
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:NELL1-SMCO4 (FusionGDB2 ID:58570) |
Fusion Gene Summary for NELL1-SMCO4 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: NELL1-SMCO4 | Fusion gene ID: 58570 | Hgene | Tgene | Gene symbol | NELL1 | SMCO4 | Gene ID | 4745 | 56935 |
| Gene name | neural EGFL like 1 | single-pass membrane protein with coiled-coil domains 4 | |
| Synonyms | IDH3GL|NRP1 | C11orf75|FN5 | |
| Cytomap | 11p15.1 | 11q21 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein kinase C-binding protein NELL1nel-related protein 1neural epidermal growth factor-like 1 | single-pass membrane and coiled-coil domain-containing protein 4UPF0443 protein C11orf75 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q92832 | Q9NRQ5 | |
| Ensembl transtripts involved in fusion gene | ENST00000298925, ENST00000325319, ENST00000357134, ENST00000532434, ENST00000529218, | ENST00000298966, ENST00000527149, ENST00000525141, | |
| Fusion gene scores | * DoF score | 12 X 12 X 3=432 | 5 X 3 X 4=60 |
| # samples | 12 | 5 | |
| ** MAII score | log2(12/432*10)=-1.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/60*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: NELL1 [Title/Abstract] AND SMCO4 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | NELL1(20907086)-SMCO4(93232514), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | NELL1 | GO:0010468 | regulation of gene expression | 21723284 |
| Hgene | NELL1 | GO:0030501 | positive regulation of bone mineralization | 21723284 |
| Hgene | NELL1 | GO:0033689 | negative regulation of osteoblast proliferation | 21723284 |
| Hgene | NELL1 | GO:0045669 | positive regulation of osteoblast differentiation | 21723284 |
| Hgene | NELL1 | GO:1903363 | negative regulation of cellular protein catabolic process | 21723284 |
 Fusion gene breakpoints across NELL1 (5'-gene) Fusion gene breakpoints across NELL1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
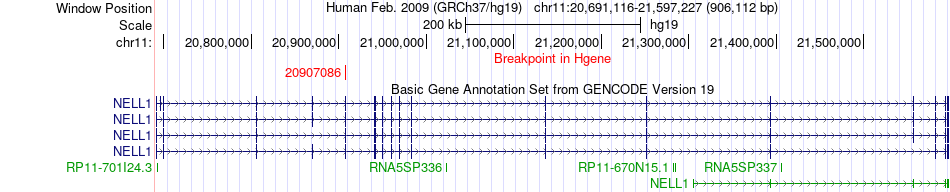 |
 Fusion gene breakpoints across SMCO4 (3'-gene) Fusion gene breakpoints across SMCO4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
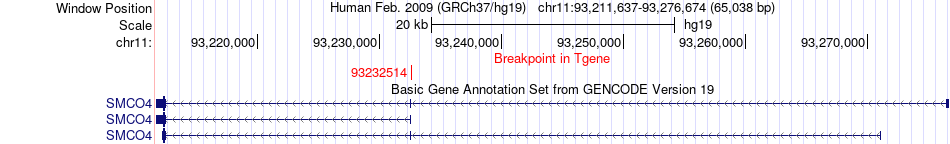 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SKCM | TCGA-ER-A19H-06A | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - |
Top |
Fusion Gene ORF analysis for NELL1-SMCO4 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000298925 | ENST00000298966 | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - |
| 5CDS-5UTR | ENST00000298925 | ENST00000527149 | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - |
| 5CDS-5UTR | ENST00000325319 | ENST00000298966 | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - |
| 5CDS-5UTR | ENST00000325319 | ENST00000527149 | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - |
| 5CDS-5UTR | ENST00000357134 | ENST00000298966 | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - |
| 5CDS-5UTR | ENST00000357134 | ENST00000527149 | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - |
| 5CDS-5UTR | ENST00000532434 | ENST00000298966 | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - |
| 5CDS-5UTR | ENST00000532434 | ENST00000527149 | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - |
| In-frame | ENST00000298925 | ENST00000525141 | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - |
| In-frame | ENST00000325319 | ENST00000525141 | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - |
| In-frame | ENST00000357134 | ENST00000525141 | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - |
| In-frame | ENST00000532434 | ENST00000525141 | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - |
| intron-3CDS | ENST00000529218 | ENST00000525141 | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - |
| intron-5UTR | ENST00000529218 | ENST00000298966 | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - |
| intron-5UTR | ENST00000529218 | ENST00000527149 | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000298925 | NELL1 | chr11 | 20907086 | + | ENST00000525141 | SMCO4 | chr11 | 93232514 | - | 1729 | 840 | 153 | 848 | 231 |
| ENST00000357134 | NELL1 | chr11 | 20907086 | + | ENST00000525141 | SMCO4 | chr11 | 93232514 | - | 1644 | 755 | 152 | 763 | 203 |
| ENST00000325319 | NELL1 | chr11 | 20907086 | + | ENST00000525141 | SMCO4 | chr11 | 93232514 | - | 1462 | 573 | 533 | 1063 | 176 |
| ENST00000532434 | NELL1 | chr11 | 20907086 | + | ENST00000525141 | SMCO4 | chr11 | 93232514 | - | 1555 | 666 | 63 | 674 | 203 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000298925 | ENST00000525141 | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - | 0.008369508 | 0.9916305 |
| ENST00000357134 | ENST00000525141 | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - | 0.004434435 | 0.9955656 |
| ENST00000325319 | ENST00000525141 | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - | 0.5853628 | 0.41463727 |
