
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:NFIA-CYP2J2 (FusionGDB2 ID:58865) |
Fusion Gene Summary for NFIA-CYP2J2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: NFIA-CYP2J2 | Fusion gene ID: 58865 | Hgene | Tgene | Gene symbol | NFIA | CYP2J2 | Gene ID | 4774 | 1573 |
| Gene name | nuclear factor I A | cytochrome P450 family 2 subfamily J member 2 | |
| Synonyms | BRMUTD|CTF|NF-I/A|NF1-A|NFI-A|NFI-L | CPJ2|CYPIIJ2 | |
| Cytomap | 1p31.3 | 1p32.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | nuclear factor 1 A-typeCCAAT-box-binding transcription factorTGGCA-binding protein | cytochrome P450 2J2albendazole monooxygenase (hydroxylating)albendazole monooxygenase (sulfoxide-forming)arachidonic acid epoxygenasecytochrome P450, family 2, subfamily J, polypeptide 2cytochrome P450, subfamily IIJ (arachidonic acid epoxygenase) po | |
| Modification date | 20200313 | 20200322 | |
| UniProtAcc | Q12857 | P51589 | |
| Ensembl transtripts involved in fusion gene | ENST00000371184, ENST00000371185, ENST00000371187, ENST00000371189, ENST00000371191, ENST00000403491, ENST00000407417, ENST00000485903, ENST00000357977, ENST00000479364, | ENST00000492633, ENST00000371204, | |
| Fusion gene scores | * DoF score | 21 X 22 X 8=3696 | 3 X 2 X 3=18 |
| # samples | 25 | 3 | |
| ** MAII score | log2(25/3696*10)=-3.88596475675397 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: NFIA [Title/Abstract] AND CYP2J2 [Title/Abstract] AND fusion [Title/Abstract] | ||
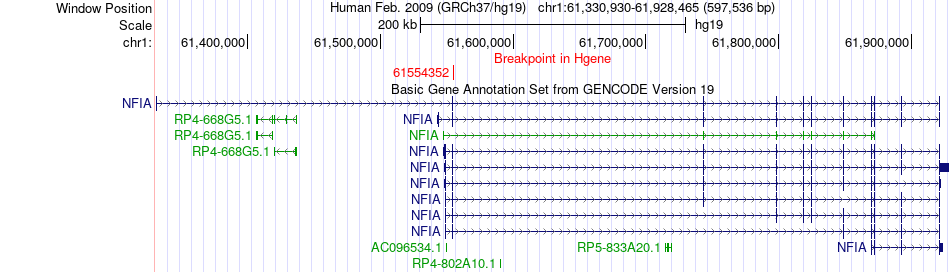
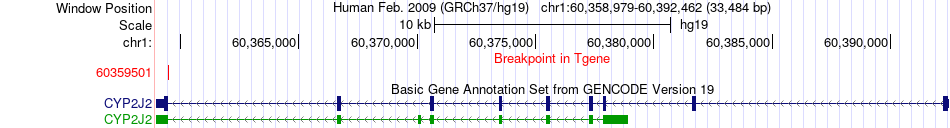
| Most frequent breakpoint | NFIA(61554352)-CYP2J2(60359501), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | NFIA | GO:0045944 | positive regulation of transcription by RNA polymerase II | 17010934 |
| Tgene | CYP2J2 | GO:0006690 | icosanoid metabolic process | 19737933 |
| Tgene | CYP2J2 | GO:0019373 | epoxygenase P450 pathway | 8631948|11901223 |
| Tgene | CYP2J2 | GO:0043651 | linoleic acid metabolic process | 11901223 |
 Fusion gene breakpoints across NFIA (5'-gene) Fusion gene breakpoints across NFIA (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across CYP2J2 (3'-gene) Fusion gene breakpoints across CYP2J2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-CH-5740-01A | NFIA | chr1 | 61554352 | - | CYP2J2 | chr1 | 60359501 | - |
| ChimerDB4 | PRAD | TCGA-CH-5740-01A | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| ChimerDB4 | PRAD | TCGA-CH-5740 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
Top |
Fusion Gene ORF analysis for NFIA-CYP2J2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000371184 | ENST00000492633 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| 5CDS-5UTR | ENST00000371185 | ENST00000492633 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| 5CDS-5UTR | ENST00000371187 | ENST00000492633 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| 5CDS-5UTR | ENST00000371189 | ENST00000492633 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| 5CDS-5UTR | ENST00000371191 | ENST00000492633 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| 