
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:NFIC-LSM7 (FusionGDB2 ID:58913) |
Fusion Gene Summary for NFIC-LSM7 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: NFIC-LSM7 | Fusion gene ID: 58913 | Hgene | Tgene | Gene symbol | NFIC | LSM7 | Gene ID | 4782 | 51690 |
| Gene name | nuclear factor I C | LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated | |
| Synonyms | CTF|CTF5|NF-I|NFI | YNL147W | |
| Cytomap | 19p13.3 | 19p13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | nuclear factor 1 C-typeCCAAT-box-binding transcription factorNF-I/CNF1-CTGGCA-binding proteinnuclear factor I/C (CCAAT-binding transcription factor) | U6 snRNA-associated Sm-like protein LSm7LSM7 U6 small nuclear RNA and mRNA degradation associatedLSM7 homolog, U6 small nuclear RNA associated | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P08651 | Q9UK45 | |
| Ensembl transtripts involved in fusion gene | ENST00000341919, ENST00000346156, ENST00000395111, ENST00000443272, ENST00000586919, ENST00000589123, ENST00000590282, ENST00000588839, | ENST00000589532, ENST00000252622, | |
| Fusion gene scores | * DoF score | 30 X 20 X 13=7800 | 4 X 3 X 4=48 |
| # samples | 36 | 5 | |
| ** MAII score | log2(36/7800*10)=-4.4374053123073 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/48*10)=0.0588936890535686 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: NFIC [Title/Abstract] AND LSM7 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | NFIC(3435205)-LSM7(2324195), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | NFIC | GO:0000122 | negative regulation of transcription by RNA polymerase II | 19706729 |
| Hgene | NFIC | GO:0045944 | positive regulation of transcription by RNA polymerase II | 1524678|19706729 |
| Tgene | LSM7 | GO:0000398 | mRNA splicing, via spliceosome | 28781166 |
 Fusion gene breakpoints across NFIC (5'-gene) Fusion gene breakpoints across NFIC (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
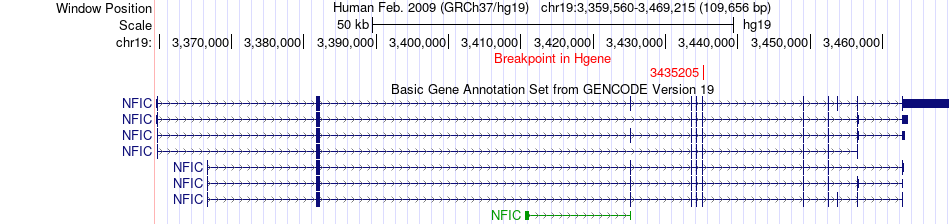 |
 Fusion gene breakpoints across LSM7 (3'-gene) Fusion gene breakpoints across LSM7 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
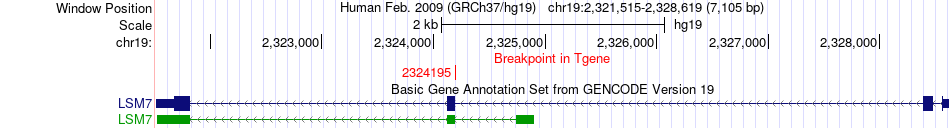 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | CESC | TCGA-DS-A1OD-01A | NFIC | chr19 | 3435205 | - | LSM7 | chr19 | 2324195 | - |
| ChimerDB4 | CESC | TCGA-DS-A1OD-01A | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - |
Top |
Fusion Gene ORF analysis for NFIC-LSM7 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000341919 | ENST00000589532 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - |
