_CEP290_chr12_88452797_ENST00000552810_length(transcript)=1585nt_BP=626nt GCTGCCGCGCCCCGCCCTTTCTCGGCCCCCGGAGGGTGACGGGGTGAAGGCGGGGGAACCGAGGTGGGGAGTCCGCCAGAGCTCCCAGAC TGCGAGCACGCGAGCCGCCGCAGCCGTCACCCGCGCCGCGTCACGGCTCCCGGGCCCGCCCTCCTCTGACCCCTCCCCTCTCTCCGTTTC CCCCTCTCCCCCTCCTCCGCCGACCGAGCAGTGACTTAAGCAACGGAGCGCGGTGAAGCTCATTTTTCTCCTTCCTCGCAGCCGCGCCAG GGAGCTCGCGGCGCGCGGCCCCTGTCCTCCGGCCCGAGATGAATCCTGCGGCAGAAGCCGAGTTCAACATCCTCCTGGCCACCGACTCCT ACAAGGTTACTCACTATAAACAATATCCACCCAACACAAGCAAAGTTTATTCCTACTTTGAATGCCGTGAAAAGAAGACAGAAAACTCCA AATTAAGGAAGGTGAAATATGAGGAAACAGTATTTTATGGGTTGCAGTACATTCTTAATAAGTACTTAAAAGGTAAAGTAGTAACCAAAG AGAAAATCCAGGAAGCCAAAGATGTCTACAAAGAACATTTCCAAGATGATGTCTTTAATGAAAAGGGATGGAACTACATTCTTGAGGAAA CTGATGCTGCAGAGAAATTACGGATAGCAAAGAATAATTTAGAGATATTAAATGAGAAGATGACAGTTCAACTAGAAGAGACTGGTAAGA GATTGCAGTTTGCAGAAAGCAGAGGTCCACAGCTTGAAGGTGCTGACAGTAAGAGCTGGAAATCCATTGTGGTTACAAGAATGTATGAAA CCAAGTTAAAAGAATTGGAAACTGATATTGCCAAAAAAAATCAAAGCATTACTGACCTTAAACAGCTTGTAAAAGAAGCAACAGAGAGAG AACAAAAAGTTAACAAATACAATGAAGACCTTGAACAACAGATTAAGATTCTTAAACATGTTCCTGAAGGTGCTGAGACAGAGCAAGGCC TTAAACGGGAGCTTCAAGTTCTTAGATTAGCTAATCATCAGCTGGATAAAGAGAAAGCAGAATTAATCCATCAGATAGAAGCTAACAAGG ACCAAAGTGGAGCTGAAAGCACCATACCTGATGCTGATCAACTAAAGGAAAAAATAAAAGATCTAGAGACACAGCTCAAAATGTCAGATC TAGAAAAGCAGCATTTGAAGGAGGAAATAAAGAAGCTGAAAAAAGAACTGGAAAATTTTGATCCTTCATTTTTTGAAGAAATTGAAGATC TTAAGTATAATTACAAGGAAGAAGTGAAGAAGAATATTCTCTTAGAAGAGAAGGTAAAAAAACTTTCAGAACAATTGGGAGTTGAATTAA CTAGCCCTGTTGCTGCTTCTGAAGAGTTTGAAGATGAAGAAGAAAGTCCTGTTAATTTCCCCATTTACTAAAGGTCACCTATAAACTTTG TTTCATTTAACTATTTATTAACTTTATAAGTTAAATATACTTGGAAATAAGCAGTTCTCCGAACTGTAGTATTTCCTTCTCACTACCTTG >57162_57162_4_NAMPT-CEP290_NAMPT_chr7_105915391_ENST00000222553_CEP290_chr12_88452797_ENST00000552810_length(amino acids)=370AA_BP=106 MNPAAEAEFNILLATDSYKVTHYKQYPPNTSKVYSYFECREKKTENSKLRKVKYEETVFYGLQYILNKYLKGKVVTKEKIQEAKDVYKEH FQDDVFNEKGWNYILEETDAAEKLRIAKNNLEILNEKMTVQLEETGKRLQFAESRGPQLEGADSKSWKSIVVTRMYETKLKELETDIAKK NQSITDLKQLVKEATEREQKVNKYNEDLEQQIKILKHVPEGAETEQGLKRELQVLRLANHQLDKEKAELIHQIEANKDQSGAESTIPDAD