5CDS-5UTR | ENST00000403491 | ENST00000492633 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| 5CDS-5UTR | ENST00000407417 | ENST00000492633 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| 5CDS-5UTR | ENST00000485903 | ENST00000492633 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| In-frame | ENST00000371184 | ENST00000371204 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| In-frame | ENST00000371185 | ENST00000371204 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| In-frame | ENST00000371187 | ENST00000371204 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| In-frame | ENST00000371189 | ENST00000371204 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| In-frame | ENST00000371191 | ENST00000371204 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| In-frame | ENST00000403491 | ENST00000371204 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| In-frame | ENST00000407417 | ENST00000371204 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| In-frame | ENST00000485903 | ENST00000371204 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| intron-3CDS | ENST00000357977 | ENST00000371204 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| intron-3CDS | ENST00000479364 | ENST00000371204 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| intron-5UTR | ENST00000357977 | ENST00000492633 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
| intron-5UTR | ENST00000479364 | ENST00000492633 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000371191 | NFIA | chr1 | 61554352 | + | ENST00000371204 | CYP2J2 | chr1 | 60359501 | - | 1262 | 740 | 112 | 918 | 268 |
| ENST00000407417 | NFIA | chr1 | 61554352 | + | ENST00000371204 | CYP2J2 | chr1 | 60359501 | - | 1409 | 887 | 310 | 1065 | 251 |
| ENST00000371189 | NFIA | chr1 | 61554352 | + | ENST00000371204 | CYP2J2 | chr1 | 60359501 | - | 1298 | 776 | 82 | 954 | 290 |
| ENST00000403491 | NFIA | chr1 | 61554352 | + | ENST00000371204 | CYP2J2 | chr1 | 60359501 | - | 1565 | 1043 | 343 | 1221 | 292 |
| ENST00000371187 | NFIA | chr1 | 61554352 | + | ENST00000371204 | CYP2J2 | chr1 | 60359501 | - | 1252 | 730 | 30 | 908 | 292 |
| ENST00000485903 | NFIA | chr1 | 61554352 | + | ENST00000371204 | CYP2J2 | chr1 | 60359501 | - | 1150 | 628 | 69 | 806 | 245 |
| ENST00000371185 | NFIA | chr1 | 61554352 | + | ENST00000371204 | CYP2J2 | chr1 | 60359501 | - | 1125 | 603 | 44 | 781 | 245 |
| ENST00000371184 | NFIA | chr1 | 61554352 | + | ENST00000371204 | CYP2J2 | chr1 | 60359501 | - | 1119 | 597 | 38 | 775 | 245 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000371191 | ENST00000371204 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - | 0.000984635 | 0.99901533 |
| ENST00000407417 | ENST00000371204 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - | 0.001671156 | 0.99832886 |
| ENST00000371189 | ENST00000371204 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - | 0.002673291 | 0.99732673 |
| ENST00000403491 | ENST00000371204 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - | 0.003595047 | 0.996405 |
