
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:NFATC2IP-HS3ST4 (FusionGDB2 ID:58794) |
Fusion Gene Summary for NFATC2IP-HS3ST4 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: NFATC2IP-HS3ST4 | Fusion gene ID: 58794 | Hgene | Tgene | Gene symbol | NFATC2IP | HS3ST4 | Gene ID | 84901 | 9951 |
| Gene name | nuclear factor of activated T cells 2 interacting protein | heparan sulfate-glucosamine 3-sulfotransferase 4 | |
| Synonyms | ESC2|NIP45|RAD60 | 3-OST-4|30ST4|3OST4|h3-OST-4 | |
| Cytomap | 16p11.2 | 16p12.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | NFATC2-interacting protein45 kDa NF-AT-interacting protein45 kDa NFAT-interacting proteinnuclear factor of activated T-cells, cytoplasmic 2-interacting proteinnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protei | heparan sulfate glucosamine 3-O-sulfotransferase 4heparan sulfate (glucosamine) 3-O-sulfotransferase 4heparan sulfate 3-O-sulfotransferase 4heparan sulfate D-glucosaminyl 3-O-sulfotransferase 4 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q8NCF5 | Q9Y661 | |
| Ensembl transtripts involved in fusion gene | ENST00000320805, ENST00000564978, ENST00000568148, ENST00000562977, | ENST00000475436, ENST00000331351, | |
| Fusion gene scores | * DoF score | 6 X 5 X 3=90 | 11 X 8 X 5=440 |
| # samples | 6 | 13 | |
| ** MAII score | log2(6/90*10)=-0.584962500721156 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(13/440*10)=-1.7589919004962 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: NFATC2IP [Title/Abstract] AND HS3ST4 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | NFATC2IP(28970421)-HS3ST4(26146933), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across NFATC2IP (5'-gene) Fusion gene breakpoints across NFATC2IP (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
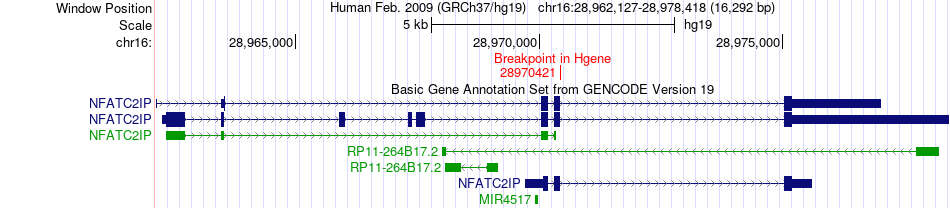 |
 Fusion gene breakpoints across HS3ST4 (3'-gene) Fusion gene breakpoints across HS3ST4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
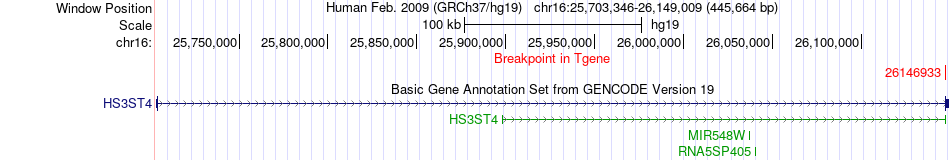 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-D8-A27M-01A | NFATC2IP | chr16 | 28970421 | - | HS3ST4 | chr16 | 26146933 | + |
| ChimerDB4 | BRCA | TCGA-D8-A27M-01A | NFATC2IP | chr16 | 28970421 | + | HS3ST4 | chr16 | 26146933 | + |