QLKEKIKDLETQLKMSDLEKQHLKEEIKKLKKELENFDPSFFEEIEDLKYNYKEEVKKNILLEEKVKKLSEQLGVELTSPVAASEEFEDE -------------------------------------------------------------- >57162_57162_5_NAMPT-CEP290_NAMPT_chr7_105915391_ENST00000354289_CEP290_chr12_88452797_ENST00000309041_length(transcript)=1150nt_BP=355nt GAGCTCGCGGCGCGCGGCCCCTGTCCTCCGGCCCGAGATGAATCCTGCGGCAGAAGCCGAGTTCAACATCCTCCTGGCCACCGACTCCTA CAAGGTTACTCACTATAAACAATATCCACCCAACACAAGCAAAGTTTATTCCTACTTTGAATGCCGTGAAAAGAAGACAGAAAACTCCAA ATTAAGGAAGGTGAAATATGAGGAAACAGTATTTTATGGGTTGCAGTACATTCTTAATAAGTACTTAAAAGGTAAAGTAGTAACCAAAGA GAAAATCCAGGAAGCCAAAGATGTCTACAAAGAACATTTCCAAGATGATGTCTTTAATGAAAAGGGATGGAACTACATTCTTGAGGAAAC TGATGCTGCAGAGAAATTACGGATAGCAAAGAATAATTTAGAGATATTAAATGAGAAGATGACAGTTCAACTAGAAGAGACTGGTAAGAG ATTGCAGTTTGCAGAAAGCAGAGGTCCACAGCTTGAAGGTGCTGACAGTAAGAGCTGGAAATCCATTGTGGTTACAAGAATGTATGAAAC CAAGTTAAAAGAATTGGAAACTGATATTGCCAAAAAAAATCAAAGCATTACTGACCTTAAACAGCTTGTAAAAGAAGCAACAGAGAGAGA ACAAAAAGTTAACAAATACAATGAAGACCTTGAACAACAGATTAAGATTCTTAAACATGTTCCTGAAGGTGCTGAGACAGAGCAAGGCCT TAAACGGGAGCTTCAAGTTCTTAGATTAGCTAATCATCAGCTGGATAAAGAGAAAGCAGAATTAATCCATCAGATAGAAGCTAACAAGGA CCAAAGTGGAGCTGAAAGCACCATACCTGATGCTGATCAACTAAAGGAAAAAATAAAAGATCTAGAGACACAGCTCAAAATGTCAGATCT AGAAAAGCAGCATTTGAAGGAGGAAATAAAGAAGCTGAAAAAAGAACTGGAAAATTTTGATCCTTCATTTTTTGAAGAAATTGAAGATCT TAAGTATAATTACAAGGAAGAAGTGAAGAAGAATATTCTCTTAGAAGAGAAGGTAAAAAAACTTTCAGAACAATTGGGAGTTGAATTAAC >57162_57162_5_NAMPT-CEP290_NAMPT_chr7_105915391_ENST00000354289_CEP290_chr12_88452797_ENST00000309041_length(amino acids)=371AA_BP=106 MNPAAEAEFNILLATDSYKVTHYKQYPPNTSKVYSYFECREKKTENSKLRKVKYEETVFYGLQYILNKYLKGKVVTKEKIQEAKDVYKEH FQDDVFNEKGWNYILEETDAAEKLRIAKNNLEILNEKMTVQLEETGKRLQFAESRGPQLEGADSKSWKSIVVTRMYETKLKELETDIAKK NQSITDLKQLVKEATEREQKVNKYNEDLEQQIKILKHVPEGAETEQGLKRELQVLRLANHQLDKEKAELIHQIEANKDQSGAESTIPDAD QLKEKIKDLETQLKMSDLEKQHLKEEIKKLKKELENFDPSFFEEIEDLKYNYKEEVKKNILLEEKVKKLSEQLGVELTSPVAASEEFEDE -------------------------------------------------------------- >57162_57162_6_NAMPT-CEP290_NAMPT_chr7_105915391_ENST00000354289_CEP290_chr12_88452797_ENST00000397838_length(transcript)=1150nt_BP=355nt