| ENST00000371187 | ENST00000371204 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - | 0.002213822 | 0.99778616 |
| ENST00000485903 | ENST00000371204 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - | 0.002106781 | 0.9978933 |
| ENST00000371185 | ENST00000371204 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - | 0.001930001 | 0.99807006 |
| ENST00000371184 | ENST00000371204 | NFIA | chr1 | 61554352 | + | CYP2J2 | chr1 | 60359501 | - | 0.00182382 | 0.9981762 |
Top |
Fusion Genomic Features for NFIA-CYP2J2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
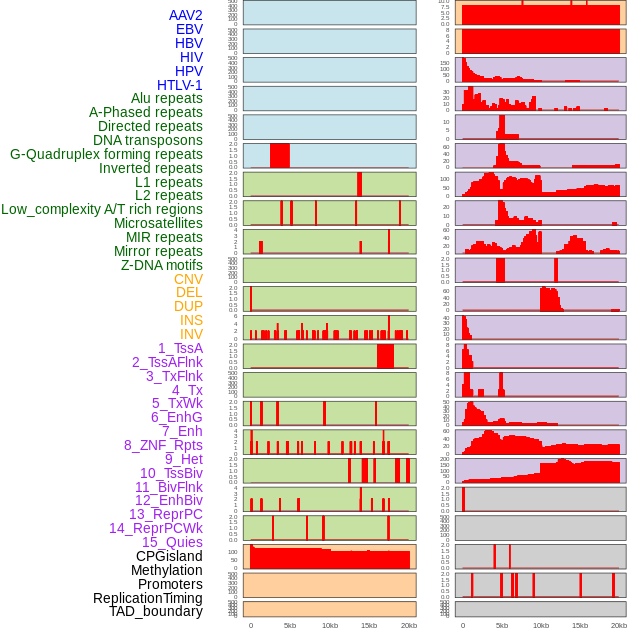 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for NFIA-CYP2J2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:61554352/chr1:60359501) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NFIA | CYP2J2 |
| FUNCTION: Recognizes and binds the palindromic sequence 5'-TTGGCNNNNNGCCAA-3' present in viral and cellular promoters and in the origin of replication of adenovirus type 2. These proteins are individually capable of activating transcription and replication. | FUNCTION: A cytochrome P450 monooxygenase involved in the metabolism of polyunsaturated fatty acids (PUFA) in the cardiovascular system (PubMed:8631948, PubMed:19965576). Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate, and reducing the second into a water molecule, with two electrons provided by NADPH via cytochrome P450 reductase (NADPH--hemoprotein reductase) (PubMed:8631948, PubMed:19965576). Catalyzes the epoxidation of double bonds of PUFA (PubMed:8631948, PubMed:19965576). Converts arachidonic acid to four regioisomeric epoxyeicosatrienoic acids (EpETrE), likely playing a major role in the epoxidation of endogenous cardiac arachidonic acid pools (PubMed:8631948). In endothelial cells, participates in eicosanoids metabolism by converting hydroperoxide species into hydroxy epoxy metabolites. In combination with 15-lipoxygenase metabolizes arachidonic acid and converts hydroperoxyicosatetraenoates (HpETEs) into hydroxy epoxy eicosatrienoates (HEETs), which are precursors of vasodilatory trihydroxyicosatrienoic acids (THETAs). This hydroperoxide isomerase activity is NADPH- and O2-independent (PubMed:19737933). Catalyzes the monooxygenation of a various xenobiotics, such as danazol, amiodarone, terfenadine, astemizole, thioridazine, tamoxifen, cyclosporin A and nabumetone (PubMed:19923256). Catalyzes hydroxylation of the anthelmintics albendazole and fenbendazole (PubMed:23959307). Catalyzes the sulfoxidation of fenbedazole (PubMed:19923256). {ECO:0000269|PubMed:19737933, ECO:0000269|PubMed:19923256, ECO:0000269|PubMed:19965576, ECO:0000269|PubMed:23959307, ECO:0000269|PubMed:8631948}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NFIA | chr1:61554352 | chr1:60359501 | ENST00000371189 | + | 3 | 12 | 1_194 | 231 | 555.0 | DNA binding | CTF/NF-I |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NFIA | chr1:61554352 | chr1:60359501 | ENST00000371187 | + | 2 | 10 | 1_194 | 186 | 499.0 | DNA binding | CTF/NF-I |
| Hgene | NFIA | chr1:61554352 | chr1:60359501 | ENST00000403491 | + | 2 | 11 | 1_194 | 186 | 510.0 | DNA binding | CTF/NF-I |
