
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:NAV2-ARL14EP (FusionGDB2 ID:57334) |
Fusion Gene Summary for NAV2-ARL14EP |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: NAV2-ARL14EP | Fusion gene ID: 57334 | Hgene | Tgene | Gene symbol | NAV2 | ARL14EP | Gene ID | 89797 | 120534 |
| Gene name | neuron navigator 2 | ADP ribosylation factor like GTPase 14 effector protein | |
| Synonyms | HELAD1|POMFIL2|RAINB1|STEERIN2|UNC53H2 | ARF7EP|C11orf46|dJ299F11.1 | |
| Cytomap | 11p15.1 | 11p14.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | neuron navigator 2helicase, APC down-regulated 1pore membrane and/or filament-interacting-like protein 2retinoic acid inducible gene in neuroblastoma 1steerin-2unc-53 homolog 2 | ARL14 effector proteinADP-ribosylation factor-like 14 effector proteinARF7 effector protein | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q8IVL1 | P0DKL9 | |
| Ensembl transtripts involved in fusion gene | ENST00000349880, ENST00000396085, ENST00000396087, ENST00000311043, ENST00000360655, ENST00000527559, ENST00000533917, ENST00000534229, ENST00000540292, | ENST00000533457, ENST00000282032, | |
| Fusion gene scores | * DoF score | 16 X 19 X 6=1824 | 2 X 3 X 2=12 |
| # samples | 19 | 2 | |
| ** MAII score | log2(19/1824*10)=-3.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(2/12*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: NAV2 [Title/Abstract] AND ARL14EP [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | NAV2(19735508)-ARL14EP(30352433), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across NAV2 (5'-gene) Fusion gene breakpoints across NAV2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
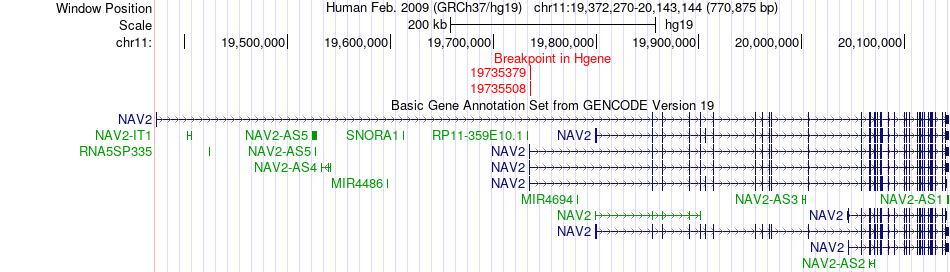 |
 Fusion gene breakpoints across ARL14EP (3'-gene) Fusion gene breakpoints across ARL14EP (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
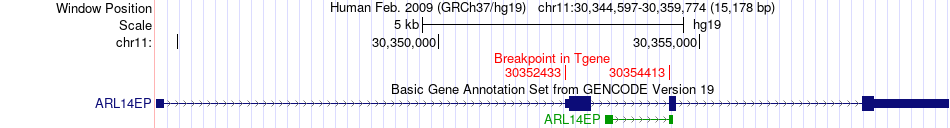 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SKCM | TCGA-WE-A8ZR-06A | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30352433 | + |
| ChimerDB4 | SKCM | TCGA-WE-A8ZR-06A | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + |
| ChimerDB4 | SKCM | TCGA-WE-A8ZR-06A | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352433 | + |
| ChimerDB4 | SKCM | TCGA-WE-A8ZR-06A | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + |
Top |
Fusion Gene ORF analysis for NAV2-ARL14EP |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000349880 | ENST00000533457 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + |
| 5CDS-3UTR | ENST00000349880 | ENST00000533457 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + |
| 5CDS-3UTR | ENST00000396085 | ENST00000533457 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + |
| 5CDS-3UTR | ENST00000396085 | ENST00000533457 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + |
| 5CDS-3UTR | ENST00000396087 | ENST00000533457 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + |
| 5CDS-3UTR | ENST00000396087 | ENST00000533457 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + |
| 5CDS-5UTR | ENST00000349880 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30352433 | + |
| 5CDS-5UTR | ENST00000349880 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352433 | + |
| 5CDS-5UTR | ENST00000396085 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30352433 | + |
| 5CDS-5UTR | ENST00000396085 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352433 | + |
| 5CDS-5UTR | ENST00000396087 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30352433 | + |
| 5CDS-5UTR | ENST00000396087 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352433 | + |
| 5CDS-intron | ENST00000349880 | ENST00000533457 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30352433 | + |
| 5CDS-intron | ENST00000349880 | ENST00000533457 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352433 | + |
| 5CDS-intron | ENST00000396085 | ENST00000533457 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30352433 | + |
| 5CDS-intron | ENST00000396085 | ENST00000533457 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352433 | + |
| 5CDS-intron | ENST00000396087 | ENST00000533457 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30352433 | + |
| 5CDS-intron | ENST00000396087 | ENST00000533457 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352433 | + |
| In-frame | ENST00000349880 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + |
| In-frame | ENST00000349880 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + |
| In-frame | ENST00000396085 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + |
| In-frame | ENST00000396085 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + |
| In-frame | ENST00000396087 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + |
| In-frame | ENST00000396087 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3CDS | ENST00000311043 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3CDS | ENST00000311043 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3CDS | ENST00000360655 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3CDS | ENST00000360655 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3CDS | ENST00000527559 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3CDS | ENST00000527559 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3CDS | ENST00000533917 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3CDS | ENST00000533917 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3CDS | ENST00000534229 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3CDS | ENST00000534229 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3CDS | ENST00000540292 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3CDS | ENST00000540292 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3UTR | ENST00000311043 | ENST00000533457 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3UTR | ENST00000311043 | ENST00000533457 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3UTR | ENST00000360655 | ENST00000533457 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3UTR | ENST00000360655 | ENST00000533457 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3UTR | ENST00000527559 | ENST00000533457 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3UTR | ENST00000527559 | ENST00000533457 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3UTR | ENST00000533917 | ENST00000533457 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3UTR | ENST00000533917 | ENST00000533457 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3UTR | ENST00000534229 | ENST00000533457 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3UTR | ENST00000534229 | ENST00000533457 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3UTR | ENST00000540292 | ENST00000533457 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + |
| intron-3UTR | ENST00000540292 | ENST00000533457 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + |
| intron-5UTR | ENST00000311043 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30352433 | + |
| intron-5UTR | ENST00000311043 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352433 | + |
| intron-5UTR | ENST00000360655 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30352433 | + |
| intron-5UTR | ENST00000360655 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352433 | + |
| intron-5UTR | ENST00000527559 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30352433 | + |
| intron-5UTR | ENST00000527559 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352433 | + |
| intron-5UTR | ENST00000533917 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30352433 | + |
| intron-5UTR | ENST00000533917 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352433 | + |
| intron-5UTR | ENST00000534229 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30352433 | + |
| intron-5UTR | ENST00000534229 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352433 | + |
| intron-5UTR | ENST00000540292 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30352433 | + |
| intron-5UTR | ENST00000540292 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352433 | + |
| intron-intron | ENST00000311043 | ENST00000533457 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30352433 | + |
| intron-intron | ENST00000311043 | ENST00000533457 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352433 | + |
| intron-intron | ENST00000360655 | ENST00000533457 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30352433 | + |
| intron-intron | ENST00000360655 | ENST00000533457 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352433 | + |
| intron-intron | ENST00000527559 | ENST00000533457 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30352433 | + |
| intron-intron | ENST00000527559 | ENST00000533457 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352433 | + |
| intron-intron | ENST00000533917 | ENST00000533457 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30352433 | + |
| intron-intron | ENST00000533917 | ENST00000533457 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352433 | + |
| intron-intron | ENST00000534229 | ENST00000533457 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30352433 | + |
| intron-intron | ENST00000534229 | ENST00000533457 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352433 | + |
| intron-intron | ENST00000540292 | ENST00000533457 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30352433 | + |
| intron-intron | ENST00000540292 | ENST00000533457 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352433 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000396085 | NAV2 | chr11 | 19735379 | + | ENST00000282032 | ARL14EP | chr11 | 30354413 | + | 1918 | 129 | 33 | 485 | 150 |
| ENST00000349880 | NAV2 | chr11 | 19735379 | + | ENST00000282032 | ARL14EP | chr11 | 30354413 | + | 1918 | 129 | 33 | 485 | 150 |
| ENST00000396087 | NAV2 | chr11 | 19735379 | + | ENST00000282032 | ARL14EP | chr11 | 30354413 | + | 1918 | 129 | 33 | 485 | 150 |
| ENST00000396085 | NAV2 | chr11 | 19735508 | + | ENST00000282032 | ARL14EP | chr11 | 30354413 | + | 2417 | 628 | 280 | 984 | 234 |
| ENST00000349880 | NAV2 | chr11 | 19735508 | + | ENST00000282032 | ARL14EP | chr11 | 30354413 | + | 2386 | 597 | 249 | 953 | 234 |
| ENST00000396087 | NAV2 | chr11 | 19735508 | + | ENST00000282032 | ARL14EP | chr11 | 30354413 | + | 2155 | 366 | 18 | 722 | 234 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000396085 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + | 0.6640558 | 0.33594424 |
| ENST00000349880 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + | 0.6640558 | 0.33594424 |
| ENST00000396087 | ENST00000282032 | NAV2 | chr11 | 19735379 | + | ARL14EP | chr11 | 30354413 | + | 0.6640558 | 0.33594424 |
| ENST00000396085 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + | 0.011278584 | 0.98872143 |
| ENST00000349880 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + | 0.010486656 | 0.98951334 |
| ENST00000396087 | ENST00000282032 | NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354413 | + | 0.006636871 | 0.99336314 |
Top |
Fusion Genomic Features for NAV2-ARL14EP |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354412 | + | 0.002143599 | 0.99785644 |
| NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352432 | + | 0.000375634 | 0.9996244 |
| NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30354412 | + | 0.002143599 | 0.99785644 |