GAGCTCGCGGCGCGCGGCCCCTGTCCTCCGGCCCGAGATGAATCCTGCGGCAGAAGCCGAGTTCAACATCCTCCTGGCCACCGACTCCTA CAAGGTTACTCACTATAAACAATATCCACCCAACACAAGCAAAGTTTATTCCTACTTTGAATGCCGTGAAAAGAAGACAGAAAACTCCAA ATTAAGGAAGGTGAAATATGAGGAAACAGTATTTTATGGGTTGCAGTACATTCTTAATAAGTACTTAAAAGGTAAAGTAGTAACCAAAGA GAAAATCCAGGAAGCCAAAGATGTCTACAAAGAACATTTCCAAGATGATGTCTTTAATGAAAAGGGATGGAACTACATTCTTGAGGAAAC TGATGCTGCAGAGAAATTACGGATAGCAAAGAATAATTTAGAGATATTAAATGAGAAGATGACAGTTCAACTAGAAGAGACTGGTAAGAG ATTGCAGTTTGCAGAAAGCAGAGGTCCACAGCTTGAAGGTGCTGACAGTAAGAGCTGGAAATCCATTGTGGTTACAAGAATGTATGAAAC CAAGTTAAAAGAATTGGAAACTGATATTGCCAAAAAAAATCAAAGCATTACTGACCTTAAACAGCTTGTAAAAGAAGCAACAGAGAGAGA ACAAAAAGTTAACAAATACAATGAAGACCTTGAACAACAGATTAAGATTCTTAAACATGTTCCTGAAGGTGCTGAGACAGAGCAAGGCCT TAAACGGGAGCTTCAAGTTCTTAGATTAGCTAATCATCAGCTGGATAAAGAGAAAGCAGAATTAATCCATCAGATAGAAGCTAACAAGGA CCAAAGTGGAGCTGAAAGCACCATACCTGATGCTGATCAACTAAAGGAAAAAATAAAAGATCTAGAGACACAGCTCAAAATGTCAGATCT AGAAAAGCAGCATTTGAAGGAGGAAATAAAGAAGCTGAAAAAAGAACTGGAAAATTTTGATCCTTCATTTTTTGAAGAAATTGAAGATCT TAAGTATAATTACAAGGAAGAAGTGAAGAAGAATATTCTCTTAGAAGAGAAGGTAAAAAAACTTTCAGAACAATTGGGAGTTGAATTAAC >57162_57162_6_NAMPT-CEP290_NAMPT_chr7_105915391_ENST00000354289_CEP290_chr12_88452797_ENST00000397838_length(amino acids)=371AA_BP=106 MNPAAEAEFNILLATDSYKVTHYKQYPPNTSKVYSYFECREKKTENSKLRKVKYEETVFYGLQYILNKYLKGKVVTKEKIQEAKDVYKEH FQDDVFNEKGWNYILEETDAAEKLRIAKNNLEILNEKMTVQLEETGKRLQFAESRGPQLEGADSKSWKSIVVTRMYETKLKELETDIAKK NQSITDLKQLVKEATEREQKVNKYNEDLEQQIKILKHVPEGAETEQGLKRELQVLRLANHQLDKEKAELIHQIEANKDQSGAESTIPDAD QLKEKIKDLETQLKMSDLEKQHLKEEIKKLKKELENFDPSFFEEIEDLKYNYKEEVKKNILLEEKVKKLSEQLGVELTSPVAASEEFEDE -------------------------------------------------------------- >57162_57162_7_NAMPT-CEP290_NAMPT_chr7_105915391_ENST00000354289_CEP290_chr12_88452797_ENST00000547691_length(transcript)=1318nt_BP=355nt GAGCTCGCGGCGCGCGGCCCCTGTCCTCCGGCCCGAGATGAATCCTGCGGCAGAAGCCGAGTTCAACATCCTCCTGGCCACCGACTCCTA CAAGGTTACTCACTATAAACAATATCCACCCAACACAAGCAAAGTTTATTCCTACTTTGAATGCCGTGAAAAGAAGACAGAAAACTCCAA ATTAAGGAAGGTGAAATATGAGGAAACAGTATTTTATGGGTTGCAGTACATTCTTAATAAGTACTTAAAAGGTAAAGTAGTAACCAAAGA