| Hgene | NFIA | chr1:61554352 | chr1:60359501 | ENST00000407417 | + | 2 | 11 | 1_194 | 178 | 502.0 | DNA binding | CTF/NF-I |
| Hgene | NFIA | chr1:61554352 | chr1:60359501 | ENST00000371187 | + | 2 | 10 | 394_402 | 186 | 499.0 | Motif | 9aaTAD |
| Hgene | NFIA | chr1:61554352 | chr1:60359501 | ENST00000371189 | + | 3 | 12 | 394_402 | 231 | 555.0 | Motif | 9aaTAD |
| Hgene | NFIA | chr1:61554352 | chr1:60359501 | ENST00000403491 | + | 2 | 11 | 394_402 | 186 | 510.0 | Motif | 9aaTAD |
| Hgene | NFIA | chr1:61554352 | chr1:60359501 | ENST00000407417 | + | 2 | 11 | 394_402 | 178 | 502.0 | Motif | 9aaTAD |
Top |
Fusion Gene Sequence for NFIA-CYP2J2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >58865_58865_1_NFIA-CYP2J2_NFIA_chr1_61554352_ENST00000371184_CYP2J2_chr1_60359501_ENST00000371204_length(transcript)=1119nt_BP=597nt CACCCAGACGCACACGCATACCCCAGCGCCCGGCAGTTATGTATTCTCCGCTCTGTCTCACCCAGGATGAATTTCATCCTTTCATCGAAG CACTTCTGCCCCACGTCCGAGCCTTTGCCTACACATGGTTCAACCTGCAGGCCCGAAAACGAAAATACTTCAAAAAACATGAAAAGCGTA TGTCAAAAGAAGAAGAGAGAGCCGTGAAGGATGAATTGCTAAGTGAAAAACCAGAGGTCAAGCAGAAGTGGGCATCTCGACTTCTGGCAA AGTTGCGGAAAGATATCCGACCCGAATATCGAGAGGATTTTGTTCTTACAGTTACAGGGAAAAAACCTCCATGTTGTGTTCTTTCCAACC CAGACCAGAAAGGCAAGATGCGAAGAATTGACTGCCTCCGCCAGGCAGATAAAGTCTGGAGGTTGGACCTTGTTATGGTGATTTTGTTTA AAGGTATTCCGCTGGAAAGTACTGATGGCGAGCGCCTTGTAAAGTCCCCACAATGCTCTAATCCAGGGCTCTGTGTCCAACCCCATCACA TAGGGGTTTCTGTTAAGGAACTCGATTTATATTTGGCATACTTTGTGCATGCAGCAGGAAAGCGGGCATGCCTCGGAGAACAGTTGGCCA GGACTGAGCTGTTTATTTTCTTCACTTCCCTTATGCAAAAATTTACCTTCAGGCCCCCAAACAATGAGAAGCTGAGCCTGAAGTTTAGAA TGGGTATCACCATTTCCCCAGTCAGTCACCGCCTCTGCGCTGTTCCTCAGGTGTAATATTGTTAAGAAAGAAAGGGGCAAGGAAAGTAAG AAGACATGGCACGTGTTCTGAAACCACTGGTGTCTGCTCAGATGTGTTGGGACAAAATGAAAGTGACTTTCAAGAAAGATCAGAGGAATT TGACTCAGAGAAAACTAGATCCAAATCCCAGCTCTACTGTCTCGTCCGAATTAGCCTTGGGAAAATCATTTATATGCTAAATAATTTACC TTTTTATCTAGGAGATGAAAAGAGGATAATGTTTCCTTCCATAAAGAAAGTTCTTGTAAGAATCAAAAGAAATGGTGAGCTTTAAGTGGT >58865_58865_1_NFIA-CYP2J2_NFIA_chr1_61554352_ENST00000371184_CYP2J2_chr1_60359501_ENST00000371204_length(amino acids)=245AA_BP=186 MYSPLCLTQDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHEKRMSKEEERAVKDELLSEKPEVKQKWASRLLAKLRKDIRPEYRED FVLTVTGKKPPCCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKGIPLESTDGERLVKSPQCSNPGLCVQPHHIGVSVKELDLYLA -------------------------------------------------------------- >58865_58865_2_NFIA-CYP2J2_NFIA_chr1_61554352_ENST00000371185_CYP2J2_chr1_60359501_ENST00000371204_length(transcript)=1125nt_BP=603nt ACCGCACACCCAGACGCACACGCATACCCCAGCGCCCGGCAGTTATGTATTCTCCGCTCTGTCTCACCCAGGATGAATTTCATCCTTTCA TCGAAGCACTTCTGCCCCACGTCCGAGCCTTTGCCTACACATGGTTCAACCTGCAGGCCCGAAAACGAAAATACTTCAAAAAACATGAAA AGCGTATGTCAAAAGAAGAAGAGAGAGCCGTGAAGGATGAATTGCTAAGTGAAAAACCAGAGGTCAAGCAGAAGTGGGCATCTCGACTTC TGGCAAAGTTGCGGAAAGATATCCGACCCGAATATCGAGAGGATTTTGTTCTTACAGTTACAGGGAAAAAACCTCCATGTTGTGTTCTTT CCAACCCAGACCAGAAAGGCAAGATGCGAAGAATTGACTGCCTCCGCCAGGCAGATAAAGTCTGGAGGTTGGACCTTGTTATGGTGATTT TGTTTAAAGGTATTCCGCTGGAAAGTACTGATGGCGAGCGCCTTGTAAAGTCCCCACAATGCTCTAATCCAGGGCTCTGTGTCCAACCCC