Top |
Fusion Gene ORF analysis for NFATC2IP-HS3ST4 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000320805 | ENST00000475436 | NFATC2IP | chr16 | 28970421 | + | HS3ST4 | chr16 | 26146933 | + |
| 5CDS-3UTR | ENST00000564978 | ENST00000475436 | NFATC2IP | chr16 | 28970421 | + | HS3ST4 | chr16 | 26146933 | + |
| 5CDS-3UTR | ENST00000568148 | ENST00000475436 | NFATC2IP | chr16 | 28970421 | + | HS3ST4 | chr16 | 26146933 | + |
| In-frame | ENST00000320805 | ENST00000331351 | NFATC2IP | chr16 | 28970421 | + | HS3ST4 | chr16 | 26146933 | + |
| In-frame | ENST00000564978 | ENST00000331351 | NFATC2IP | chr16 | 28970421 | + | HS3ST4 | chr16 | 26146933 | + |
| In-frame | ENST00000568148 | ENST00000331351 | NFATC2IP | chr16 | 28970421 | + | HS3ST4 | chr16 | 26146933 | + |
| intron-3CDS | ENST00000562977 | ENST00000331351 | NFATC2IP | chr16 | 28970421 | + | HS3ST4 | chr16 | 26146933 | + |
| intron-3UTR | ENST00000562977 | ENST00000475436 | NFATC2IP | chr16 | 28970421 | + | HS3ST4 | chr16 | 26146933 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000564978 | NFATC2IP | chr16 | 28970421 | + | ENST00000331351 | HS3ST4 | chr16 | 26146933 | + | 2433 | 356 | 363 | 992 | 209 |
| ENST00000320805 | NFATC2IP | chr16 | 28970421 | + | ENST00000331351 | HS3ST4 | chr16 | 26146933 | + | 3253 | 1176 | 75 | 1247 | 390 |
| ENST00000568148 | NFATC2IP | chr16 | 28970421 | + | ENST00000331351 | HS3ST4 | chr16 | 26146933 | + | 2659 | 582 | 589 | 1218 | 209 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000564978 | ENST00000331351 | NFATC2IP | chr16 | 28970421 | + | HS3ST4 | chr16 | 26146933 | + | 0.005600702 | 0.99439925 |
| ENST00000320805 | ENST00000331351 | NFATC2IP | chr16 | 28970421 | + | HS3ST4 | chr16 | 26146933 | + | 0.00608756 | 0.99391246 |
| ENST00000568148 | ENST00000331351 | NFATC2IP | chr16 | 28970421 | + | HS3ST4 | chr16 | 26146933 | + | 0.005715617 | 0.99428433 |
Top |
Fusion Genomic Features for NFATC2IP-HS3ST4 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| NFATC2IP | chr16 | 28970421 | + | HS3ST4 | chr16 | 26146932 | + | 5.08E-09 | 1 |
| NFATC2IP | chr16 | 28970421 | + | HS3ST4 | chr16 | 26146932 | + | 5.08E-09 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