GAAAATCCAGGAAGCCAAAGATGTCTACAAAGAACATTTCCAAGATGATGTCTTTAATGAAAAGGGATGGAACTACATTCTTGAGGAAAC TGATGCTGCAGAGAAATTACGGATAGCAAAGAATAATTTAGAGATATTAAATGAGAAGATGACAGTTCAACTAGAAGAGACTGGTAAGAG ATTGCAGTTTGCAGAAAGCAGAGGTCCACAGCTTGAAGGTGCTGACAGTAAGAGCTGGAAATCCATTGTGGTTACAAGAATGTATGAAAC CAAGTTAAAAGAATTGGAAACTGATATTGCCAAAAAAAATCAAAGCATTACTGACCTTAAACAGCTTGTAAAAGAAGCAACAGAGAGAGA ACAAAAAGTTAACAAATACAATGAAGACCTTGAACAACAGATTAAGATTCTTAAACATGTTCCTGAAGGTGCTGAGACAGAGCAAGGCCT TAAACGGGAGCTTCAAGTTCTTAGATTAGCTAATCATCAGCTGGATAAAGAGAAAGCAGAATTAATCCATCAGATAGAAGCTAACAAGGA CCAAAGTGGAGCTGAAAGCACCATACCTGATGCTGATCAACTAAAGGAAAAAATAAAAGATCTAGAGACACAGCTCAAAATGTCAGATCT AGAAAAGCAGCATTTGAAGGAGGAAATAAAGAAGCTGAAAAAAGAACTGGAAAATTTTGATCCTTCATTTTTTGAAGAAATTGAAGATCT TAAGTATAATTACAAGGAAGAAGTGAAGAAGAATATTCTCTTAGAAGAGAAGGTAAAAAAACTTTCAGAACAATTGGGAGTTGAATTAAC TAGCCCTGTTGCTGCTTCTGAAGAGTTTGAAGATGAAGAAGAAAGTCCTGTTAATTTCCCCATTTACTAAAGGTCACCTATAAACTTTGT TTCATTTAACTATTTATTAACTTTATAAGTTAAATATACTTGGAAATAAGCAGTTCTCCGAACTGTAGTATTTCCTTCTCACTACCTTGT >57162_57162_7_NAMPT-CEP290_NAMPT_chr7_105915391_ENST00000354289_CEP290_chr12_88452797_ENST00000547691_length(amino acids)=370AA_BP=106 MNPAAEAEFNILLATDSYKVTHYKQYPPNTSKVYSYFECREKKTENSKLRKVKYEETVFYGLQYILNKYLKGKVVTKEKIQEAKDVYKEH FQDDVFNEKGWNYILEETDAAEKLRIAKNNLEILNEKMTVQLEETGKRLQFAESRGPQLEGADSKSWKSIVVTRMYETKLKELETDIAKK NQSITDLKQLVKEATEREQKVNKYNEDLEQQIKILKHVPEGAETEQGLKRELQVLRLANHQLDKEKAELIHQIEANKDQSGAESTIPDAD QLKEKIKDLETQLKMSDLEKQHLKEEIKKLKKELENFDPSFFEEIEDLKYNYKEEVKKNILLEEKVKKLSEQLGVELTSPVAASEEFEDE -------------------------------------------------------------- >57162_57162_8_NAMPT-CEP290_NAMPT_chr7_105915391_ENST00000354289_CEP290_chr12_88452797_ENST00000552810_length(transcript)=1314nt_BP=355nt GAGCTCGCGGCGCGCGGCCCCTGTCCTCCGGCCCGAGATGAATCCTGCGGCAGAAGCCGAGTTCAACATCCTCCTGGCCACCGACTCCTA CAAGGTTACTCACTATAAACAATATCCACCCAACACAAGCAAAGTTTATTCCTACTTTGAATGCCGTGAAAAGAAGACAGAAAACTCCAA ATTAAGGAAGGTGAAATATGAGGAAACAGTATTTTATGGGTTGCAGTACATTCTTAATAAGTACTTAAAAGGTAAAGTAGTAACCAAAGA GAAAATCCAGGAAGCCAAAGATGTCTACAAAGAACATTTCCAAGATGATGTCTTTAATGAAAAGGGATGGAACTACATTCTTGAGGAAAC