| NAV2 | chr11 | 19735508 | + | ARL14EP | chr11 | 30352432 | + | 0.000375634 | 0.9996244 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
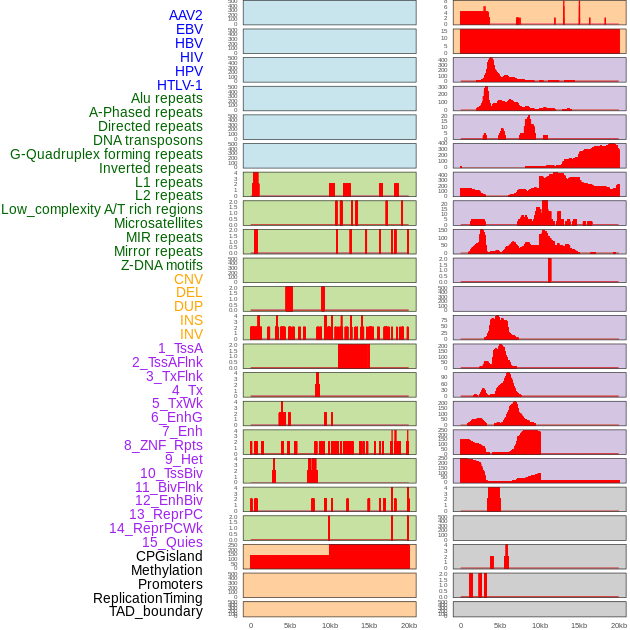 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
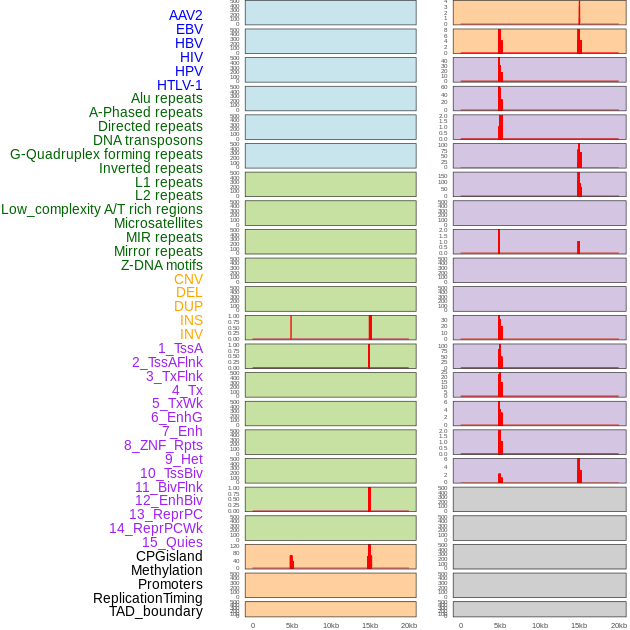 |
Top |
Fusion Protein Features for NAV2-ARL14EP |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:19735508/chr11:30352433) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NAV2 | ARL14EP |
| FUNCTION: Possesses 3' to 5' helicase activity and exonuclease activity. Involved in neuronal development, specifically in the development of different sensory organs. {ECO:0000269|PubMed:12214280, ECO:0000269|PubMed:15158073}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ARL14EP | chr11:19735379 | chr11:30354413 | ENST00000282032 | 1 | 4 | 209_237 | 142 | 261.0 | Compositional bias | Note=Cys-rich | |
| Tgene | ARL14EP | chr11:19735508 | chr11:30354413 | ENST00000282032 | 1 | 4 | 209_237 | 142 | 261.0 | Compositional bias | Note=Cys-rich |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 1686_1773 | 0 | 1494.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 1897_1964 | 0 | 1494.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 498_531 | 0 | 1494.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 743_771 | 0 | 1494.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 1686_1773 | 0 | 2430.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 1897_1964 | 0 | 2430.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 498_531 | 0 | 2430.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 743_771 | 0 | 2430.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 1686_1773 | 0 | 2366.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 1897_1964 | 0 | 2366.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 498_531 | 0 | 2366.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 743_771 | 0 | 2366.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 1686_1773 | 0 | 2433.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 1897_1964 | 0 | 2433.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 498_531 | 0 | 2433.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 743_771 | 0 | 2433.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 1686_1773 | 0 | 2489.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 1897_1964 | 0 | 2489.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 498_531 | 0 | 2489.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 743_771 | 0 | 2489.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 1686_1773 | 0 | 2418.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 1897_1964 | 0 | 2418.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 498_531 | 0 | 2418.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 743_771 | 0 | 2418.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 1686_1773 | 0 | 1494.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 1897_1964 | 0 | 1494.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 498_531 | 0 | 1494.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 743_771 | 0 | 1494.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 1686_1773 | 0 | 2420.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 1897_1964 | 0 | 2420.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 498_531 | 0 | 2420.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 743_771 | 0 | 2420.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 1686_1773 | 0 | 1494.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 1897_1964 | 0 | 1494.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 498_531 | 0 | 1494.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 743_771 | 0 | 1494.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 1686_1773 | 89 | 2430.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 1897_1964 | 89 | 2430.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 498_531 | 89 | 2430.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 743_771 | 89 | 2430.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 1686_1773 | 0 | 2366.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 1897_1964 | 0 | 2366.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 498_531 | 0 | 2366.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 743_771 | 0 | 2366.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 1686_1773 | 89 | 2433.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 1897_1964 | 89 | 2433.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 498_531 | 89 | 2433.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 743_771 | 89 | 2433.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 1686_1773 | 89 | 2489.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 1897_1964 | 89 | 2489.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 498_531 | 89 | 2489.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 743_771 | 89 | 2489.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 1686_1773 | 0 | 2418.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 1897_1964 | 0 | 2418.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 498_531 | 0 | 2418.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 743_771 | 0 | 2418.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 1686_1773 | 0 | 1494.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 1897_1964 | 0 | 1494.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 498_531 | 0 | 1494.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 743_771 | 0 | 1494.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 1686_1773 | 0 | 2420.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 1897_1964 | 0 | 2420.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 498_531 | 0 | 2420.