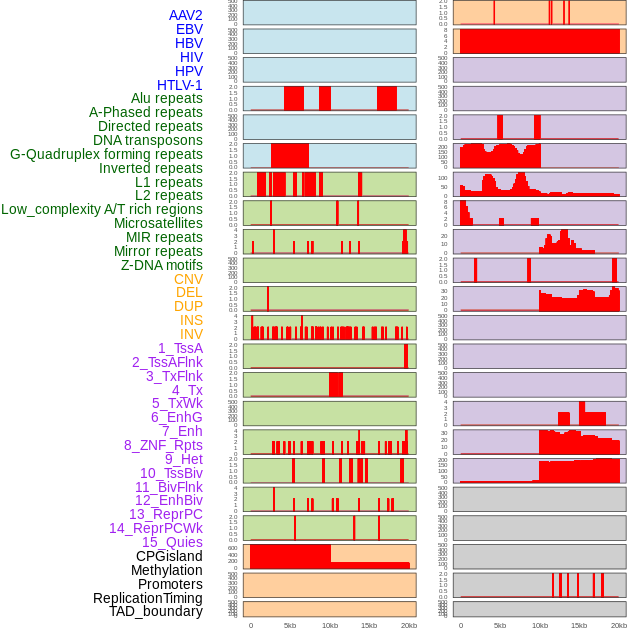 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
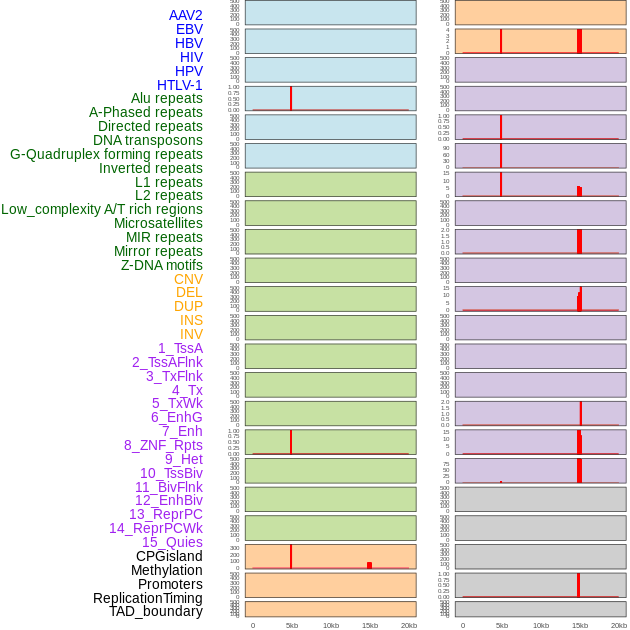 |
Top |
Fusion Protein Features for NFATC2IP-HS3ST4 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:28970421/chr16:26146933) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NFATC2IP | HS3ST4 |
| FUNCTION: In T-helper 2 (Th2) cells, regulates the magnitude of NFAT-driven transcription of a specific subset of cytokine genes, including IL3, IL4, IL5 and IL13, but not IL2. Recruits PRMT1 to the IL4 promoter; this leads to enhancement of histone H4 'Arg-3'-methylation and facilitates subsequent histone acetylation at the IL4 locus, thus promotes robust cytokine expression (By similarity). Down-regulates formation of poly-SUMO chains by UBE2I/UBC9 (By similarity). {ECO:0000250}. | FUNCTION: Sulfotransferase that utilizes 3'-phospho-5'-adenylyl sulfate (PAPS) to catalyze the transfer of a sulfo group to an N-unsubstituted glucosamine linked to a 2-O-sulfo iduronic acid unit on heparan sulfate. Unlike 3-OST-1, does not convert non-anticoagulant heparan sulfate to anticoagulant heparan sulfate (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NFATC2IP | chr16:28970421 | chr16:26146933 | ENST00000320805 | + | 7 | 8 | 209_231 | 367 | 420.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NFATC2IP | chr16:28970421 | chr16:26146933 | ENST00000320805 | + | 7 | 8 | 108_112 | 367 | 420.0 | Compositional bias | Note=Poly-Arg |
| Tgene | HS3ST4 | chr16:28970421 | chr16:26146933 | ENST00000331351 | 0 | 2 | 413_417 | 244 | 457.0 | Nucleotide binding | PAPS | |
| Tgene | HS3ST4 | chr16:28970421 | chr16:26146933 | ENST00000331351 | 0 | 2 | 260_263 | 244 | 457.0 | Region | Substrate binding | |
| Tgene | HS3ST4 | chr16:28970421 | chr16:26146933 | ENST00000331351 | 0 | 2 | 328_329 | 244 | 457.0 | Region | Substrate binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NFATC2IP | chr16:28970421 | chr16:26146933 | ENST00000568148 | + | 2 | 3 | 209_231 | 75 | 128.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NFATC2IP | chr16:28970421 | chr16:26146933 | ENST00000568148 | + | 2 | 3 | 108_112 | 75 | 128.0 | Compositional bias | Note=Poly-Arg |
| Hgene | NFATC2IP | chr16:28970421 | chr16:26146933 | ENST00000320805 | + | 7 | 8 | 348_419 | 367 | 420.0 | Domain | Ubiquitin-like |