ATCACATAGGGGTTTCTGTTAAGGAACTCGATTTATATTTGGCATACTTTGTGCATGCAGCAGGAAAGCGGGCATGCCTCGGAGAACAGT TGGCCAGGACTGAGCTGTTTATTTTCTTCACTTCCCTTATGCAAAAATTTACCTTCAGGCCCCCAAACAATGAGAAGCTGAGCCTGAAGT TTAGAATGGGTATCACCATTTCCCCAGTCAGTCACCGCCTCTGCGCTGTTCCTCAGGTGTAATATTGTTAAGAAAGAAAGGGGCAAGGAA AGTAAGAAGACATGGCACGTGTTCTGAAACCACTGGTGTCTGCTCAGATGTGTTGGGACAAAATGAAAGTGACTTTCAAGAAAGATCAGA GGAATTTGACTCAGAGAAAACTAGATCCAAATCCCAGCTCTACTGTCTCGTCCGAATTAGCCTTGGGAAAATCATTTATATGCTAAATAA TTTACCTTTTTATCTAGGAGATGAAAAGAGGATAATGTTTCCTTCCATAAAGAAAGTTCTTGTAAGAATCAAAAGAAATGGTGAGCTTTA >58865_58865_2_NFIA-CYP2J2_NFIA_chr1_61554352_ENST00000371185_CYP2J2_chr1_60359501_ENST00000371204_length(amino acids)=245AA_BP=186 MYSPLCLTQDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHEKRMSKEEERAVKDELLSEKPEVKQKWASRLLAKLRKDIRPEYRED FVLTVTGKKPPCCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKGIPLESTDGERLVKSPQCSNPGLCVQPHHIGVSVKELDLYLA -------------------------------------------------------------- >58865_58865_3_NFIA-CYP2J2_NFIA_chr1_61554352_ENST00000371187_CYP2J2_chr1_60359501_ENST00000371204_length(transcript)=1252nt_BP=730nt GACTTGGAAATGTGAACGCAAGAAGCAGGCTTGATTTTTTTTTCTCCCCCCTTCTCTCTCTCTCTCTCTCTCTCTCTTCCTCTCTCCCTC TTTCTCCTCTCTCACCCACACTCACGCACACCTCCAAACCGCACACCCAGACGCACACGCATACCCCAGCGCCCGGCAGTTATGTATTCT CCGCTCTGTCTCACCCAGGATGAATTTCATCCTTTCATCGAAGCACTTCTGCCCCACGTCCGAGCCTTTGCCTACACATGGTTCAACCTG CAGGCCCGAAAACGAAAATACTTCAAAAAACATGAAAAGCGTATGTCAAAAGAAGAAGAGAGAGCCGTGAAGGATGAATTGCTAAGTGAA AAACCAGAGGTCAAGCAGAAGTGGGCATCTCGACTTCTGGCAAAGTTGCGGAAAGATATCCGACCCGAATATCGAGAGGATTTTGTTCTT ACAGTTACAGGGAAAAAACCTCCATGTTGTGTTCTTTCCAACCCAGACCAGAAAGGCAAGATGCGAAGAATTGACTGCCTCCGCCAGGCA GATAAAGTCTGGAGGTTGGACCTTGTTATGGTGATTTTGTTTAAAGGTATTCCGCTGGAAAGTACTGATGGCGAGCGCCTTGTAAAGTCC CCACAATGCTCTAATCCAGGGCTCTGTGTCCAACCCCATCACATAGGGGTTTCTGTTAAGGAACTCGATTTATATTTGGCATACTTTGTG CATGCAGCAGGAAAGCGGGCATGCCTCGGAGAACAGTTGGCCAGGACTGAGCTGTTTATTTTCTTCACTTCCCTTATGCAAAAATTTACC TTCAGGCCCCCAAACAATGAGAAGCTGAGCCTGAAGTTTAGAATGGGTATCACCATTTCCCCAGTCAGTCACCGCCTCTGCGCTGTTCCT CAGGTGTAATATTGTTAAGAAAGAAAGGGGCAAGGAAAGTAAGAAGACATGGCACGTGTTCTGAAACCACTGGTGTCTGCTCAGATGTGT TGGGACAAAATGAAAGTGACTTTCAAGAAAGATCAGAGGAATTTGACTCAGAGAAAACTAGATCCAAATCCCAGCTCTACTGTCTCGTCC GAATTAGCCTTGGGAAAATCATTTATATGCTAAATAATTTACCTTTTTATCTAGGAGATGAAAAGAGGATAATGTTTCCTTCCATAAAGA >58865_58865_3_NFIA-CYP2J2_NFIA_chr1_61554352_ENST00000371187_CYP2J2_chr1_60359501_ENST00000371204_length(amino acids)=292AA_BP=233 MIFFSPPFSLSLSLSLPLSLFLLSHPHSRTPPNRTPRRTRIPQRPAVMYSPLCLTQDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKK HEKRMSKEEERAVKDELLSEKPEVKQKWASRLLAKLRKDIRPEYREDFVLTVTGKKPPCCVLSNPDQKGKMRRIDCLRQADKVWRLDLVM VILFKGIPLESTDGERLVKSPQCSNPGLCVQPHHIGVSVKELDLYLAYFVHAAGKRACLGEQLARTELFIFFTSLMQKFTFRPPNNEKLS -------------------------------------------------------------- >58865_58865_4_NFIA-CYP2J2_NFIA_chr1_61554352_ENST00000371189_CYP2J2_chr1_60359501_ENST00000371204_length(transcript)=1298nt_BP=776nt ACAATGCTTTGAGCCTTACCGCATTTCAGTGGGGGAAAAAAAGTTACCTAGGAGGTCTGATTTTTCAAAGAAATTTGCATACATGCAAAT GTGCCGCCCGGCCTCCTCCTCGGTTCTCTACGTGCCCACGCGGTGGCCCGGGGGGTGCGGGGCAACCTGGCAAAGTTGCCCAAGTCCTCC ACCGAGACGCACACGCATACCCCAGCGCCCGGCAGTTATGTATTCTCCGCTCTGTCTCACCCAGGATGAATTTCATCCTTTCATCGAAGC