| ENST00000532434 | ENST00000525141 | NELL1 | chr11 | 20907086 | + | SMCO4 | chr11 | 93232514 | - | 0.004338557 | 0.99566144 |
Top |
Fusion Genomic Features for NELL1-SMCO4 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
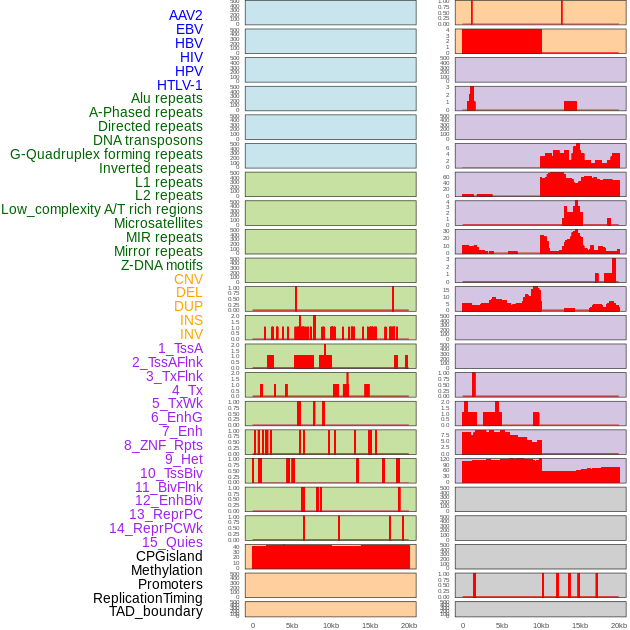 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for NELL1-SMCO4 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:20907086/chr11:93232514) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NELL1 | SMCO4 |
| FUNCTION: Plays a role in the control of cell growth and differentiation. Promotes osteoblast cell differentiation and terminal mineralization. {ECO:0000269|PubMed:21723284}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | SMCO4 | chr11:20907086 | chr11:93232514 | ENST00000298966 | 0 | 3 | 9_31 | 0 | 60.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | SMCO4 | chr11:20907086 | chr11:93232514 | ENST00000525141 | -1 | 2 | 9_31 | 0 | 60.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | SMCO4 | chr11:20907086 | chr11:93232514 | ENST00000527149 | 0 | 3 | 9_31 | 0 | 60.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | SMCO4 | chr11:20907086 | chr11:93232514 | ENST00000298966 | 0 | 3 | 32_52 | 0 | 60.0 | Transmembrane | Helical | |
| Tgene | SMCO4 | chr11:20907086 | chr11:93232514 | ENST00000525141 | -1 | 2 | 32_52 | 0 | 60.0 | Transmembrane | Helical | |
| Tgene | SMCO4 | chr11:20907086 | chr11:93232514 | ENST00000527149 | 0 | 3 | 32_52 | 0 | 60.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NELL1 | chr11:20907086 | chr11:93232514 | ENST00000357134 | + | 5 | 20 | 271_332 | 201 | 811.0 | Domain | VWFC 1 |
| Hgene | NELL1 | chr11:20907086 | chr11:93232514 | ENST00000357134 | + | 5 | 20 | 434_475 | 201 | 811.0 | Domain | EGF-like 1%3B calcium-binding |
| Hgene | NELL1 | chr11:20907086 | chr11:93232514 | ENST00000357134 | + | 5 | 20 | 476_516 | 201 | 811.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | NELL1 | chr11:20907086 | chr11:93232514 | ENST00000357134 | + | 5 | 20 | 517_547 | 201 | 811.0 | Domain | EGF-like 3 |
| Hgene | NELL1 | chr11:20907086 | chr11:93232514 | ENST00000357134 | + | 5 | 20 | 549_587 | 201 | 811.0 | Domain | EGF-like 4%3B calcium-binding |
| Hgene | NELL1 | chr11:20907086 | chr11:93232514 | ENST00000357134 | + | 5 | 20 | 596_631 | 201 | 811.0 | Domain | EGF-like 5%3B calcium-binding |
| Hgene | NELL1 | chr11:20907086 | chr11:93232514 | ENST00000357134 | + | 5 | 20 | 64_227 | 201 | 811.0 | Domain | Note=Laminin G-like |