| Hgene | NFATC2IP | chr16:28970421 | chr16:26146933 | ENST00000568148 | + | 2 | 3 | 348_419 | 75 | 128.0 | Domain | Ubiquitin-like |
| Tgene | HS3ST4 | chr16:28970421 | chr16:26146933 | ENST00000331351 | 0 | 2 | 207_211 | 244 | 457.0 | Nucleotide binding | PAPS | |
| Tgene | HS3ST4 | chr16:28970421 | chr16:26146933 | ENST00000331351 | 0 | 2 | 229_235 | 244 | 457.0 | Region | Substrate binding | |
| Tgene | HS3ST4 | chr16:28970421 | chr16:26146933 | ENST00000331351 | 0 | 2 | 1_34 | 244 | 457.0 | Topological domain | Cytoplasmic | |
| Tgene | HS3ST4 | chr16:28970421 | chr16:26146933 | ENST00000331351 | 0 | 2 | 60_456 | 244 | 457.0 | Topological domain | Lumenal | |
| Tgene | HS3ST4 | chr16:28970421 | chr16:26146933 | ENST00000331351 | 0 | 2 | 35_59 | 244 | 457.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
Top |
Fusion Gene Sequence for NFATC2IP-HS3ST4 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >58794_58794_1_NFATC2IP-HS3ST4_NFATC2IP_chr16_28970421_ENST00000320805_HS3ST4_chr16_26146933_ENST00000331351_length(transcript)=3253nt_BP=1176nt TGCAAGGCGAAAGTCGCTGAAGGGGGCGGGGCGAGGCGAGAGGAGGCGGGCGTGTGTTGTGGAGGAAAGTGTGCCATGGCGGAGCCTGTG GGGAAGCGGGGCCGCTGGTCCGGAGGTAGCGGTGCCGGCCGAGGGGGTCGGGGCGGCTGGGGCGGTCGGGGCCGGCGTCCTCGGGCCCAG CGGTCTCCATCCCGGGGCACGCTGGACGTAGTGTCTGTGGACTTGGTCACCGACAGCGATGAGGAAATTCTGGAGGTCGCCACCGCTCGC GGTGCCGCGGACGAGGTTGAGGTGGAGCCCCCGGAGCCCCCGGGGCCGGTCGCGTCCCGGGATAACAGCAACAGTGACAGCGAAGGGGAG GACAGGCGGCCCGCAGGACCCCCGCGGGAGCCGGTCAGGCGGCGGCGGCGGCTGGTGCTGGATCCGGGGGAGGCGCCGCTGGTTCCGGTG TACTCGGGGAAGGTTAAAAGCAGCCTTCGCCTTATCCCAGATGATCTATCCCTCCTGAAACTCTACCCTCCAGGGGATGAGGAAGAGGCA GAGCTGGCAGATTCGAGTGGTCTCTACCATGAGGGCTCCCCATCACCAGGCTCTCCCTGGAAGACAAAGCTGAGGACTAAGGATAAAGAA GAGAAGAAAAAGACAGAGTTTCTGGATCTGGACAACTCTCCTCTGTCCCCACCTTCACCAAGGACCAAAAGCAGAACGCATACTCGGGCA CTCAAGAAGTTAAGTGAGGTGAACAAGCGCCTCCAGGATCTCCGTTCCTGTCTGAGCCCCAAGCCACCTCAGGGTCAAGAGCAACAGGGC CAAGAGGATGAAGTGGTCTTGGTGGAAGGGCCCACCCTCCCAGAGACCCCCCGACTCTTCCCACTCAAAATCCGTTGCCGGGCTGACCTG GTCAGATTGCCCCTCAGGATGTCGGAGCCCCTGCAGAGTGTGGTGGACCACATGGCCACCCACCTTGGGGTGTCCCCAAGCAGGATCCTT TTGCTTTTTGGAGAGACAGAGCTATCACCTACTGCCACTCCCAGGACCCTAAAGCTCGGAGTGGCTGACATCATTGACTGTGTGGTACTA ACAAGTTCTCCAGAGGCCACAGAGACGTCCCAACAGCTCCAGCTCCGGGTGCAGGGAAAGGAGAAACACCAGACACTGGAAGTCTCACTG