| 5CDS-5UTR | ENST00000346156 | ENST00000589532 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - |
| 5CDS-5UTR | ENST00000395111 | ENST00000589532 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - |
| 5CDS-5UTR | ENST00000443272 | ENST00000589532 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - |
| 5CDS-5UTR | ENST00000586919 | ENST00000589532 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - |
| 5CDS-5UTR | ENST00000589123 | ENST00000589532 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - |
| 5CDS-5UTR | ENST00000590282 | ENST00000589532 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - |
| In-frame | ENST00000341919 | ENST00000252622 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - |
| In-frame | ENST00000346156 | ENST00000252622 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - |
| In-frame | ENST00000395111 | ENST00000252622 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - |
| In-frame | ENST00000443272 | ENST00000252622 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - |
| In-frame | ENST00000586919 | ENST00000252622 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - |
| In-frame | ENST00000589123 | ENST00000252622 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - |
| In-frame | ENST00000590282 | ENST00000252622 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - |
| intron-3CDS | ENST00000588839 | ENST00000252622 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - |
| intron-5UTR | ENST00000588839 | ENST00000589532 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000589123 | NFIC | chr19 | 3435205 | + | ENST00000252622 | LSM7 | chr19 | 2324195 | - | 1429 | 1051 | 48 | 1265 | 405 |
| ENST00000346156 | NFIC | chr19 | 3435205 | + | ENST00000252622 | LSM7 | chr19 | 2324195 | - | 1302 | 924 | 65 | 1138 | 357 |
| ENST00000395111 | NFIC | chr19 | 3435205 | + | ENST00000252622 | LSM7 | chr19 | 2324195 | - | 1345 | 967 | 36 | 1181 | 381 |
| ENST00000586919 | NFIC | chr19 | 3435205 | + | ENST00000252622 | LSM7 | chr19 | 2324195 | - | 1237 | 859 | 0 | 1073 | 357 |
| ENST00000341919 | NFIC | chr19 | 3435205 | + | ENST00000252622 | LSM7 | chr19 | 2324195 | - | 1424 | 1046 | 88 | 1260 | 390 |
| ENST00000590282 | NFIC | chr19 | 3435205 | + | ENST00000252622 | LSM7 | chr19 | 2324195 | - | 1390 | 1012 | 54 | 1226 | 390 |
| ENST00000443272 | NFIC | chr19 | 3435205 | + | ENST00000252622 | LSM7 | chr19 | 2324195 | - | 1387 | 1009 | 51 | 1223 | 390 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000589123 | ENST00000252622 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - | 0.010279759 | 0.98972017 |
| ENST00000346156 | ENST00000252622 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - | 0.008790519 | 0.9912095 |