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 743_771 | 0 | 2420.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 1363_1683 | 0 | 1494.0 | Compositional bias | Note=Ser-rich |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 1824_1829 | 0 | 1494.0 | Compositional bias | Note=Poly-Lys |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 192_255 | 0 | 1494.0 | Compositional bias | Note=Gln-rich |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 324_328 | 0 | 1494.0 | Compositional bias | Note=Poly-Pro |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 633_638 | 0 | 1494.0 | Compositional bias | Note=Poly-Ser |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 1363_1683 | 0 | 2430.0 | Compositional bias | Note=Ser-rich |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 1824_1829 | 0 | 2430.0 | Compositional bias | Note=Poly-Lys |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 192_255 | 0 | 2430.0 | Compositional bias | Note=Gln-rich |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 324_328 | 0 | 2430.0 | Compositional bias | Note=Poly-Pro |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 633_638 | 0 | 2430.0 | Compositional bias | Note=Poly-Ser |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 1363_1683 | 0 | 2366.0 | Compositional bias | Note=Ser-rich |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 1824_1829 | 0 | 2366.0 | Compositional bias | Note=Poly-Lys |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 192_255 | 0 | 2366.0 | Compositional bias | Note=Gln-rich |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 324_328 | 0 | 2366.0 | Compositional bias | Note=Poly-Pro |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 633_638 | 0 | 2366.0 | Compositional bias | Note=Poly-Ser |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 1363_1683 | 0 | 2433.0 | Compositional bias | Note=Ser-rich |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 1824_1829 | 0 | 2433.0 | Compositional bias | Note=Poly-Lys |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 192_255 | 0 | 2433.0 | Compositional bias | Note=Gln-rich |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 324_328 | 0 | 2433.0 | Compositional bias | Note=Poly-Pro |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 633_638 | 0 | 2433.0 | Compositional bias | Note=Poly-Ser |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 1363_1683 | 0 | 2489.0 | Compositional bias | Note=Ser-rich |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 1824_1829 | 0 | 2489.0 | Compositional bias | Note=Poly-Lys |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 192_255 | 0 | 2489.0 | Compositional bias | Note=Gln-rich |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 324_328 | 0 | 2489.0 | Compositional bias | Note=Poly-Pro |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 633_638 | 0 | 2489.0 | Compositional bias | Note=Poly-Ser |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 1363_1683 | 0 | 2418.0 | Compositional bias | Note=Ser-rich |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 1824_1829 | 0 | 2418.0 | Compositional bias | Note=Poly-Lys |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 192_255 | 0 | 2418.0 | Compositional bias | Note=Gln-rich |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 324_328 | 0 | 2418.0 | Compositional bias | Note=Poly-Pro |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 633_638 | 0 | 2418.0 | Compositional bias | Note=Poly-Ser |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 1363_1683 | 0 | 1494.0 | Compositional bias | Note=Ser-rich |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 1824_1829 | 0 | 1494.0 | Compositional bias | Note=Poly-Lys |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 192_255 | 0 | 1494.0 | Compositional bias | Note=Gln-rich |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 324_328 | 0 | 1494.0 | Compositional bias | Note=Poly-Pro |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 633_638 | 0 | 1494.0 | Compositional bias | Note=Poly-Ser |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 1363_1683 | 0 | 2420.0 | Compositional bias | Note=Ser-rich |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 1824_1829 | 0 | 2420.0 | Compositional bias | Note=Poly-Lys |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 192_255 | 0 | 2420.0 | Compositional bias | Note=Gln-rich |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 324_328 | 0 | 2420.0 | Compositional bias | Note=Poly-Pro |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 633_638 | 0 | 2420.0 | Compositional bias | Note=Poly-Ser |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 1363_1683 | 0 | 1494.0 | Compositional bias | Note=Ser-rich |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 1824_1829 | 0 | 1494.0 | Compositional bias | Note=Poly-Lys |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 192_255 | 0 | 1494.0 | Compositional bias | Note=Gln-rich |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 324_328 | 0 | 1494.0 | Compositional bias | Note=Poly-Pro |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 633_638 | 0 | 1494.0 | Compositional bias | Note=Poly-Ser |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 1363_1683 | 89 | 2430.0 | Compositional bias | Note=Ser-rich |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 1824_1829 | 89 | 2430.0 | Compositional bias | Note=Poly-Lys |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 192_255 | 89 | 2430.0 | Compositional bias | Note=Gln-rich |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 324_328 | 89 | 2430.0 | Compositional bias | Note=Poly-Pro |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 633_638 | 89 | 2430.0 | Compositional bias | Note=Poly-Ser |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 1363_1683 | 0 | 2366.0 | Compositional bias | Note=Ser-rich |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 1824_1829 | 0 | 2366.0 | Compositional bias | Note=Poly-Lys |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 192_255 | 0 | 2366.0 | Compositional bias | Note=Gln-rich |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 324_328 | 0 | 2366.0 | Compositional bias | Note=Poly-Pro |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 633_638 | 0 | 2366.0 | Compositional bias | Note=Poly-Ser |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 1363_1683 | 89 | 2433.0 | Compositional bias | Note=Ser-rich |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 1824_1829 | 89 | 2433.0 | Compositional bias | Note=Poly-Lys |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 192_255 | 89 | 2433.0 | Compositional bias | Note=Gln-rich |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 324_328 | 89 | 2433.0 | Compositional bias | Note=Poly-Pro |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 633_638 | 89 | 2433.0 | Compositional bias | Note=Poly-Ser |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 1363_1683 | 89 | 2489.0 | Compositional bias | Note=Ser-rich |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 1824_1829 | 89 | 2489.0 | Compositional bias | Note=Poly-Lys |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 192_255 | 89 | 2489.0 | Compositional bias | Note=Gln-rich |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 324_328 | 89 | 2489.0 | Compositional bias | Note=Poly-Pro |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 633_638 | 89 | 2489.0 | Compositional bias | Note=Poly-Ser |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 1363_1683 | 0 | 2418.0 | Compositional bias | Note=Ser-rich |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 1824_1829 | 0 | 2418.0 | Compositional bias | Note=Poly-Lys |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 192_255 | 0 | 2418.0 | Compositional bias | Note=Gln-rich |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 324_328 | 0 | 2418.0 | Compositional bias | Note=Poly-Pro |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 633_638 | 0 | 2418.0 | Compositional bias | Note=Poly-Ser |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 1363_1683 | 0 | 1494.0 | Compositional bias | Note=Ser-rich |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 1824_1829 | 0 | 1494.0 | Compositional bias | Note=Poly-Lys |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 192_255 | 0 | 1494.0 | Compositional bias | Note=Gln-rich |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 324_328 | 0 | 1494.0 | Compositional bias | Note=Poly-Pro |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 633_638 | 0 | 1494.0 | Compositional bias | Note=Poly-Ser |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 1363_1683 | 0 | 2420.0 | Compositional bias | Note=Ser-rich |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 1824_1829 | 0 | 2420.0 | Compositional bias | Note=Poly-Lys |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 192_255 | 0 | 2420.0 | Compositional bias | Note=Gln-rich |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 324_328 | 0 | 2420.0 | Compositional bias | Note=Poly-Pro |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 633_638 | 0 | 2420.0 | Compositional bias | Note=Poly-Ser |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 85_192 | 0 | 1494.0 | Domain | Calponin-homology (CH) |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 85_192 | 0 | 2430.0 | Domain | Calponin-homology (CH) |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 85_192 | 0 | 2366.0 | Domain | Calponin-homology (CH) |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 85_192 | 0 | 2433.0 | Domain | Calponin-homology (CH) |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 85_192 | 0 | 2489.0 | Domain | Calponin-homology (CH) |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 85_192 | 0 | 2418.0 | Domain | Calponin-homology (CH) |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 85_192 | 0 | 1494.0 | Domain | Calponin-homology (CH) |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 85_192 | 0 | 2420.0 | Domain | Calponin-homology (CH) |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 85_192 | 0 | 1494.0 | Domain | Calponin-homology (CH) |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 85_192 | 89 | 2430.0 | Domain | Calponin-homology (CH) |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 85_192 | 0 | 2366.0 | Domain | Calponin-homology (CH) |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 85_192 | 89 | 2433.0 | Domain | Calponin-homology (CH) |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 85_192 | 89 | 2489.0 | Domain | Calponin-homology (CH) |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 85_192 | 0 | 2418.0 | Domain | Calponin-homology (CH) |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 85_192 | 0 | 1494.0 | Domain | Calponin-homology (CH) |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 85_192 | 0 | 2420.0 | Domain | Calponin-homology (CH) |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 2157_2164 | 0 | 1494.0 | Nucleotide binding | ATP |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 2157_2164 | 0 | 2430.0 | Nucleotide binding | ATP |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 2157_2164 | 0 | 2366.0 | Nucleotide binding | ATP |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 2157_2164 | 0 | 2433.0 | Nucleotide binding | ATP |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 2157_2164 | 0 | 2489.0 | Nucleotide binding | ATP |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 2157_2164 | 0 | 2418.0 | Nucleotide binding | ATP |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 2157_2164 | 0 | 1494.0 | Nucleotide binding | ATP |