TCTCGAAAATGTGATGCCCAAGACTTTGGATGGGCAAATAACCATGGAGAAGACTCCAAGTTACTTTGTGACAAATGAGGCTCCCAAGCG CATTCACTCCATGGCCAAGGACATCAAACTGATTGTGGTGGTGAGAAACCCCGTGACCAGGGCCATCTCTGACTACACGCAGACACTGTC AAAGAAACCCGAGATCCCCACCTTTGAGGTGCTGGCCTTCAAAAACCGGACCCTCGGGCTGATCGATGCTTCCTGGAGTGCCATTCGAAT AGGGATCTATGCGCTGCATCTGGAAAACTGGCTCCAGTATTTCCCCCTCTCCCAGATCCTCTTTGTCAGTGGTGAGCGACTCATTGTGGA CCCCGCCGGGGAAATGGCCAAAGTACAGGATTTTCTAGGCCTCAAACGTGTTGTGACTGAGAAGCATTTCTATTTCAACAAAACCAAGGG GTTCCCTTGCCTAAAGAAGCCAGAAGACAGCAGTGCCCCGAGGTGCTTAGGCAAGAGCAAAGGTCGGACTCATCCTCGCATTGACCCAGA TGTCATCCACAGACTGAGGAAATTCTACAAACCCTTCAACTTGATGTTTTACCAAATGACTGGTCAAGATTTTCAGTGGGAACAGGAAGA GGGTGATAAATGAGGCTAGAGAGGCAGAGGAAGGCTAGTCAATAAGCTAAGGAGGCTCCTTGCCTGAGTCCTTGAATACCCCAGCTTCTG CAGCTTCACTTGCTGGAGTGCCAAGTAGATCTCCTCCTCCTTCATGCAGCCAGGATTGCCTCCAGTGCTGTTAGCTTAGGCAAACAGGTG GATCCCATGGCATCCCCATGGAGGAACCAGGCCCATCTGGGCAGCAGCATCTGGTTGACCAGATGGCCACCAGAACCCACTGTTCATTCT TATCTTCTGCTAGTTAATATAGCCTGAAGACAGAGGATAAATAGTTGTCAATGTCAGAGACAGTGCTATTAATGTATATGTGAGCGACAA AAAAGGTCTGCTTTATAGGGGTTCTCACTCTAGCTTGGGGAGCCAGGGTTCTAGCCCTGTATCTGTCATGGGCACCTGCTGTCTAAACCT CTGCTTGGGCTTCTCCCCAGAATGCACTTTGTGGCTGAGTGCTCCAGGACTCCTAGGGAGCAAGGTCCTCCCTCTAAGGTGTTTCTAGTC TTCTCTTTAAAGGTCTCATCCCACAACCCCTGACTTCCTCCCTCCCCACATCATGAAGGCAGAGGCATGCACATTCCTCACTGAAAAAGA AAACACACACCCACCCACACACACACACACAGAAGAAAATGAAAGCTGACACACCTCGAAGCCTTCTTTCCAAGAGCCCTCTAAATGGGG TTGGGTCTCACTCTTCATGAGTATCCTGGGTTGTGCAGAAGCTTAGCATATGCCCTTGTGTTCGGATCAGGCCCACAGGGCTGCTCAAAG AGTAGAGTAATTGTAACCGAGGTCAGAGCTCTGGGGTTGGCAGAGATGAGTGGCCATATCTGGGGGTAAAAGAAGAAATCCTGTCCTCTT GGTGGGAGGTTACCTTACCTGAAGACCATCTCTCCCAAGCACTGTAGTTCTGAGCATGTTTTTGGGGTGGACTCTGTCCCCTAGGGTCCC TAGAAGGGCAAAGACCAGAGAGTTGACAAGTCTGTTATTAGGAATAATCCTTAGCCATGTAATGGAGAAAGGAGCAGTCAGCATTCTTCC AATTTGCCCCACCACCACCTCCTCGGGCTTCATTTTCTCTATTTAGAGATGGCAGAGAGTGAGGTAGTGGCGAGAAAGCTGACTCCATTC ATCAGATCCAGTTTATGAGGGTTGGGGGTGAGCAAGGGCTGTCTGCAGAAACCCCCATCAAGAGCTGCTGAATGAAGTGTCCCTTCCCAT CAGTTTGATTCAATTAAAATGCATCATTTGACATAAAGCACTTGTTCACAGATCTCCAAAACCAGGAATTGTTCTAGTAAAACTGGAAAT TTGTATGAGTGGGGGGAGTTAAATCTGTTCAGCTGTTATTAAACTGTCATTTCTCCCGCTAAATGAAAACCGTGTTGTTATAAAGCTTAA >58794_58794_1_NFATC2IP-HS3ST4_NFATC2IP_chr16_28970421_ENST00000320805_HS3ST4_chr16_26146933_ENST00000331351_length(amino acids)=390AA_BP=363 