ACTTCTGCCCCACGTCCGAGCCTTTGCCTACACATGGTTCAACCTGCAGGCCCGAAAACGAAAATACTTCAAAAAACATGAAAAGCGTAT GTCAAAAGAAGAAGAGAGAGCCGTGAAGGATGAATTGCTAAGTGAAAAACCAGAGGTCAAGCAGAAGTGGGCATCTCGACTTCTGGCAAA GTTGCGGAAAGATATCCGACCCGAATATCGAGAGGATTTTGTTCTTACAGTTACAGGGAAAAAACCTCCATGTTGTGTTCTTTCCAACCC AGACCAGAAAGGCAAGATGCGAAGAATTGACTGCCTCCGCCAGGCAGATAAAGTCTGGAGGTTGGACCTTGTTATGGTGATTTTGTTTAA AGGTATTCCGCTGGAAAGTACTGATGGCGAGCGCCTTGTAAAGTCCCCACAATGCTCTAATCCAGGGCTCTGTGTCCAACCCCATCACAT AGGGGTTTCTGTTAAGGAACTCGATTTATATTTGGCATACTTTGTGCATGCAGCAGGAAAGCGGGCATGCCTCGGAGAACAGTTGGCCAG GACTGAGCTGTTTATTTTCTTCACTTCCCTTATGCAAAAATTTACCTTCAGGCCCCCAAACAATGAGAAGCTGAGCCTGAAGTTTAGAAT GGGTATCACCATTTCCCCAGTCAGTCACCGCCTCTGCGCTGTTCCTCAGGTGTAATATTGTTAAGAAAGAAAGGGGCAAGGAAAGTAAGA AGACATGGCACGTGTTCTGAAACCACTGGTGTCTGCTCAGATGTGTTGGGACAAAATGAAAGTGACTTTCAAGAAAGATCAGAGGAATTT GACTCAGAGAAAACTAGATCCAAATCCCAGCTCTACTGTCTCGTCCGAATTAGCCTTGGGAAAATCATTTATATGCTAAATAATTTACCT TTTTATCTAGGAGATGAAAAGAGGATAATGTTTCCTTCCATAAAGAAAGTTCTTGTAAGAATCAAAAGAAATGGTGAGCTTTAAGTGGTT >58865_58865_4_NFIA-CYP2J2_NFIA_chr1_61554352_ENST00000371189_CYP2J2_chr1_60359501_ENST00000371204_length(amino acids)=290AA_BP=231 MQMCRPASSSVLYVPTRWPGGCGATWQSCPSPPPRRTRIPQRPAVMYSPLCLTQDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHE KRMSKEEERAVKDELLSEKPEVKQKWASRLLAKLRKDIRPEYREDFVLTVTGKKPPCCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVI LFKGIPLESTDGERLVKSPQCSNPGLCVQPHHIGVSVKELDLYLAYFVHAAGKRACLGEQLARTELFIFFTSLMQKFTFRPPNNEKLSLK -------------------------------------------------------------- >58865_58865_5_NFIA-CYP2J2_NFIA_chr1_61554352_ENST00000371191_CYP2J2_chr1_60359501_ENST00000371204_length(transcript)=1262nt_BP=740nt TTTTTTTTTTACCCATGATTCCATATGGTATGGACCGGACTACATGTTGCCCCACATGCCACTGCAAATGTTTTCTCAATTAGGTGTCTT ATCTCCCGTGAGTTAGCATCAGATGAAGCTTGCTGACAGCGTAATGGCAGGGAAAGCTTCCGACGGCTCCATCAAATGGCAGCTCTGCTA CGACATCTCGGCCAGAACTTGGTGGATGGATGAATTTCATCCTTTCATCGAAGCACTTCTGCCCCACGTCCGAGCCTTTGCCTACACATG GTTCAACCTGCAGGCCCGAAAACGAAAATACTTCAAAAAACATGAAAAGCGTATGTCAAAAGAAGAAGAGAGAGCCGTGAAGGATGAATT GCTAAGTGAAAAACCAGAGGTCAAGCAGAAGTGGGCATCTCGACTTCTGGCAAAGTTGCGGAAAGATATCCGACCCGAATATCGAGAGGA TTTTGTTCTTACAGTTACAGGGAAAAAACCTCCATGTTGTGTTCTTTCCAACCCAGACCAGAAAGGCAAGATGCGAAGAATTGACTGCCT CCGCCAGGCAGATAAAGTCTGGAGGTTGGACCTTGTTATGGTGATTTTGTTTAAAGGTATTCCGCTGGAAAGTACTGATGGCGAGCGCCT TGTAAAGTCCCCACAATGCTCTAATCCAGGGCTCTGTGTCCAACCCCATCACATAGGGGTTTCTGTTAAGGAACTCGATTTATATTTGGC ATACTTTGTGCATGCAGCAGGAAAGCGGGCATGCCTCGGAGAACAGTTGGCCAGGACTGAGCTGTTTATTTTCTTCACTTCCCTTATGCA AAAATTTACCTTCAGGCCCCCAAACAATGAGAAGCTGAGCCTGAAGTTTAGAATGGGTATCACCATTTCCCCAGTCAGTCACCGCCTCTG CGCTGTTCCTCAGGTGTAATATTGTTAAGAAAGAAAGGGGCAAGGAAAGTAAGAAGACATGGCACGTGTTCTGAAACCACTGGTGTCTGC TCAGATGTGTTGGGACAAAATGAAAGTGACTTTCAAGAAAGATCAGAGGAATTTGACTCAGAGAAAACTAGATCCAAATCCCAGCTCTAC TGTCTCGTCCGAATTAGCCTTGGGAAAATCATTTATATGCTAAATAATTTACCTTTTTATCTAGGAGATGAAAAGAGGATAATGTTTCCT TCCATAAAGAAAGTTCTTGTAAGAATCAAAAGAAATGGTGAGCTTTAAGTGGTTTGTAAACCATAAAACACATCATAAAAGTTCTATCTA >58865_58865_5_NFIA-CYP2J2_NFIA_chr1_61554352_ENST00000371191_CYP2J2_chr1_60359501_ENST00000371204_length(amino acids)=268AA_BP=209 