| Hgene | NAV2 | chr11:19735379 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 2157_2164 | 0 | 2420.0 | Nucleotide binding | ATP |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000311043 | + | 1 | 27 | 2157_2164 | 0 | 1494.0 | Nucleotide binding | ATP |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000349880 | + | 1 | 38 | 2157_2164 | 89 | 2430.0 | Nucleotide binding | ATP |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000360655 | + | 1 | 38 | 2157_2164 | 0 | 2366.0 | Nucleotide binding | ATP |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396085 | + | 1 | 39 | 2157_2164 | 89 | 2433.0 | Nucleotide binding | ATP |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000396087 | + | 1 | 41 | 2157_2164 | 89 | 2489.0 | Nucleotide binding | ATP |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000527559 | + | 1 | 42 | 2157_2164 | 0 | 2418.0 | Nucleotide binding | ATP |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000533917 | + | 1 | 28 | 2157_2164 | 0 | 1494.0 | Nucleotide binding | ATP |
| Hgene | NAV2 | chr11:19735508 | chr11:30354413 | ENST00000540292 | + | 1 | 42 | 2157_2164 | 0 | 2420.0 | Nucleotide binding | ATP |
Top |
Fusion Gene Sequence for NAV2-ARL14EP |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >57334_57334_1_NAV2-ARL14EP_NAV2_chr11_19735379_ENST00000349880_ARL14EP_chr11_30354413_ENST00000282032_length(transcript)=1918nt_BP=129nt GTGAGCAAGACCACCTATCCTAGCCAGATCCCCCTGAAATCGCAGGTGCTGCAGGGGCTGCAGGAGCCAGCGGGGGAGGGGCTCCCGCTG CGGAAGAGCGGCTCGGTGGAAAACGGGTTCGATACCCAGGGAAGAACTGCTAAAGCTTTGAGGTCATTACAATTTACGAATCCAGGAAGG CAAACTGAATTTGCTCCAGAAACTGGTAAAAGAGAAAAAAGAAGGCTTACAAAAAATGCAACCGCTGGTTCAGACAGACAAGTGATACCA GCAAAGAGTAAGGTCTATGATAGCCAGGGTCTCCTGATTTTTAGTGGGATGGACCTCTGTGACTGCCTGGATGAAGACTGCTTAGGATGT TTCTATGCTTGTCCTGCCTGTGGTTCTACCAAGTGTGGAGCTGAATGCCGCTGTGACCGCAAGTGGCTGTATGAGCAAATTGAAATTGAA GGAGGAGAAATAATTCATAATAAACATGCTGGATAATCTGCGGTACCAAACTATGGAGCCTTTAAAGGTCTTTATTTCTAAAAATCTGTT ACTCTAAGATACATTTTAAGCTTGATTATCATATGACAAAGATTTTAAAACCATCTCAGTGTGCCCTAATTTTTCATCATGGGTGCTTTA AGATTCACTATTTGATATAAATTCAGATAGGCTATTTTTCAGTAGTCAGCGTTAAGCCTGTCTGGATCAATATAAACAAGTAGGGTGTAG GCAGTCCTCTATTTGCATGTTTCCCATGGGCACAAATTTCAGTGACCTAGATTTAGTTTAAATACCAGTTTCCTTACCAGGAAGGAAAGA AAACTGGTAAGGAAACTGTTGTTGTTAAAATCTAGGTTAAAATTTTAGTTAGCACATTGTAACTGAGTAATTACATGAAGTACAAACCTC TCTGCTAGCTCTTCAGTCTACAAATCGCTATGTAAATAACAGATATGCTTCATGATTGTGACCAGTCATGTTATTTCTTTCAAATTCTTC CAGTGGTTTGTCCCTGTGCATCTGTTAATTCAGTTCACGTACAGCAGAGCATGTAGTTATGCTGTCTCTCTGTCATCTACTTGACATTCT ATAGAAGTGAACACTCGAAAGAACTGGTCAACAAAGATGAAAGTGCAGCAAAGCAATGAAAAATGATAACACTGGAAGTGAAATTTTAAT CAAACATAAATGAATTTGTAGAAGAAGTCACTGACCATGGGAATGTTGTTCTTGCTGCTGTGTATTCATAGGAGCTTAGTGAAGGCAAAC TTACCAACACAAATAAGCAAAGTGGTTGCAATAAAGACAGATACGTCCCAGAGGAAGTGATGGTTAAAAAAAAAAAACTTTACATTAAAA GATATTTAATGTGAATATAGAAATATTTCACAACATTGAAAGCACAAGCAATAAAAGGGTGGAAGCTGACCCAAACTTAGAAGGAGTATG GCAGTTTGCCGTGAACATAAAAGATGCTTACTCAGTATGGTAAGTTACTTGCCAGAAAAAAAGGCAAGCCCTATTCAAACTACTTTGTTT TTACAATGAAATAAAACACTTTAATTCTATTTCTAATGTTTTAAATTACAGTGTACTAAATATTTTTAAACATTTTTGCATTTTTTTCTA CATTTATAACTGGTCTTAAGAGTTTTTAATATTTGAGCAGAACCTTAAAGGTCATGGAACAATCATCATTTTCCCTATTGATTATTAAGA TTGCAGGACCATTTTTATGGTCCCACAGTACCATGTGAAATGAGGATCGCTTGTAGCTGGGAACAGTGCTTACTTCAGGGAAAACTGATT TTGATAGAACCGACTAGTTGAGTATTAAACTGTTTTGGAAATAATTTATATATCTAAGTGCTGAAATATATTTCTCTTCTCCCCACTTAT >57334_57334_1_NAV2-ARL14EP_NAV2_chr11_19735379_ENST00000349880_ARL14EP_chr11_30354413_ENST00000282032_length(amino acids)=150AA_BP=31 MKSQVLQGLQEPAGEGLPLRKSGSVENGFDTQGRTAKALRSLQFTNPGRQTEFAPETGKREKRRLTKNATAGSDRQVIPAKSKVYDSQGL -------------------------------------------------------------- >57334_57334_2_NAV2-ARL14EP_NAV2_chr11_19735379_ENST00000396085_ARL14EP_chr11_30354413_ENST00000282032_length(transcript)=1918nt_BP=129nt GTGAGCAAGACCACCTATCCTAGCCAGATCCCCCTGAAATCGCAGGTGCTGCAGGGGCTGCAGGAGCCAGCGGGGGAGGGGCTCCCGCTG CGGAAGAGCGGCTCGGTGGAAAACGGGTTCGATACCCAGGGAAGAACTGCTAAAGCTTTGAGGTCATTACAATTTACGAATCCAGGAAGG CAAACTGAATTTGCTCCAGAAACTGGTAAAAGAGAAAAAAGAAGGCTTACAAAAAATGCAACCGCTGGTTCAGACAGACAAGTGATACCA GCAAAGAGTAAGGTCTATGATAGCCAGGGTCTCCTGATTTTTAGTGGGATGGACCTCTGTGACTGCCTGGATGAAGACTGCTTAGGATGT TTCTATGCTTGTCCTGCCTGTGGTTCTACCAAGTGTGGAGCTGAATGCCGCTGTGACCGCAAGTGGCTGTATGAGCAAATTGAAATTGAA GGAGGAGAAATAATTCATAATAAACATGCTGGATAATCTGCGGTACCAAACTATGGAGCCTTTAAAGGTCTTTATTTCTAAAAATCTGTT ACTCTAAGATACATTTTAAGCTTGATTATCATATGACAAAGATTTTAAAACCATCTCAGTGTGCCCTAATTTTTCATCATGGGTGCTTTA AGATTCACTATTTGATATAAATTCAGATAGGCTATTTTTCAGTAGTCAGCGTTAAGCCTGTCTGGATCAATATAAACAAGTAGGGTGTAG GCAGTCCTCTATTTGCATGTTTCCCATGGGCACAAATTTCAGTGACCTAGATTTAGTTTAAATACCAGTTTCCTTACCAGGAAGGAAAGA AAACTGGTAAGGAAACTGTTGTTGTTAAAATCTAGGTTAAAATTTTAGTTAGCACATTGTAACTGAGTAATTACATGAAGTACAAACCTC TCTGCTAGCTCTTCAGTCTACAAATCGCTATGTAAATAACAGATATGCTTCATGATTGTGACCAGTCATGTTATTTCTTTCAAATTCTTC CAGTGGTTTGTCCCTGTGCATCTGTTAATTCAGTTCACGTACAGCAGAGCATGTAGTTATGCTGTCTCTCTGTCATCTACTTGACATTCT ATAGAAGTGAACACTCGAAAGAACTGGTCAACAAAGATGAAAGTGCAGCAAAGCAATGAAAAATGATAACACTGGAAGTGAAATTTTAAT CAAACATAAATGAATTTGTAGAAGAAGTCACTGACCATGGGAATGTTGTTCTTGCTGCTGTGTATTCATAGGAGCTTAGTGAAGGCAAAC TTACCAACACAAATAAGCAAAGTGGTTGCAATAAAGACAGATACGTCCCAGAGGAAGTGATGGTTAAAAAAAAAAAACTTTACATTAAAA GATATTTAATGTGAATATAGAAATATTTCACAACATTGAAAGCACAAGCAATAAAAGGGTGGAAGCTGACCCAAACTTAGAAGGAGTATG GCAGTTTGCCGTGAACATAAAAGATGCTTACTCAGTATGGTAAGTTACTTGCCAGAAAAAAAGGCAAGCCCTATTCAAACTACTTTGTTT TTACAATGAAATAAAACACTTTAATTCTATTTCTAATGTTTTAAATTACAGTGTACTAAATATTTTTAAACATTTTTGCATTTTTTTCTA CATTTATAACTGGTCTTAAGAGTTTTTAATATTTGAGCAGAACCTTAAAGGTCATGGAACAATCATCATTTTCCCTATTGATTATTAAGA