MAEPVGKRGRWSGGSGAGRGGRGGWGGRGRRPRAQRSPSRGTLDVVSVDLVTDSDEEILEVATARGAADEVEVEPPEPPGPVASRDNSNS DSEGEDRRPAGPPREPVRRRRRLVLDPGEAPLVPVYSGKVKSSLRLIPDDLSLLKLYPPGDEEEAELADSSGLYHEGSPSPGSPWKTKLR TKDKEEKKKTEFLDLDNSPLSPPSPRTKSRTHTRALKKLSEVNKRLQDLRSCLSPKPPQGQEQQGQEDEVVLVEGPTLPETPRLFPLKIR CRADLVRLPLRMSEPLQSVVDHMATHLGVSPSRILLLFGETELSPTATPRTLKLGVADIIDCVVLTSSPEATETSQQLQLRVQGKEKHQT -------------------------------------------------------------- >58794_58794_2_NFATC2IP-HS3ST4_NFATC2IP_chr16_28970421_ENST00000564978_HS3ST4_chr16_26146933_ENST00000331351_length(transcript)=2433nt_BP=356nt TGGAATCTTTGGCGCCTTGGTTTCAGAGGTTAAAAGCAGCCTTCGCCTTATCCCAGATGATCTATCCCTCCTGAAACTCTACCCTCCAGG GGATGAGGAAGTCGGAGCCCCTGCAGAGTGTGGTGGACCACATGGCCACCCACCTTGGGGTGTCCCCAAGCAGGATCCTTTTGCTTTTTG GAGAGACAGAGCTATCACCTACTGCCACTCCCAGGACCCTAAAGCTCGGAGTGGCTGACATCATTGACTGTGTGGTACTAACAAGTTCTC CAGAGGCCACAGAGACGTCCCAACAGCTCCAGCTCCGGGTGCAGGGAAAGGAGAAACACCAGACACTGGAAGTCTCACTGTCTCGAAAAT GTGATGCCCAAGACTTTGGATGGGCAAATAACCATGGAGAAGACTCCAAGTTACTTTGTGACAAATGAGGCTCCCAAGCGCATTCACTCC ATGGCCAAGGACATCAAACTGATTGTGGTGGTGAGAAACCCCGTGACCAGGGCCATCTCTGACTACACGCAGACACTGTCAAAGAAACCC GAGATCCCCACCTTTGAGGTGCTGGCCTTCAAAAACCGGACCCTCGGGCTGATCGATGCTTCCTGGAGTGCCATTCGAATAGGGATCTAT GCGCTGCATCTGGAAAACTGGCTCCAGTATTTCCCCCTCTCCCAGATCCTCTTTGTCAGTGGTGAGCGACTCATTGTGGACCCCGCCGGG GAAATGGCCAAAGTACAGGATTTTCTAGGCCTCAAACGTGTTGTGACTGAGAAGCATTTCTATTTCAACAAAACCAAGGGGTTCCCTTGC CTAAAGAAGCCAGAAGACAGCAGTGCCCCGAGGTGCTTAGGCAAGAGCAAAGGTCGGACTCATCCTCGCATTGACCCAGATGTCATCCAC AGACTGAGGAAATTCTACAAACCCTTCAACTTGATGTTTTACCAAATGACTGGTCAAGATTTTCAGTGGGAACAGGAAGAGGGTGATAAA TGAGGCTAGAGAGGCAGAGGAAGGCTAGTCAATAAGCTAAGGAGGCTCCTTGCCTGAGTCCTTGAATACCCCAGCTTCTGCAGCTTCACT TGCTGGAGTGCCAAGTAGATCTCCTCCTCCTTCATGCAGCCAGGATTGCCTCCAGTGCTGTTAGCTTAGGCAAACAGGTGGATCCCATGG CATCCCCATGGAGGAACCAGGCCCATCTGGGCAGCAGCATCTGGTTGACCAGATGGCCACCAGAACCCACTGTTCATTCTTATCTTCTGC TAGTTAATATAGCCTGAAGACAGAGGATAAATAGTTGTCAATGTCAGAGACAGTGCTATTAATGTATATGTGAGCGACAAAAAAGGTCTG CTTTATAGGGGTTCTCACTCTAGCTTGGGGAGCCAGGGTTCTAGCCCTGTATCTGTCATGGGCACCTGCTGTCTAAACCTCTGCTTGGGC TTCTCCCCAGAATGCACTTTGTGGCTGAGTGCTCCAGGACTCCTAGGGAGCAAGGTCCTCCCTCTAAGGTGTTTCTAGTCTTCTCTTTAA AGGTCTCATCCCACAACCCCTGACTTCCTCCCTCCCCACATCATGAAGGCAGAGGCATGCACATTCCTCACTGAAAAAGAAAACACACAC CCACCCACACACACACACACAGAAGAAAATGAAAGCTGACACACCTCGAAGCCTTCTTTCCAAGAGCCCTCTAAATGGGGTTGGGTCTCA