MKLADSVMAGKASDGSIKWQLCYDISARTWWMDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHEKRMSKEEERAVKDELLSEKPEV KQKWASRLLAKLRKDIRPEYREDFVLTVTGKKPPCCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKGIPLESTDGERLVKSPQCS -------------------------------------------------------------- >58865_58865_6_NFIA-CYP2J2_NFIA_chr1_61554352_ENST00000403491_CYP2J2_chr1_60359501_ENST00000371204_length(transcript)=1565nt_BP=1043nt GGCCGCGGAGGCTCGGGACCCGGCTGGCCGCGCGGCGCCGCAGCCGCCCCCTCCCCCACACCCCCTCCCCCCCGCGGCGGCGGCGCGAGC GGGCGGCGGCTGTGCGGTGCGGTGCAGAGCGGAGGCGGAGGCGGGCGCGCGGGCAGCTCGCGGGCACCCGGCCGGGCCGGCGCGGGAGCG GGAAAGGGTGCGCTATGCCTTTAACACCCGCGTACAGTAGGCATGTATAGTGGAGTGTAGGGAAACTCTAGGCGGGGTTAAAGTTCAGCT CATGGAGCGGCAATAGCGCTGGCTGGCTGGCTGCAGTTGAGCCGACTTGGAAATGTGAACGCAAGAAGCAGGCTTGATTTTTTTTTCTCC CCCCTTCTCTCTCTCTCTCTCTCTCTCTCTTCCTCTCTCCCTCTTTCTCCTCTCTCACCCACACTCACGCACACCTCCAAACCGCACACC CAGACGCACACGCATACCCCAGCGCCCGGCAGTTATGTATTCTCCGCTCTGTCTCACCCAGGATGAATTTCATCCTTTCATCGAAGCACT TCTGCCCCACGTCCGAGCCTTTGCCTACACATGGTTCAACCTGCAGGCCCGAAAACGAAAATACTTCAAAAAACATGAAAAGCGTATGTC AAAAGAAGAAGAGAGAGCCGTGAAGGATGAATTGCTAAGTGAAAAACCAGAGGTCAAGCAGAAGTGGGCATCTCGACTTCTGGCAAAGTT GCGGAAAGATATCCGACCCGAATATCGAGAGGATTTTGTTCTTACAGTTACAGGGAAAAAACCTCCATGTTGTGTTCTTTCCAACCCAGA CCAGAAAGGCAAGATGCGAAGAATTGACTGCCTCCGCCAGGCAGATAAAGTCTGGAGGTTGGACCTTGTTATGGTGATTTTGTTTAAAGG TATTCCGCTGGAAAGTACTGATGGCGAGCGCCTTGTAAAGTCCCCACAATGCTCTAATCCAGGGCTCTGTGTCCAACCCCATCACATAGG GGTTTCTGTTAAGGAACTCGATTTATATTTGGCATACTTTGTGCATGCAGCAGGAAAGCGGGCATGCCTCGGAGAACAGTTGGCCAGGAC TGAGCTGTTTATTTTCTTCACTTCCCTTATGCAAAAATTTACCTTCAGGCCCCCAAACAATGAGAAGCTGAGCCTGAAGTTTAGAATGGG TATCACCATTTCCCCAGTCAGTCACCGCCTCTGCGCTGTTCCTCAGGTGTAATATTGTTAAGAAAGAAAGGGGCAAGGAAAGTAAGAAGA CATGGCACGTGTTCTGAAACCACTGGTGTCTGCTCAGATGTGTTGGGACAAAATGAAAGTGACTTTCAAGAAAGATCAGAGGAATTTGAC TCAGAGAAAACTAGATCCAAATCCCAGCTCTACTGTCTCGTCCGAATTAGCCTTGGGAAAATCATTTATATGCTAAATAATTTACCTTTT TATCTAGGAGATGAAAAGAGGATAATGTTTCCTTCCATAAAGAAAGTTCTTGTAAGAATCAAAAGAAATGGTGAGCTTTAAGTGGTTTGT >58865_58865_6_NFIA-CYP2J2_NFIA_chr1_61554352_ENST00000403491_CYP2J2_chr1_60359501_ENST00000371204_length(amino acids)=292AA_BP=233 MIFFSPPFSLSLSLSLPLSLFLLSHPHSRTPPNRTPRRTRIPQRPAVMYSPLCLTQDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKK HEKRMSKEEERAVKDELLSEKPEVKQKWASRLLAKLRKDIRPEYREDFVLTVTGKKPPCCVLSNPDQKGKMRRIDCLRQADKVWRLDLVM VILFKGIPLESTDGERLVKSPQCSNPGLCVQPHHIGVSVKELDLYLAYFVHAAGKRACLGEQLARTELFIFFTSLMQKFTFRPPNNEKLS -------------------------------------------------------------- >58865_58865_7_NFIA-CYP2J2_NFIA_chr1_61554352_ENST00000407417_CYP2J2_chr1_60359501_ENST00000371204_length(transcript)=1409nt_BP=887nt GTACAATCCAAAGCACGCAAGGCAAAACCAGAGCGAATCTGAGAACCAGCCAGCCCGGGTTGGATCGCTTTCAAAAGAGAGAGTGAGAGC GAGCGAGCGAGCGAGCGAGAGCGAGACAGTGAGAGAGGGAGAGAGCGCGCCTTGCAATTGCAAAAGCCTTCTGATTTGGAGTTAAAAAAA AAATTCTCCAAGAAGCCTGCAATTGGCCAACAGCTTTTTAAAAAATCTCTTCCGGGTTTTCAACGGTTCCAAGTTTATTTAGATATTTTT TAAATGCAAAAAAAAGGGGACAACAAATGAAGCAATTCGTCTGAGCATTTTAAAGTTTCTCACGTACTCTTTTTACATTGCAATGGATGA ATTTCATCCTTTCATCGAAGCACTTCTGCCCCACGTCCGAGCCTTTGCCTACACATGGTTCAACCTGCAGGCCCGAAAACGAAAATACTT CAAAAAACATGAAAAGCGTATGTCAAAAGAAGAAGAGAGAGCCGTGAAGGATGAATTGCTAAGTGAAAAACCAGAGGTCAAGCAGAAGTG