| ENST00000395111 | ENST00000252622 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - | 0.009458602 | 0.9905414 |
| ENST00000586919 | ENST00000252622 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - | 0.008293469 | 0.9917066 |
| ENST00000341919 | ENST00000252622 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - | 0.012872961 | 0.98712707 |
| ENST00000590282 | ENST00000252622 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - | 0.0150033 | 0.9849967 |
| ENST00000443272 | ENST00000252622 | NFIC | chr19 | 3435205 | + | LSM7 | chr19 | 2324195 | - | 0.015144719 | 0.98485523 |
Top |
Fusion Genomic Features for NFIC-LSM7 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
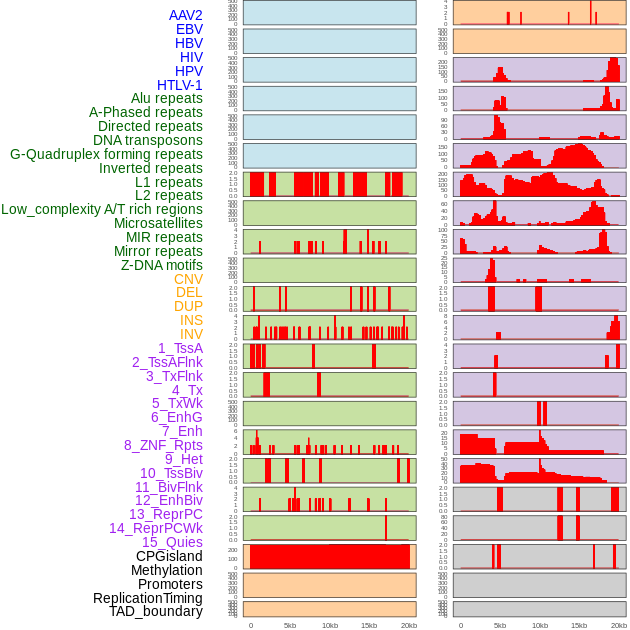 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for NFIC-LSM7 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:3435205/chr19:2324195) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NFIC | LSM7 |
| FUNCTION: Recognizes and binds the palindromic sequence 5'-TTGGCNNNNNGCCAA-3' present in viral and cellular promoters and in the origin of replication of adenovirus type 2. These proteins are individually capable of activating transcription and replication. | FUNCTION: Plays role in pre-mRNA splicing as component of the U4/U6-U5 tri-snRNP complex that is involved in spliceosome assembly, and as component of the precatalytic spliceosome (spliceosome B complex) (PubMed:28781166). The heptameric LSM2-8 complex binds specifically to the 3'-terminal U-tract of U6 snRNA (PubMed:10523320). {ECO:0000269|PubMed:10523320, ECO:0000269|PubMed:28781166}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NFIC | chr19:3435205 | chr19:2324195 | ENST00000341919 | + | 6 | 9 | 1_195 | 319 | 429.0 | DNA binding | CTF/NF-I |
| Hgene | NFIC | chr19:3435205 | chr19:2324195 | ENST00000346156 | + | 5 | 9 | 1_195 | 286 | 669.0 | DNA binding | CTF/NF-I |
| Hgene | NFIC | chr19:3435205 | chr19:2324195 | ENST00000395111 | + | 6 | 10 | 1_195 | 310 | 548.3333333333334 | DNA binding | CTF/NF-I |
| Hgene | NFIC | chr19:3435205 | chr19:2324195 | ENST00000443272 | + | 6 | 11 | 1_195 | 319 | 509.0 | DNA binding | CTF/NF-I |
| Hgene | NFIC | chr19:3435205 | chr19:2324195 | ENST00000586919 | + | 5 | 8 | 1_195 | 286 | 407.0 | DNA binding | CTF/NF-I |
| Hgene | NFIC | chr19:3435205 | chr19:2324195 | ENST00000589123 | + | 6 | 11 | 1_195 | 310 | 500.0 | DNA binding | CTF/NF-I |
| Hgene | NFIC | chr19:3435205 | chr19:2324195 | ENST00000590282 | + | 6 | 10 | 1_195 | 319 | 461.6666666666667 | DNA binding | CTF/NF-I |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NFIC | chr19:3435205 | chr19:2324195 | ENST00000341919 | + | 6 | 9 | 404_412 | 319 | 429.0 | Motif | 9aaTAD |