TGATGCTGCAGAGAAATTACGGATAGCAAAGAATAATTTAGAGATATTAAATGAGAAGATGACAGTTCAACTAGAAGAGACTGGTAAGAG ATTGCAGTTTGCAGAAAGCAGAGGTCCACAGCTTGAAGGTGCTGACAGTAAGAGCTGGAAATCCATTGTGGTTACAAGAATGTATGAAAC CAAGTTAAAAGAATTGGAAACTGATATTGCCAAAAAAAATCAAAGCATTACTGACCTTAAACAGCTTGTAAAAGAAGCAACAGAGAGAGA ACAAAAAGTTAACAAATACAATGAAGACCTTGAACAACAGATTAAGATTCTTAAACATGTTCCTGAAGGTGCTGAGACAGAGCAAGGCCT TAAACGGGAGCTTCAAGTTCTTAGATTAGCTAATCATCAGCTGGATAAAGAGAAAGCAGAATTAATCCATCAGATAGAAGCTAACAAGGA CCAAAGTGGAGCTGAAAGCACCATACCTGATGCTGATCAACTAAAGGAAAAAATAAAAGATCTAGAGACACAGCTCAAAATGTCAGATCT AGAAAAGCAGCATTTGAAGGAGGAAATAAAGAAGCTGAAAAAAGAACTGGAAAATTTTGATCCTTCATTTTTTGAAGAAATTGAAGATCT TAAGTATAATTACAAGGAAGAAGTGAAGAAGAATATTCTCTTAGAAGAGAAGGTAAAAAAACTTTCAGAACAATTGGGAGTTGAATTAAC TAGCCCTGTTGCTGCTTCTGAAGAGTTTGAAGATGAAGAAGAAAGTCCTGTTAATTTCCCCATTTACTAAAGGTCACCTATAAACTTTGT TTCATTTAACTATTTATTAACTTTATAAGTTAAATATACTTGGAAATAAGCAGTTCTCCGAACTGTAGTATTTCCTTCTCACTACCTTGT >57162_57162_8_NAMPT-CEP290_NAMPT_chr7_105915391_ENST00000354289_CEP290_chr12_88452797_ENST00000552810_length(amino acids)=370AA_BP=106 MNPAAEAEFNILLATDSYKVTHYKQYPPNTSKVYSYFECREKKTENSKLRKVKYEETVFYGLQYILNKYLKGKVVTKEKIQEAKDVYKEH FQDDVFNEKGWNYILEETDAAEKLRIAKNNLEILNEKMTVQLEETGKRLQFAESRGPQLEGADSKSWKSIVVTRMYETKLKELETDIAKK NQSITDLKQLVKEATEREQKVNKYNEDLEQQIKILKHVPEGAETEQGLKRELQVLRLANHQLDKEKAELIHQIEANKDQSGAESTIPDAD QLKEKIKDLETQLKMSDLEKQHLKEEIKKLKKELENFDPSFFEEIEDLKYNYKEEVKKNILLEEKVKKLSEQLGVELTSPVAASEEFEDE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for NAMPT-CEP290 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000397838 | 39 | 46 | 696_896 | 1275.0 | 1540.0 | IQCB1 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000547691 | 22 | 29 | 696_896 | 1275.0 | 1540.0 | IQCB1 | |
| Tgene | CEP290 | chr7:105915391 | chr12:88452797 | ENST00000552810 | 47 | 54 | 696_896 | 2215.0 | 2480.0 | IQCB1 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for NAMPT-CEP290 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for NAMPT-CEP290 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