GGCATCTCGACTTCTGGCAAAGTTGCGGAAAGATATCCGACCCGAATATCGAGAGGATTTTGTTCTTACAGTTACAGGGAAAAAACCTCC ATGTTGTGTTCTTTCCAACCCAGACCAGAAAGGCAAGATGCGAAGAATTGACTGCCTCCGCCAGGCAGATAAAGTCTGGAGGTTGGACCT TGTTATGGTGATTTTGTTTAAAGGTATTCCGCTGGAAAGTACTGATGGCGAGCGCCTTGTAAAGTCCCCACAATGCTCTAATCCAGGGCT CTGTGTCCAACCCCATCACATAGGGGTTTCTGTTAAGGAACTCGATTTATATTTGGCATACTTTGTGCATGCAGCAGGAAAGCGGGCATG CCTCGGAGAACAGTTGGCCAGGACTGAGCTGTTTATTTTCTTCACTTCCCTTATGCAAAAATTTACCTTCAGGCCCCCAAACAATGAGAA GCTGAGCCTGAAGTTTAGAATGGGTATCACCATTTCCCCAGTCAGTCACCGCCTCTGCGCTGTTCCTCAGGTGTAATATTGTTAAGAAAG AAAGGGGCAAGGAAAGTAAGAAGACATGGCACGTGTTCTGAAACCACTGGTGTCTGCTCAGATGTGTTGGGACAAAATGAAAGTGACTTT CAAGAAAGATCAGAGGAATTTGACTCAGAGAAAACTAGATCCAAATCCCAGCTCTACTGTCTCGTCCGAATTAGCCTTGGGAAAATCATT TATATGCTAAATAATTTACCTTTTTATCTAGGAGATGAAAAGAGGATAATGTTTCCTTCCATAAAGAAAGTTCTTGTAAGAATCAAAAGA >58865_58865_7_NFIA-CYP2J2_NFIA_chr1_61554352_ENST00000407417_CYP2J2_chr1_60359501_ENST00000371204_length(amino acids)=251AA_BP=192 MSILKFLTYSFYIAMDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHEKRMSKEEERAVKDELLSEKPEVKQKWASRLLAKLRKDIR PEYREDFVLTVTGKKPPCCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKGIPLESTDGERLVKSPQCSNPGLCVQPHHIGVSVKE -------------------------------------------------------------- >58865_58865_8_NFIA-CYP2J2_NFIA_chr1_61554352_ENST00000485903_CYP2J2_chr1_60359501_ENST00000371204_length(transcript)=1150nt_BP=628nt CACCCACACTCACGCACACCTCCAAACCGCACACCCAGACGCACACGCATACCCCAGCGCCCGGCAGTTATGTATTCTCCGCTCTGTCTC ACCCAGGATGAATTTCATCCTTTCATCGAAGCACTTCTGCCCCACGTCCGAGCCTTTGCCTACACATGGTTCAACCTGCAGGCCCGAAAA CGAAAATACTTCAAAAAACATGAAAAGCGTATGTCAAAAGAAGAAGAGAGAGCCGTGAAGGATGAATTGCTAAGTGAAAAACCAGAGGTC AAGCAGAAGTGGGCATCTCGACTTCTGGCAAAGTTGCGGAAAGATATCCGACCCGAATATCGAGAGGATTTTGTTCTTACAGTTACAGGG AAAAAACCTCCATGTTGTGTTCTTTCCAACCCAGACCAGAAAGGCAAGATGCGAAGAATTGACTGCCTCCGCCAGGCAGATAAAGTCTGG AGGTTGGACCTTGTTATGGTGATTTTGTTTAAAGGTATTCCGCTGGAAAGTACTGATGGCGAGCGCCTTGTAAAGTCCCCACAATGCTCT AATCCAGGGCTCTGTGTCCAACCCCATCACATAGGGGTTTCTGTTAAGGAACTCGATTTATATTTGGCATACTTTGTGCATGCAGCAGGA AAGCGGGCATGCCTCGGAGAACAGTTGGCCAGGACTGAGCTGTTTATTTTCTTCACTTCCCTTATGCAAAAATTTACCTTCAGGCCCCCA AACAATGAGAAGCTGAGCCTGAAGTTTAGAATGGGTATCACCATTTCCCCAGTCAGTCACCGCCTCTGCGCTGTTCCTCAGGTGTAATAT TGTTAAGAAAGAAAGGGGCAAGGAAAGTAAGAAGACATGGCACGTGTTCTGAAACCACTGGTGTCTGCTCAGATGTGTTGGGACAAAATG AAAGTGACTTTCAAGAAAGATCAGAGGAATTTGACTCAGAGAAAACTAGATCCAAATCCCAGCTCTACTGTCTCGTCCGAATTAGCCTTG GGAAAATCATTTATATGCTAAATAATTTACCTTTTTATCTAGGAGATGAAAAGAGGATAATGTTTCCTTCCATAAAGAAAGTTCTTGTAA >58865_58865_8_NFIA-CYP2J2_NFIA_chr1_61554352_ENST00000485903_CYP2J2_chr1_60359501_ENST00000371204_length(amino acids)=245AA_BP=186 MYSPLCLTQDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHEKRMSKEEERAVKDELLSEKPEVKQKWASRLLAKLRKDIRPEYRED FVLTVTGKKPPCCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKGIPLESTDGERLVKSPQCSNPGLCVQPHHIGVSVKELDLYLA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for NFIA-CYP2J2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for NFIA-CYP2J2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for NFIA-CYP2J2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