CTCTTCATGAGTATCCTGGGTTGTGCAGAAGCTTAGCATATGCCCTTGTGTTCGGATCAGGCCCACAGGGCTGCTCAAAGAGTAGAGTAA TTGTAACCGAGGTCAGAGCTCTGGGGTTGGCAGAGATGAGTGGCCATATCTGGGGGTAAAAGAAGAAATCCTGTCCTCTTGGTGGGAGGT TACCTTACCTGAAGACCATCTCTCCCAAGCACTGTAGTTCTGAGCATGTTTTTGGGGTGGACTCTGTCCCCTAGGGTCCCTAGAAGGGCA AAGACCAGAGAGTTGACAAGTCTGTTATTAGGAATAATCCTTAGCCATGTAATGGAGAAAGGAGCAGTCAGCATTCTTCCAATTTGCCCC ACCACCACCTCCTCGGGCTTCATTTTCTCTATTTAGAGATGGCAGAGAGTGAGGTAGTGGCGAGAAAGCTGACTCCATTCATCAGATCCA GTTTATGAGGGTTGGGGGTGAGCAAGGGCTGTCTGCAGAAACCCCCATCAAGAGCTGCTGAATGAAGTGTCCCTTCCCATCAGTTTGATT CAATTAAAATGCATCATTTGACATAAAGCACTTGTTCACAGATCTCCAAAACCAGGAATTGTTCTAGTAAAACTGGAAATTTGTATGAGT GGGGGGAGTTAAATCTGTTCAGCTGTTATTAAACTGTCATTTCTCCCGCTAAATGAAAACCGTGTTGTTATAAAGCTTAATGCAACCTGA >58794_58794_2_NFATC2IP-HS3ST4_NFATC2IP_chr16_28970421_ENST00000564978_HS3ST4_chr16_26146933_ENST00000331351_length(amino acids)=209AA_BP=0 MPKTLDGQITMEKTPSYFVTNEAPKRIHSMAKDIKLIVVVRNPVTRAISDYTQTLSKKPEIPTFEVLAFKNRTLGLIDASWSAIRIGIYA LHLENWLQYFPLSQILFVSGERLIVDPAGEMAKVQDFLGLKRVVTEKHFYFNKTKGFPCLKKPEDSSAPRCLGKSKGRTHPRIDPDVIHR -------------------------------------------------------------- >58794_58794_3_NFATC2IP-HS3ST4_NFATC2IP_chr16_28970421_ENST00000568148_HS3ST4_chr16_26146933_ENST00000331351_length(transcript)=2659nt_BP=582nt CATACTGGCCCACGTTCCCACACAGTGCCAATCAGCTGGAGCTGCGTAACAGCCAGCCTGTCTAGGTGGCATGTGAATTCCAGTTTGCTG CAATCCCCACCACCCCTTTCTTTGTTCTAACCCATGGCTGCTATATTTCTTTGTGTTACTCGCCTGTACCCCTTGCTGACACGATTTTAT TCTATACCTTGCCAGGTAAATATGATGAAACTCACAGCTGAGGAGCTTAGCAAGTAGCTAAGGCCAGAGCTTGTGTTTGGGTGGTGTGGC TGGGGGCATGGATTTCAAGGCAGGAAGACCTCTTTTGCTTGTTGTCCCCATCCCTAGTCGGAGCCCCTGCAGAGTGTGGTGGACCACATG GCCACCCACCTTGGGGTGTCCCCAAGCAGGATCCTTTTGCTTTTTGGAGAGACAGAGCTATCACCTACTGCCACTCCCAGGACCCTAAAG CTCGGAGTGGCTGACATCATTGACTGTGTGGTACTAACAAGTTCTCCAGAGGCCACAGAGACGTCCCAACAGCTCCAGCTCCGGGTGCAG GGAAAGGAGAAACACCAGACACTGGAAGTCTCACTGTCTCGAAAATGTGATGCCCAAGACTTTGGATGGGCAAATAACCATGGAGAAGAC TCCAAGTTACTTTGTGACAAATGAGGCTCCCAAGCGCATTCACTCCATGGCCAAGGACATCAAACTGATTGTGGTGGTGAGAAACCCCGT GACCAGGGCCATCTCTGACTACACGCAGACACTGTCAAAGAAACCCGAGATCCCCACCTTTGAGGTGCTGGCCTTCAAAAACCGGACCCT CGGGCTGATCGATGCTTCCTGGAGTGCCATTCGAATAGGGATCTATGCGCTGCATCTGGAAAACTGGCTCCAGTATTTCCCCCTCTCCCA GATCCTCTTTGTCAGTGGTGAGCGACTCATTGTGGACCCCGCCGGGGAAATGGCCAAAGTACAGGATTTTCTAGGCCTCAAACGTGTTGT