| Hgene | NELL1 | chr11:20907086 | chr11:93232514 | ENST00000357134 | + | 5 | 20 | 692_750 | 201 | 811.0 | Domain | VWFC 2 |
| Hgene | NELL1 | chr11:20907086 | chr11:93232514 | ENST00000532434 | + | 5 | 19 | 271_332 | 201 | 764.0 | Domain | VWFC 1 |
| Hgene | NELL1 | chr11:20907086 | chr11:93232514 | ENST00000532434 | + | 5 | 19 | 434_475 | 201 | 764.0 | Domain | EGF-like 1%3B calcium-binding |
| Hgene | NELL1 | chr11:20907086 | chr11:93232514 | ENST00000532434 | + | 5 | 19 | 476_516 | 201 | 764.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | NELL1 | chr11:20907086 | chr11:93232514 | ENST00000532434 | + | 5 | 19 | 517_547 | 201 | 764.0 | Domain | EGF-like 3 |
| Hgene | NELL1 | chr11:20907086 | chr11:93232514 | ENST00000532434 | + | 5 | 19 | 549_587 | 201 | 764.0 | Domain | EGF-like 4%3B calcium-binding |
| Hgene | NELL1 | chr11:20907086 | chr11:93232514 | ENST00000532434 | + | 5 | 19 | 596_631 | 201 | 764.0 | Domain | EGF-like 5%3B calcium-binding |
| Hgene | NELL1 | chr11:20907086 | chr11:93232514 | ENST00000532434 | + | 5 | 19 | 64_227 | 201 | 764.0 | Domain | Note=Laminin G-like |
| Hgene | NELL1 | chr11:20907086 | chr11:93232514 | ENST00000532434 | + | 5 | 19 | 692_750 | 201 | 764.0 | Domain | VWFC 2 |
Top |
Fusion Gene Sequence for NELL1-SMCO4 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >58570_58570_1_NELL1-SMCO4_NELL1_chr11_20907086_ENST00000298925_SMCO4_chr11_93232514_ENST00000525141_length(transcript)=1729nt_BP=840nt ATATGCGAGCGCAGCACCCGGCGCTGCCGAGCCACCTCCCCCGCCGCCCGCTAGCAAGTTTGGCGGCTCCAAGCCAGGCGCGCCTCAGGA TCCAGGCTCATTTGCTTCCACCTAGCTTCGGTGCCCCCTGCTAGGCGGGGACCCTCGAGAGCGATGCCGATGGATTTGATTTTAGTTGTG TGGTTCTGTGTGTGCACTGCCAGGACAGCAGAAGATATGAAGCCACCCGTGTTATCGCTGATATGGAGAACTAGAGTTCTGAAGTCTCTG CTTCAGCAATCCCTTCAGGGAGTGGTGGGCTTTGGGATGGACCCTGACCTTCAGATGGATATCGTCACCGAGCTTGACCTTGTGAACACC ACCCTTGGAGTTGCTCAGGTGTCTGGAATGCACAATGCCAGCAAAGCATTTTTATTTCAAGACATAGAAAGAGAGATCCATGCAGCTCCT CATGTGAGTGAGAAATTAATTCAGCTGTTCCGGAACAAGAGTGAATTCACCATTTTGGCCACTGTACAGCAGAAGCCATCCACTTCAGGA GTGATACTGTCCATTCGAGAACTGGAGCACAGCTATTTTGAACTGGAGAGCAGTGGCCTGAGGGATGAGATTCGGTATCACTACATACAC AATGGGAAGCCAAGGACAGAGGCACTTCCTTACCGCATGGCAGATGGACAATGGCACAAGGTTGCACTGTCAGTTAGCGCCTCTCATCTC CTGCTCCATGTCGACTGTAACAGGATTTATGAGCGTGTGATAGACCCTCCAGATACCAACCTTCCCCCAGGAATCAATTTATGGCTTGGC CAGCGCAACCAAAAGCATGGCTTATTCAAACAGGAATAACTCAAGTCACCTGTACTGGAAATCAGTTTGCTGAAATTAATCAACGATTCT TGAAGTTGAAGAAAAGTTGTTCTCTCTACAGGAGGTTCCAGCCTTGGCAAGAGGAGTGTGGCCCTTCCTGGAATCCCTCTGGACACACCC