TTGCAGGACCATTTTTATGGTCCCACAGTACCATGTGAAATGAGGATCGCTTGTAGCTGGGAACAGTGCTTACTTCAGGGAAAACTGATT TTGATAGAACCGACTAGTTGAGTATTAAACTGTTTTGGAAATAATTTATATATCTAAGTGCTGAAATATATTTCTCTTCTCCCCACTTAT >57334_57334_2_NAV2-ARL14EP_NAV2_chr11_19735379_ENST00000396085_ARL14EP_chr11_30354413_ENST00000282032_length(amino acids)=150AA_BP=31 MKSQVLQGLQEPAGEGLPLRKSGSVENGFDTQGRTAKALRSLQFTNPGRQTEFAPETGKREKRRLTKNATAGSDRQVIPAKSKVYDSQGL -------------------------------------------------------------- >57334_57334_3_NAV2-ARL14EP_NAV2_chr11_19735379_ENST00000396087_ARL14EP_chr11_30354413_ENST00000282032_length(transcript)=1918nt_BP=129nt GTGAGCAAGACCACCTATCCTAGCCAGATCCCCCTGAAATCGCAGGTGCTGCAGGGGCTGCAGGAGCCAGCGGGGGAGGGGCTCCCGCTG CGGAAGAGCGGCTCGGTGGAAAACGGGTTCGATACCCAGGGAAGAACTGCTAAAGCTTTGAGGTCATTACAATTTACGAATCCAGGAAGG CAAACTGAATTTGCTCCAGAAACTGGTAAAAGAGAAAAAAGAAGGCTTACAAAAAATGCAACCGCTGGTTCAGACAGACAAGTGATACCA GCAAAGAGTAAGGTCTATGATAGCCAGGGTCTCCTGATTTTTAGTGGGATGGACCTCTGTGACTGCCTGGATGAAGACTGCTTAGGATGT TTCTATGCTTGTCCTGCCTGTGGTTCTACCAAGTGTGGAGCTGAATGCCGCTGTGACCGCAAGTGGCTGTATGAGCAAATTGAAATTGAA GGAGGAGAAATAATTCATAATAAACATGCTGGATAATCTGCGGTACCAAACTATGGAGCCTTTAAAGGTCTTTATTTCTAAAAATCTGTT ACTCTAAGATACATTTTAAGCTTGATTATCATATGACAAAGATTTTAAAACCATCTCAGTGTGCCCTAATTTTTCATCATGGGTGCTTTA AGATTCACTATTTGATATAAATTCAGATAGGCTATTTTTCAGTAGTCAGCGTTAAGCCTGTCTGGATCAATATAAACAAGTAGGGTGTAG GCAGTCCTCTATTTGCATGTTTCCCATGGGCACAAATTTCAGTGACCTAGATTTAGTTTAAATACCAGTTTCCTTACCAGGAAGGAAAGA AAACTGGTAAGGAAACTGTTGTTGTTAAAATCTAGGTTAAAATTTTAGTTAGCACATTGTAACTGAGTAATTACATGAAGTACAAACCTC TCTGCTAGCTCTTCAGTCTACAAATCGCTATGTAAATAACAGATATGCTTCATGATTGTGACCAGTCATGTTATTTCTTTCAAATTCTTC CAGTGGTTTGTCCCTGTGCATCTGTTAATTCAGTTCACGTACAGCAGAGCATGTAGTTATGCTGTCTCTCTGTCATCTACTTGACATTCT ATAGAAGTGAACACTCGAAAGAACTGGTCAACAAAGATGAAAGTGCAGCAAAGCAATGAAAAATGATAACACTGGAAGTGAAATTTTAAT CAAACATAAATGAATTTGTAGAAGAAGTCACTGACCATGGGAATGTTGTTCTTGCTGCTGTGTATTCATAGGAGCTTAGTGAAGGCAAAC TTACCAACACAAATAAGCAAAGTGGTTGCAATAAAGACAGATACGTCCCAGAGGAAGTGATGGTTAAAAAAAAAAAACTTTACATTAAAA GATATTTAATGTGAATATAGAAATATTTCACAACATTGAAAGCACAAGCAATAAAAGGGTGGAAGCTGACCCAAACTTAGAAGGAGTATG GCAGTTTGCCGTGAACATAAAAGATGCTTACTCAGTATGGTAAGTTACTTGCCAGAAAAAAAGGCAAGCCCTATTCAAACTACTTTGTTT TTACAATGAAATAAAACACTTTAATTCTATTTCTAATGTTTTAAATTACAGTGTACTAAATATTTTTAAACATTTTTGCATTTTTTTCTA CATTTATAACTGGTCTTAAGAGTTTTTAATATTTGAGCAGAACCTTAAAGGTCATGGAACAATCATCATTTTCCCTATTGATTATTAAGA TTGCAGGACCATTTTTATGGTCCCACAGTACCATGTGAAATGAGGATCGCTTGTAGCTGGGAACAGTGCTTACTTCAGGGAAAACTGATT TTGATAGAACCGACTAGTTGAGTATTAAACTGTTTTGGAAATAATTTATATATCTAAGTGCTGAAATATATTTCTCTTCTCCCCACTTAT >57334_57334_3_NAV2-ARL14EP_NAV2_chr11_19735379_ENST00000396087_ARL14EP_chr11_30354413_ENST00000282032_length(amino acids)=150AA_BP=31 MKSQVLQGLQEPAGEGLPLRKSGSVENGFDTQGRTAKALRSLQFTNPGRQTEFAPETGKREKRRLTKNATAGSDRQVIPAKSKVYDSQGL -------------------------------------------------------------- >57334_57334_4_NAV2-ARL14EP_NAV2_chr11_19735508_ENST00000349880_ARL14EP_chr11_30354413_ENST00000282032_length(transcript)=2386nt_BP=597nt AGGAGGAAATTTGCAAGAGGCGGCAGCCCCTGAGCGCCCAGAGCTCTTGAAAGGCCACCCAGGAGAGGTGTGAGACCCGGCGGCAGCATC CGTCCAGGTGGGACCCGCTGAGCGCCGTGGCCAGTCCCCCATTCCCATCCCGGCACCCCAAAGGCGCGCTCGCCCGATTGCTTCGAGTTC CCCGACCTGGGGATTTTTTTTTTAGCCGCTGGTGGTGGGCGCCTCGTGGGCTAAGGCCCGGCGCCTGCTCTGCTACCCGCGCTGCCTTTA GCGGTCGCCCCCGCCGCCGCTGCCAGGGACGTGCTGGGAAAGCCCAAGCCCCGGGAGAAGATGCCGGCCATCCTGGTCGCCTCCAAAATG AAGTCGGGACTGCCCAAACCCGTGCACAGCGCCGCGCCCATCCTGCACGTGCCCCCGGCCCGGGCGGGCCCCCAGCCCTGCTACCTGAAG TTGGGAAGCAAGGTGGAGGTGAGCAAGACCACCTATCCTAGCCAGATCCCCCTGAAATCGCAGGTGCTGCAGGGGCTGCAGGAGCCAGCG GGGGAGGGGCTCCCGCTGCGGAAGAGCGGCTCGGTGGAAAACGGGTTCGATACCCAGGGAAGAACTGCTAAAGCTTTGAGGTCATTACAA TTTACGAATCCAGGAAGGCAAACTGAATTTGCTCCAGAAACTGGTAAAAGAGAAAAAAGAAGGCTTACAAAAAATGCAACCGCTGGTTCA GACAGACAAGTGATACCAGCAAAGAGTAAGGTCTATGATAGCCAGGGTCTCCTGATTTTTAGTGGGATGGACCTCTGTGACTGCCTGGAT GAAGACTGCTTAGGATGTTTCTATGCTTGTCCTGCCTGTGGTTCTACCAAGTGTGGAGCTGAATGCCGCTGTGACCGCAAGTGGCTGTAT GAGCAAATTGAAATTGAAGGAGGAGAAATAATTCATAATAAACATGCTGGATAATCTGCGGTACCAAACTATGGAGCCTTTAAAGGTCTT TATTTCTAAAAATCTGTTACTCTAAGATACATTTTAAGCTTGATTATCATATGACAAAGATTTTAAAACCATCTCAGTGTGCCCTAATTT TTCATCATGGGTGCTTTAAGATTCACTATTTGATATAAATTCAGATAGGCTATTTTTCAGTAGTCAGCGTTAAGCCTGTCTGGATCAATA TAAACAAGTAGGGTGTAGGCAGTCCTCTATTTGCATGTTTCCCATGGGCACAAATTTCAGTGACCTAGATTTAGTTTAAATACCAGTTTC CTTACCAGGAAGGAAAGAAAACTGGTAAGGAAACTGTTGTTGTTAAAATCTAGGTTAAAATTTTAGTTAGCACATTGTAACTGAGTAATT ACATGAAGTACAAACCTCTCTGCTAGCTCTTCAGTCTACAAATCGCTATGTAAATAACAGATATGCTTCATGATTGTGACCAGTCATGTT ATTTCTTTCAAATTCTTCCAGTGGTTTGTCCCTGTGCATCTGTTAATTCAGTTCACGTACAGCAGAGCATGTAGTTATGCTGTCTCTCTG TCATCTACTTGACATTCTATAGAAGTGAACACTCGAAAGAACTGGTCAACAAAGATGAAAGTGCAGCAAAGCAATGAAAAATGATAACAC TGGAAGTGAAATTTTAATCAAACATAAATGAATTTGTAGAAGAAGTCACTGACCATGGGAATGTTGTTCTTGCTGCTGTGTATTCATAGG AGCTTAGTGAAGGCAAACTTACCAACACAAATAAGCAAAGTGGTTGCAATAAAGACAGATACGTCCCAGAGGAAGTGATGGTTAAAAAAA AAAAACTTTACATTAAAAGATATTTAATGTGAATATAGAAATATTTCACAACATTGAAAGCACAAGCAATAAAAGGGTGGAAGCTGACCC AAACTTAGAAGGAGTATGGCAGTTTGCCGTGAACATAAAAGATGCTTACTCAGTATGGTAAGTTACTTGCCAGAAAAAAAGGCAAGCCCT ATTCAAACTACTTTGTTTTTACAATGAAATAAAACACTTTAATTCTATTTCTAATGTTTTAAATTACAGTGTACTAAATATTTTTAAACA TTTTTGCATTTTTTTCTACATTTATAACTGGTCTTAAGAGTTTTTAATATTTGAGCAGAACCTTAAAGGTCATGGAACAATCATCATTTT CCCTATTGATTATTAAGATTGCAGGACCATTTTTATGGTCCCACAGTACCATGTGAAATGAGGATCGCTTGTAGCTGGGAACAGTGCTTA CTTCAGGGAAAACTGATTTTGATAGAACCGACTAGTTGAGTATTAAACTGTTTTGGAAATAATTTATATATCTAAGTGCTGAAATATATT >57334_57334_4_NAV2-ARL14EP_NAV2_chr11_19735508_ENST00000349880_ARL14EP_chr11_30354413_ENST00000282032_length(amino acids)=234AA_BP=115 MLPALPLAVAPAAAARDVLGKPKPREKMPAILVASKMKSGLPKPVHSAAPILHVPPARAGPQPCYLKLGSKVEVSKTTYPSQIPLKSQVL QGLQEPAGEGLPLRKSGSVENGFDTQGRTAKALRSLQFTNPGRQTEFAPETGKREKRRLTKNATAGSDRQVIPAKSKVYDSQGLLIFSGM -------------------------------------------------------------- >57334_57334_5_NAV2-ARL14EP_NAV2_chr11_19735508_ENST00000396085_ARL14EP_chr11_30354413_ENST00000282032_length(transcript)=2417nt_BP=628nt GTTAATATGGGCGTCCAAGCCTCTGTTTCCCAGGAGGAAATTTGCAAGAGGCGGCAGCCCCTGAGCGCCCAGAGCTCTTGAAAGGCCACC CAGGAGAGGTGTGAGACCCGGCGGCAGCATCCGTCCAGGTGGGACCCGCTGAGCGCCGTGGCCAGTCCCCCATTCCCATCCCGGCACCCC AAAGGCGCGCTCGCCCGATTGCTTCGAGTTCCCCGACCTGGGGATTTTTTTTTTAGCCGCTGGTGGTGGGCGCCTCGTGGGCTAAGGCCC GGCGCCTGCTCTGCTACCCGCGCTGCCTTTAGCGGTCGCCCCCGCCGCCGCTGCCAGGGACGTGCTGGGAAAGCCCAAGCCCCGGGAGAA GATGCCGGCCATCCTGGTCGCCTCCAAAATGAAGTCGGGACTGCCCAAACCCGTGCACAGCGCCGCGCCCATCCTGCACGTGCCCCCGGC CCGGGCGGGCCCCCAGCCCTGCTACCTGAAGTTGGGAAGCAAGGTGGAGGTGAGCAAGACCACCTATCCTAGCCAGATCCCCCTGAAATC GCAGGTGCTGCAGGGGCTGCAGGAGCCAGCGGGGGAGGGGCTCCCGCTGCGGAAGAGCGGCTCGGTGGAAAACGGGTTCGATACCCAGGG AAGAACTGCTAAAGCTTTGAGGTCATTACAATTTACGAATCCAGGAAGGCAAACTGAATTTGCTCCAGAAACTGGTAAAAGAGAAAAAAG AAGGCTTACAAAAAATGCAACCGCTGGTTCAGACAGACAAGTGATACCAGCAAAGAGTAAGGTCTATGATAGCCAGGGTCTCCTGATTTT TAGTGGGATGGACCTCTGTGACTGCCTGGATGAAGACTGCTTAGGATGTTTCTATGCTTGTCCTGCCTGTGGTTCTACCAAGTGTGGAGC TGAATGCCGCTGTGACCGCAAGTGGCTGTATGAGCAAATTGAAATTGAAGGAGGAGAAATAATTCATAATAAACATGCTGGATAATCTGC GGTACCAAACTATGGAGCCTTTAAAGGTCTTTATTTCTAAAAATCTGTTACTCTAAGATACATTTTAAGCTTGATTATCATATGACAAAG ATTTTAAAACCATCTCAGTGTGCCCTAATTTTTCATCATGGGTGCTTTAAGATTCACTATTTGATATAAATTCAGATAGGCTATTTTTCA GTAGTCAGCGTTAAGCCTGTCTGGATCAATATAAACAAGTAGGGTGTAGGCAGTCCTCTATTTGCATGTTTCCCATGGGCACAAATTTCA GTGACCTAGATTTAGTTTAAATACCAGTTTCCTTACCAGGAAGGAAAGAAAACTGGTAAGGAAACTGTTGTTGTTAAAATCTAGGTTAAA ATTTTAGTTAGCACATTGTAACTGAGTAATTACATGAAGTACAAACCTCTCTGCTAGCTCTTCAGTCTACAAATCGCTATGTAAATAACA GATATGCTTCATGATTGTGACCAGTCATGTTATTTCTTTCAAATTCTTCCAGTGGTTTGTCCCTGTGCATCTGTTAATTCAGTTCACGTA CAGCAGAGCATGTAGTTATGCTGTCTCTCTGTCATCTACTTGACATTCTATAGAAGTGAACACTCGAAAGAACTGGTCAACAAAGATGAA AGTGCAGCAAAGCAATGAAAAATGATAACACTGGAAGTGAAATTTTAATCAAACATAAATGAATTTGTAGAAGAAGTCACTGACCATGGG AATGTTGTTCTTGCTGCTGTGTATTCATAGGAGCTTAGTGAAGGCAAACTTACCAACACAAATAAGCAAAGTGGTTGCAATAAAGACAGA TACGTCCCAGAGGAAGTGATGGTTAAAAAAAAAAAACTTTACATTAAAAGATATTTAATGTGAATATAGAAATATTTCACAACATTGAAA GCACAAGCAATAAAAGGGTGGAAGCTGACCCAAACTTAGAAGGAGTATGGCAGTTTGCCGTGAACATAAAAGATGCTTACTCAGTATGGT AAGTTACTTGCCAGAAAAAAAGGCAAGCCCTATTCAAACTACTTTGTTTTTACAATGAAATAAAACACTTTAATTCTATTTCTAATGTTT TAAATTACAGTGTACTAAATATTTTTAAACATTTTTGCATTTTTTTCTACATTTATAACTGGTCTTAAGAGTTTTTAATATTTGAGCAGA ACCTTAAAGGTCATGGAACAATCATCATTTTCCCTATTGATTATTAAGATTGCAGGACCATTTTTATGGTCCCACAGTACCATGTGAAAT GAGGATCGCTTGTAGCTGGGAACAGTGCTTACTTCAGGGAAAACTGATTTTGATAGAACCGACTAGTTGAGTATTAAACTGTTTTGGAAA >57334_57334_5_NAV2-ARL14EP_NAV2_chr11_19735508_ENST00000396085_ARL14EP_chr11_30354413_ENST00000282032_length(amino acids)=234AA_BP=115 MLPALPLAVAPAAAARDVLGKPKPREKMPAILVASKMKSGLPKPVHSAAPILHVPPARAGPQPCYLKLGSKVEVSKTTYPSQIPLKSQVL QGLQEPAGEGLPLRKSGSVENGFDTQGRTAKALRSLQFTNPGRQTEFAPETGKREKRRLTKNATAGSDRQVIPAKSKVYDSQGLLIFSGM -------------------------------------------------------------- >57334_57334_6_NAV2-ARL14EP_NAV2_chr11_19735508_ENST00000396087_ARL14EP_chr11_30354413_ENST00000282032_length(transcript)=2155nt_BP=366nt TAAGGCCCGGCGCCTGCTCTGCTACCCGCGCTGCCTTTAGCGGTCGCCCCCGCCGCCGCTGCCAGGGACGTGCTGGGAAAGCCCAAGCCC CGGGAGAAGATGCCGGCCATCCTGGTCGCCTCCAAAATGAAGTCGGGACTGCCCAAACCCGTGCACAGCGCCGCGCCCATCCTGCACGTG CCCCCGGCCCGGGCGGGCCCCCAGCCCTGCTACCTGAAGTTGGGAAGCAAGGTGGAGGTGAGCAAGACCACCTATCCTAGCCAGATCCCC CTGAAATCGCAGGTGCTGCAGGGGCTGCAGGAGCCAGCGGGGGAGGGGCTCCCGCTGCGGAAGAGCGGCTCGGTGGAAAACGGGTTCGAT ACCCAGGGAAGAACTGCTAAAGCTTTGAGGTCATTACAATTTACGAATCCAGGAAGGCAAACTGAATTTGCTCCAGAAACTGGTAAAAGA GAAAAAAGAAGGCTTACAAAAAATGCAACCGCTGGTTCAGACAGACAAGTGATACCAGCAAAGAGTAAGGTCTATGATAGCCAGGGTCTC CTGATTTTTAGTGGGATGGACCTCTGTGACTGCCTGGATGAAGACTGCTTAGGATGTTTCTATGCTTGTCCTGCCTGTGGTTCTACCAAG TGTGGAGCTGAATGCCGCTGTGACCGCAAGTGGCTGTATGAGCAAATTGAAATTGAAGGAGGAGAAATAATTCATAATAAACATGCTGGA TAATCTGCGGTACCAAACTATGGAGCCTTTAAAGGTCTTTATTTCTAAAAATCTGTTACTCTAAGATACATTTTAAGCTTGATTATCATA TGACAAAGATTTTAAAACCATCTCAGTGTGCCCTAATTTTTCATCATGGGTGCTTTAAGATTCACTATTTGATATAAATTCAGATAGGCT ATTTTTCAGTAGTCAGCGTTAAGCCTGTCTGGATCAATATAAACAAGTAGGGTGTAGGCAGTCCTCTATTTGCATGTTTCCCATGGGCAC AAATTTCAGTGACCTAGATTTAGTTTAAATACCAGTTTCCTTACCAGGAAGGAAAGAAAACTGGTAAGGAAACTGTTGTTGTTAAAATCT AGGTTAAAATTTTAGTTAGCACATTGTAACTGAGTAATTACATGAAGTACAAACCTCTCTGCTAGCTCTTCAGTCTACAAATCGCTATGT AAATAACAGATATGCTTCATGATTGTGACCAGTCATGTTATTTCTTTCAAATTCTTCCAGTGGTTTGTCCCTGTGCATCTGTTAATTCAG TTCACGTACAGCAGAGCATGTAGTTATGCTGTCTCTCTGTCATCTACTTGACATTCTATAGAAGTGAACACTCGAAAGAACTGGTCAACA AAGATGAAAGTGCAGCAAAGCAATGAAAAATGATAACACTGGAAGTGAAATTTTAATCAAACATAAATGAATTTGTAGAAGAAGTCACTG ACCATGGGAATGTTGTTCTTGCTGCTGTGTATTCATAGGAGCTTAGTGAAGGCAAACTTACCAACACAAATAAGCAAAGTGGTTGCAATA AAGACAGATACGTCCCAGAGGAAGTGATGGTTAAAAAAAAAAAACTTTACATTAAAAGATATTTAATGTGAATATAGAAATATTTCACAA CATTGAAAGCACAAGCAATAAAAGGGTGGAAGCTGACCCAAACTTAGAAGGAGTATGGCAGTTTGCCGTGAACATAAAAGATGCTTACTC AGTATGGTAAGTTACTTGCCAGAAAAAAAGGCAAGCCCTATTCAAACTACTTTGTTTTTACAATGAAATAAAACACTTTAATTCTATTTC TAATGTTTTAAATTACAGTGTACTAAATATTTTTAAACATTTTTGCATTTTTTTCTACATTTATAACTGGTCTTAAGAGTTTTTAATATT TGAGCAGAACCTTAAAGGTCATGGAACAATCATCATTTTCCCTATTGATTATTAAGATTGCAGGACCATTTTTATGGTCCCACAGTACCA TGTGAAATGAGGATCGCTTGTAGCTGGGAACAGTGCTTACTTCAGGGAAAACTGATTTTGATAGAACCGACTAGTTGAGTATTAAACTGT >57334_57334_6_NAV2-ARL14EP_NAV2_chr11_19735508_ENST00000396087_ARL14EP_chr11_30354413_ENST00000282032_length(amino acids)=234AA_BP=115 MLPALPLAVAPAAAARDVLGKPKPREKMPAILVASKMKSGLPKPVHSAAPILHVPPARAGPQPCYLKLGSKVEVSKTTYPSQIPLKSQVL QGLQEPAGEGLPLRKSGSVENGFDTQGRTAKALRSLQFTNPGRQTEFAPETGKREKRRLTKNATAGSDRQVIPAKSKVYDSQGLLIFSGM -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for NAV2-ARL14EP |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for NAV2-ARL14EP |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for NAV2-ARL14EP |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