GACTGAGAAGCATTTCTATTTCAACAAAACCAAGGGGTTCCCTTGCCTAAAGAAGCCAGAAGACAGCAGTGCCCCGAGGTGCTTAGGCAA GAGCAAAGGTCGGACTCATCCTCGCATTGACCCAGATGTCATCCACAGACTGAGGAAATTCTACAAACCCTTCAACTTGATGTTTTACCA AATGACTGGTCAAGATTTTCAGTGGGAACAGGAAGAGGGTGATAAATGAGGCTAGAGAGGCAGAGGAAGGCTAGTCAATAAGCTAAGGAG GCTCCTTGCCTGAGTCCTTGAATACCCCAGCTTCTGCAGCTTCACTTGCTGGAGTGCCAAGTAGATCTCCTCCTCCTTCATGCAGCCAGG ATTGCCTCCAGTGCTGTTAGCTTAGGCAAACAGGTGGATCCCATGGCATCCCCATGGAGGAACCAGGCCCATCTGGGCAGCAGCATCTGG TTGACCAGATGGCCACCAGAACCCACTGTTCATTCTTATCTTCTGCTAGTTAATATAGCCTGAAGACAGAGGATAAATAGTTGTCAATGT CAGAGACAGTGCTATTAATGTATATGTGAGCGACAAAAAAGGTCTGCTTTATAGGGGTTCTCACTCTAGCTTGGGGAGCCAGGGTTCTAG CCCTGTATCTGTCATGGGCACCTGCTGTCTAAACCTCTGCTTGGGCTTCTCCCCAGAATGCACTTTGTGGCTGAGTGCTCCAGGACTCCT AGGGAGCAAGGTCCTCCCTCTAAGGTGTTTCTAGTCTTCTCTTTAAAGGTCTCATCCCACAACCCCTGACTTCCTCCCTCCCCACATCAT GAAGGCAGAGGCATGCACATTCCTCACTGAAAAAGAAAACACACACCCACCCACACACACACACACAGAAGAAAATGAAAGCTGACACAC CTCGAAGCCTTCTTTCCAAGAGCCCTCTAAATGGGGTTGGGTCTCACTCTTCATGAGTATCCTGGGTTGTGCAGAAGCTTAGCATATGCC CTTGTGTTCGGATCAGGCCCACAGGGCTGCTCAAAGAGTAGAGTAATTGTAACCGAGGTCAGAGCTCTGGGGTTGGCAGAGATGAGTGGC CATATCTGGGGGTAAAAGAAGAAATCCTGTCCTCTTGGTGGGAGGTTACCTTACCTGAAGACCATCTCTCCCAAGCACTGTAGTTCTGAG CATGTTTTTGGGGTGGACTCTGTCCCCTAGGGTCCCTAGAAGGGCAAAGACCAGAGAGTTGACAAGTCTGTTATTAGGAATAATCCTTAG CCATGTAATGGAGAAAGGAGCAGTCAGCATTCTTCCAATTTGCCCCACCACCACCTCCTCGGGCTTCATTTTCTCTATTTAGAGATGGCA GAGAGTGAGGTAGTGGCGAGAAAGCTGACTCCATTCATCAGATCCAGTTTATGAGGGTTGGGGGTGAGCAAGGGCTGTCTGCAGAAACCC CCATCAAGAGCTGCTGAATGAAGTGTCCCTTCCCATCAGTTTGATTCAATTAAAATGCATCATTTGACATAAAGCACTTGTTCACAGATC TCCAAAACCAGGAATTGTTCTAGTAAAACTGGAAATTTGTATGAGTGGGGGGAGTTAAATCTGTTCAGCTGTTATTAAACTGTCATTTCT >58794_58794_3_NFATC2IP-HS3ST4_NFATC2IP_chr16_28970421_ENST00000568148_HS3ST4_chr16_26146933_ENST00000331351_length(amino acids)=209AA_BP=0 MPKTLDGQITMEKTPSYFVTNEAPKRIHSMAKDIKLIVVVRNPVTRAISDYTQTLSKKPEIPTFEVLAFKNRTLGLIDASWSAIRIGIYA LHLENWLQYFPLSQILFVSGERLIVDPAGEMAKVQDFLGLKRVVTEKHFYFNKTKGFPCLKKPEDSSAPRCLGKSKGRTHPRIDPDVIHR -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for NFATC2IP-HS3ST4 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for NFATC2IP-HS3ST4 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for NFATC2IP-HS3ST4 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