TCCTAGCATCCTCTAGGAAAGATGCGGCAGCTCAAAGGGAAGCCCAAGAAGGAGACCTCCAAGGACAAGAAGGAGCGGAAGCAAGCCATG CAGGAGGCCCGGCAGCAGATCACTACAGTGGTGCTGCCCACGCTGGCCGTGGTCGTGCTCTTGATCGTGGTGTTTGTGTACGTGGCCACG CGCCCCACCATCACCGAGTGAGCCCCGCAGCCGGCTGCGGACCCCATCGGCAGGGAGAGGAGGCGCGGGAGGGGGACGCAAACAAAAAAT GGCTTTCATATTCAGAGATGTTCATGTTGCTGAGCTGTAAGCAGGAGCACCCTGTCTTCTCTGGTCTTTGACTTGATTAAAGTATCTCCG CTTTCTTGGGAGGGAATAGGGGATGTTTTATCAGTGAATGTGCCATACACCTTATGGTCCACTTCATGTGCCTTTCAGACTTCAAAGCGC GCGCGCATGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGCTTCTTTTTCTCTCCTAAAAATCGATAAGTAGCTCCACCTGAA GAGGGATGGAACCTCTGGGTCAGGAAACAGCTGGAATCCACACTCACCTCATTCCCATTGTTTGGATCATGCTTCTTTCCAACACGTGTT CACAATCTCCAAAGGGACTGTATTTCTTCTCTGTGCTTAATGTGATTTGAAATATGTTGAATCAAAGTGAAATATTTATTTTTTGAATAA >58570_58570_1_NELL1-SMCO4_NELL1_chr11_20907086_ENST00000298925_SMCO4_chr11_93232514_ENST00000525141_length(amino acids)=231AA_BP= MPMDLILVVWFCVCTARTAEDMKPPVLSLIWRTRVLKSLLQQSLQGVVGFGMDPDLQMDIVTELDLVNTTLGVAQVSGMHNASKAFLFQD IEREIHAAPHVSEKLIQLFRNKSEFTILATVQQKPSTSGVILSIRELEHSYFELESSGLRDEIRYHYIHNGKPRTEALPYRMADGQWHKV -------------------------------------------------------------- >58570_58570_2_NELL1-SMCO4_NELL1_chr11_20907086_ENST00000325319_SMCO4_chr11_93232514_ENST00000525141_length(transcript)=1462nt_BP=573nt AGCACCCGGCGCTGCCGAGCCACCTCCCCCGCCGCCCGCTAGCAAGTTTGGCGGCTCCAAGCCAGGCGCGCCTCAGGATCCAGGCTCATT TGCTTCCACCTAGCTTCGGTGCCCCCTGCTAGGCGGGGACCCTCGAGAGCGATGCCGATGGATTTGATTTTAGTTGTGTGGTTCTGTGTG TGCACTGCCAGGACAGTGGTGGGCTTTGGGATGGACCCTGACCTTCAGATGGATATCGTCACCGAGCTTGACCTTGTGAACACCACCCTT GGAGTTGCTCAGGTGTCTGGAATGCACAATGCCAGCAAAGCATTTTTATTTCAAGACATAGAAAGAGAGATCCATGCAGCTCCTCATGTG AGTGAGAAATTAATTCAGCTGTTCCGGAACAAGAGTGAATTCACCATTTTGGCCACTGTACAGCAGAAGCCATCCACTTCAGGAGTGATA CTGTCCATTCGAGAACTGGAGCACAGGATTTATGAGCGTGTGATAGACCCTCCAGATACCAACCTTCCCCCAGGAATCAATTTATGGCTT GGCCAGCGCAACCAAAAGCATGGCTTATTCAAACAGGAATAACTCAAGTCACCTGTACTGGAAATCAGTTTGCTGAAATTAATCAACGAT TCTTGAAGTTGAAGAAAAGTTGTTCTCTCTACAGGAGGTTCCAGCCTTGGCAAGAGGAGTGTGGCCCTTCCTGGAATCCCTCTGGACACA CCCTCCTAGCATCCTCTAGGAAAGATGCGGCAGCTCAAAGGGAAGCCCAAGAAGGAGACCTCCAAGGACAAGAAGGAGCGGAAGCAAGCC ATGCAGGAGGCCCGGCAGCAGATCACTACAGTGGTGCTGCCCACGCTGGCCGTGGTCGTGCTCTTGATCGTGGTGTTTGTGTACGTGGCC ACGCGCCCCACCATCACCGAGTGAGCCCCGCAGCCGGCTGCGGACCCCATCGGCAGGGAGAGGAGGCGCGGGAGGGGGACGCAAACAAAA AATGGCTTTCATATTCAGAGATGTTCATGTTGCTGAGCTGTAAGCAGGAGCACCCTGTCTTCTCTGGTCTTTGACTTGATTAAAGTATCT CCGCTTTCTTGGGAGGGAATAGGGGATGTTTTATCAGTGAATGTGCCATACACCTTATGGTCCACTTCATGTGCCTTTCAGACTTCAAAG CGCGCGCGCATGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGCTTCTTTTTCTCTCCTAAAAATCGATAAGTAGCTCCACCT GAAGAGGGATGGAACCTCTGGGTCAGGAAACAGCTGGAATCCACACTCACCTCATTCCCATTGTTTGGATCATGCTTCTTTCCAACACGT GTTCACAATCTCCAAAGGGACTGTATTTCTTCTCTGTGCTTAATGTGATTTGAAATATGTTGAATCAAAGTGAAATATTTATTTTTTGAA >58570_58570_2_NELL1-SMCO4_NELL1_chr11_20907086_ENST00000325319_SMCO4_chr11_93232514_ENST00000525141_length(amino acids)=176AA_BP=13 MAWPAQPKAWLIQTGITQVTCTGNQFAEINQRFLKLKKSCSLYRRFQPWQEECGPSWNPSGHTLLASSRKDAAAQREAQEGDLQGQEGAE -------------------------------------------------------------- >58570_58570_3_NELL1-SMCO4_NELL1_chr11_20907086_ENST00000357134_SMCO4_chr11_93232514_ENST00000525141_length(transcript)=1644nt_BP=755nt TATGCGAGCGCAGCACCCGGCGCTGCCGAGCCACCTCCCCCGCCGCCCGCTAGCAAGTTTGGCGGCTCCAAGCCAGGCGCGCCTCAGGAT CCAGGCTCATTTGCTTCCACCTAGCTTCGGTGCCCCCTGCTAGGCGGGGACCCTCGAGAGCGATGCCGATGGATTTGATTTTAGTTGTGT GGTTCTGTGTGTGCACTGCCAGGACAGTGGTGGGCTTTGGGATGGACCCTGACCTTCAGATGGATATCGTCACCGAGCTTGACCTTGTGA ACACCACCCTTGGAGTTGCTCAGGTGTCTGGAATGCACAATGCCAGCAAAGCATTTTTATTTCAAGACATAGAAAGAGAGATCCATGCAG CTCCTCATGTGAGTGAGAAATTAATTCAGCTGTTCCGGAACAAGAGTGAATTCACCATTTTGGCCACTGTACAGCAGAAGCCATCCACTT CAGGAGTGATACTGTCCATTCGAGAACTGGAGCACAGCTATTTTGAACTGGAGAGCAGTGGCCTGAGGGATGAGATTCGGTATCACTACA TACACAATGGGAAGCCAAGGACAGAGGCACTTCCTTACCGCATGGCAGATGGACAATGGCACAAGGTTGCACTGTCAGTTAGCGCCTCTC ATCTCCTGCTCCATGTCGACTGTAACAGGATTTATGAGCGTGTGATAGACCCTCCAGATACCAACCTTCCCCCAGGAATCAATTTATGGC TTGGCCAGCGCAACCAAAAGCATGGCTTATTCAAACAGGAATAACTCAAGTCACCTGTACTGGAAATCAGTTTGCTGAAATTAATCAACG ATTCTTGAAGTTGAAGAAAAGTTGTTCTCTCTACAGGAGGTTCCAGCCTTGGCAAGAGGAGTGTGGCCCTTCCTGGAATCCCTCTGGACA CACCCTCCTAGCATCCTCTAGGAAAGATGCGGCAGCTCAAAGGGAAGCCCAAGAAGGAGACCTCCAAGGACAAGAAGGAGCGGAAGCAAG CCATGCAGGAGGCCCGGCAGCAGATCACTACAGTGGTGCTGCCCACGCTGGCCGTGGTCGTGCTCTTGATCGTGGTGTTTGTGTACGTGG CCACGCGCCCCACCATCACCGAGTGAGCCCCGCAGCCGGCTGCGGACCCCATCGGCAGGGAGAGGAGGCGCGGGAGGGGGACGCAAACAA AAAATGGCTTTCATATTCAGAGATGTTCATGTTGCTGAGCTGTAAGCAGGAGCACCCTGTCTTCTCTGGTCTTTGACTTGATTAAAGTAT CTCCGCTTTCTTGGGAGGGAATAGGGGATGTTTTATCAGTGAATGTGCCATACACCTTATGGTCCACTTCATGTGCCTTTCAGACTTCAA AGCGCGCGCGCATGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGCTTCTTTTTCTCTCCTAAAAATCGATAAGTAGCTCCAC CTGAAGAGGGATGGAACCTCTGGGTCAGGAAACAGCTGGAATCCACACTCACCTCATTCCCATTGTTTGGATCATGCTTCTTTCCAACAC GTGTTCACAATCTCCAAAGGGACTGTATTTCTTCTCTGTGCTTAATGTGATTTGAAATATGTTGAATCAAAGTGAAATATTTATTTTTTG >58570_58570_3_NELL1-SMCO4_NELL1_chr11_20907086_ENST00000357134_SMCO4_chr11_93232514_ENST00000525141_length(amino acids)=203AA_BP= MPMDLILVVWFCVCTARTVVGFGMDPDLQMDIVTELDLVNTTLGVAQVSGMHNASKAFLFQDIEREIHAAPHVSEKLIQLFRNKSEFTIL ATVQQKPSTSGVILSIRELEHSYFELESSGLRDEIRYHYIHNGKPRTEALPYRMADGQWHKVALSVSASHLLLHVDCNRIYERVIDPPDT -------------------------------------------------------------- >58570_58570_4_NELL1-SMCO4_NELL1_chr11_20907086_ENST00000532434_SMCO4_chr11_93232514_ENST00000525141_length(transcript)=1555nt_BP=666nt TCCAGGCTCATTTGCTTCCACCTAGCTTCGGTGCCCCCTGCTAGGCGGGGACCCTCGAGAGCGATGCCGATGGATTTGATTTTAGTTGTG TGGTTCTGTGTGTGCACTGCCAGGACAGTGGTGGGCTTTGGGATGGACCCTGACCTTCAGATGGATATCGTCACCGAGCTTGACCTTGTG AACACCACCCTTGGAGTTGCTCAGGTGTCTGGAATGCACAATGCCAGCAAAGCATTTTTATTTCAAGACATAGAAAGAGAGATCCATGCA GCTCCTCATGTGAGTGAGAAATTAATTCAGCTGTTCCGGAACAAGAGTGAATTCACCATTTTGGCCACTGTACAGCAGAAGCCATCCACT TCAGGAGTGATACTGTCCATTCGAGAACTGGAGCACAGCTATTTTGAACTGGAGAGCAGTGGCCTGAGGGATGAGATTCGGTATCACTAC ATACACAATGGGAAGCCAAGGACAGAGGCACTTCCTTACCGCATGGCAGATGGACAATGGCACAAGGTTGCACTGTCAGTTAGCGCCTCT CATCTCCTGCTCCATGTCGACTGTAACAGGATTTATGAGCGTGTGATAGACCCTCCAGATACCAACCTTCCCCCAGGAATCAATTTATGG CTTGGCCAGCGCAACCAAAAGCATGGCTTATTCAAACAGGAATAACTCAAGTCACCTGTACTGGAAATCAGTTTGCTGAAATTAATCAAC GATTCTTGAAGTTGAAGAAAAGTTGTTCTCTCTACAGGAGGTTCCAGCCTTGGCAAGAGGAGTGTGGCCCTTCCTGGAATCCCTCTGGAC ACACCCTCCTAGCATCCTCTAGGAAAGATGCGGCAGCTCAAAGGGAAGCCCAAGAAGGAGACCTCCAAGGACAAGAAGGAGCGGAAGCAA GCCATGCAGGAGGCCCGGCAGCAGATCACTACAGTGGTGCTGCCCACGCTGGCCGTGGTCGTGCTCTTGATCGTGGTGTTTGTGTACGTG GCCACGCGCCCCACCATCACCGAGTGAGCCCCGCAGCCGGCTGCGGACCCCATCGGCAGGGAGAGGAGGCGCGGGAGGGGGACGCAAACA AAAAATGGCTTTCATATTCAGAGATGTTCATGTTGCTGAGCTGTAAGCAGGAGCACCCTGTCTTCTCTGGTCTTTGACTTGATTAAAGTA TCTCCGCTTTCTTGGGAGGGAATAGGGGATGTTTTATCAGTGAATGTGCCATACACCTTATGGTCCACTTCATGTGCCTTTCAGACTTCA AAGCGCGCGCGCATGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGCTTCTTTTTCTCTCCTAAAAATCGATAAGTAGCTCCA CCTGAAGAGGGATGGAACCTCTGGGTCAGGAAACAGCTGGAATCCACACTCACCTCATTCCCATTGTTTGGATCATGCTTCTTTCCAACA CGTGTTCACAATCTCCAAAGGGACTGTATTTCTTCTCTGTGCTTAATGTGATTTGAAATATGTTGAATCAAAGTGAAATATTTATTTTTT >58570_58570_4_NELL1-SMCO4_NELL1_chr11_20907086_ENST00000532434_SMCO4_chr11_93232514_ENST00000525141_length(amino acids)=203AA_BP= MPMDLILVVWFCVCTARTVVGFGMDPDLQMDIVTELDLVNTTLGVAQVSGMHNASKAFLFQDIEREIHAAPHVSEKLIQLFRNKSEFTIL ATVQQKPSTSGVILSIRELEHSYFELESSGLRDEIRYHYIHNGKPRTEALPYRMADGQWHKVALSVSASHLLLHVDCNRIYERVIDPPDT -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for NELL1-SMCO4 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for NELL1-SMCO4 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for NELL1-SMCO4 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