| Hgene | NFIC | chr19:3435205 | chr19:2324195 | ENST00000346156 | + | 5 | 9 | 404_412 | 286 | 669.0 | Motif | 9aaTAD |
| Hgene | NFIC | chr19:3435205 | chr19:2324195 | ENST00000395111 | + | 6 | 10 | 404_412 | 310 | 548.3333333333334 | Motif | 9aaTAD |
| Hgene | NFIC | chr19:3435205 | chr19:2324195 | ENST00000443272 | + | 6 | 11 | 404_412 | 319 | 509.0 | Motif | 9aaTAD |
| Hgene | NFIC | chr19:3435205 | chr19:2324195 | ENST00000586919 | + | 5 | 8 | 404_412 | 286 | 407.0 | Motif | 9aaTAD |
| Hgene | NFIC | chr19:3435205 | chr19:2324195 | ENST00000589123 | + | 6 | 11 | 404_412 | 310 | 500.0 | Motif | 9aaTAD |
| Hgene | NFIC | chr19:3435205 | chr19:2324195 | ENST00000590282 | + | 6 | 10 | 404_412 | 319 | 461.6666666666667 | Motif | 9aaTAD |
Top |
Fusion Gene Sequence for NFIC-LSM7 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >58913_58913_1_NFIC-LSM7_NFIC_chr19_3435205_ENST00000341919_LSM7_chr19_2324195_ENST00000252622_length(transcript)=1424nt_BP=1046nt GGGGGGGCGGGGGGGTGGTTTGGAAAAATGACTCAGTAAGTTCAGCGCGCCCGCTCCGGCCGGCCCTGCGCCTCCCGCCGCGCCCGGGAT GTATTCGTCCCCGCTCTGCCTCACCCAGGATGAGTTCCACCCGTTCATCGAGGCCCTGCTGCCTCACGTCCGCGCCTTCGCCTACACCTG GTTCAACCTGCAGGCGCGGAAGCGCAAGTACTTCAAGAAGCACGAGAAGCGGATGTCGAAGGACGAGGAGCGTGCGGTCAAGGACGAGCT GCTGGGCGAGAAGCCCGAGGTCAAGCAGAAGTGGGCGTCGCGGCTGCTGGCCAAGCTGCGCAAGGACATCCGGCCCGAGTGCCGCGAGGA CTTCGTGCTGAGCATCACCGGCAAGAAGGCGCCGGGCTGCGTGCTCTCCAACCCCGACCAGAAGGGCAAGATGCGGCGCATCGACTGTCT CCGGCAGGCGGACAAGGTGTGGCGGCTGGACCTGGTCATGGTCATCCTGTTCAAGGGCATCCCGCTGGAGAGCACCGACGGCGAGCGCCT GGTCAAGGCTGCGCAGTGCGGTCACCCGGTCCTGTGCGTGCAGCCGCACCACATTGGCGTGGCCGTCAAGGAGCTGGACCTCTACCTGGC CTACTTCGTGCGTGAGCGAGATGCAGAGCAAAGCGGCAGTCCCCGGACAGGGATGGGCTCTGACCAGGAGGACAGCAAGCCCATCACGCT GGACACGACCGACTTCCAGGAGAGCTTTGTCACCTCCGGCGTGTTCAGCGTCACTGAGCTCATCCAAGTGTCCCGGACACCCGTGGTGAC TGGAACAGGACCCAACTTCTCCCTGGGGGAGCTGCAGGGGCACCTGGCATACGACCTGAACCCAGCCAGCACTGGCCTCAGAAGAACGCT GCCCAGCACCTCCTCCAGTGGGAGCAAGCGGCACAAATCGGGCTCGATGGAGGAAGACGTGGACACGAGCCCTGGCGGCGATTACTACAC TTCGCCCAGCTCGCCCACGAGTAGCAGCCGCAACTGGACGGAGGACATGGAAGGAGCCAGTGGAATCCTGAAGGGCTTCGACCCACTCCT CAACCTTGTGCTGGACGGCACCATTGAGTACATGCGAGACCCTGACGACCAGTACAAGCTCACGGAGGACACCCGGCAGCTGGGCCTCGT GGTGTGCCGGGGCACGTCCGTGGTGCTAATCTGCCCGCAGGACGGCATGGAGGCCATCCCCAACCCCTTCATCCAGCAGCAGGACGCCTA GCCTGGCCGGGGGCGCGGGGGGTGCAGGGCAGGCCCGAGCAGCTCGGTTTCCCGCGGACTTGGCTGCTGCTCCCACCGCAGTACCGCCTC >58913_58913_1_NFIC-LSM7_NFIC_chr19_3435205_ENST00000341919_LSM7_chr19_2324195_ENST00000252622_length(amino acids)=390AA_BP=319 MYSSPLCLTQDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHEKRMSKDEERAVKDELLGEKPEVKQKWASRLLAKLRKDIRPECRE DFVLSITGKKAPGCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKGIPLESTDGERLVKAAQCGHPVLCVQPHHIGVAVKELDLYL AYFVRERDAEQSGSPRTGMGSDQEDSKPITLDTTDFQESFVTSGVFSVTELIQVSRTPVVTGTGPNFSLGELQGHLAYDLNPASTGLRRT LPSTSSSGSKRHKSGSMEEDVDTSPGGDYYTSPSSPTSSSRNWTEDMEGASGILKGFDPLLNLVLDGTIEYMRDPDDQYKLTEDTRQLGL -------------------------------------------------------------- >58913_58913_2_NFIC-LSM7_NFIC_chr19_3435205_ENST00000346156_LSM7_chr19_2324195_ENST00000252622_length(transcript)=1302nt_BP=924nt CCCCCCCTCGCCGGGGACCGAGCGCGCTCGCTCCGGCGCCGGCCTCGCCTCCTCGCAGCAGCGCCATGGATGAGTTCCACCCGTTCATCG AGGCCCTGCTGCCTCACGTCCGCGCCTTCGCCTACACCTGGTTCAACCTGCAGGCGCGGAAGCGCAAGTACTTCAAGAAGCACGAGAAGC GGATGTCGAAGGACGAGGAGCGTGCGGTCAAGGACGAGCTGCTGGGCGAGAAGCCCGAGGTCAAGCAGAAGTGGGCGTCGCGGCTGCTGG CCAAGCTGCGCAAGGACATCCGGCCCGAGTGCCGCGAGGACTTCGTGCTGAGCATCACCGGCAAGAAGGCGCCGGGCTGCGTGCTCTCCA ACCCCGACCAGAAGGGCAAGATGCGGCGCATCGACTGTCTCCGGCAGGCGGACAAGGTGTGGCGGCTGGACCTGGTCATGGTCATCCTGT TCAAGGGCATCCCGCTGGAGAGCACCGACGGCGAGCGCCTGGTCAAGGCTGCGCAGTGCGGTCACCCGGTCCTGTGCGTGCAGCCGCACC ACATTGGCGTGGCCGTCAAGGAGCTGGACCTCTACCTGGCCTACTTCGTGCGTGAGCGAGACACGACCGACTTCCAGGAGAGCTTTGTCA CCTCCGGCGTGTTCAGCGTCACTGAGCTCATCCAAGTGTCCCGGACACCCGTGGTGACTGGAACAGGACCCAACTTCTCCCTGGGGGAGC TGCAGGGGCACCTGGCATACGACCTGAACCCAGCCAGCACTGGCCTCAGAAGAACGCTGCCCAGCACCTCCTCCAGTGGGAGCAAGCGGC ACAAATCGGGCTCGATGGAGGAAGACGTGGACACGAGCCCTGGCGGCGATTACTACACTTCGCCCAGCTCGCCCACGAGTAGCAGCCGCA ACTGGACGGAGGACATGGAAGGAGCCAGTGGAATCCTGAAGGGCTTCGACCCACTCCTCAACCTTGTGCTGGACGGCACCATTGAGTACA TGCGAGACCCTGACGACCAGTACAAGCTCACGGAGGACACCCGGCAGCTGGGCCTCGTGGTGTGCCGGGGCACGTCCGTGGTGCTAATCT GCCCGCAGGACGGCATGGAGGCCATCCCCAACCCCTTCATCCAGCAGCAGGACGCCTAGCCTGGCCGGGGGCGCGGGGGGTGCAGGGCAG GCCCGAGCAGCTCGGTTTCCCGCGGACTTGGCTGCTGCTCCCACCGCAGTACCGCCTCCTGGAACGGAAGCATTTTCCTTTTTGTATAGG >58913_58913_2_NFIC-LSM7_NFIC_chr19_3435205_ENST00000346156_LSM7_chr19_2324195_ENST00000252622_length(amino acids)=357AA_BP=286 MDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHEKRMSKDEERAVKDELLGEKPEVKQKWASRLLAKLRKDIRPECREDFVLSITGK KAPGCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKGIPLESTDGERLVKAAQCGHPVLCVQPHHIGVAVKELDLYLAYFVRERDT TDFQESFVTSGVFSVTELIQVSRTPVVTGTGPNFSLGELQGHLAYDLNPASTGLRRTLPSTSSSGSKRHKSGSMEEDVDTSPGGDYYTSP -------------------------------------------------------------- >58913_58913_3_NFIC-LSM7_NFIC_chr19_3435205_ENST00000395111_LSM7_chr19_2324195_ENST00000252622_length(transcript)=1345nt_BP=967nt GCTCCGGCGCCGGCCTCGCCTCCTCGCAGCAGCGCCATGGATGAGTTCCACCCGTTCATCGAGGCCCTGCTGCCTCACGTCCGCGCCTTC GCCTACACCTGGTTCAACCTGCAGGCGCGGAAGCGCAAGTACTTCAAGAAGCACGAGAAGCGGATGTCGAAGGACGAGGAGCGTGCGGTC AAGGACGAGCTGCTGGGCGAGAAGCCCGAGGTCAAGCAGAAGTGGGCGTCGCGGCTGCTGGCCAAGCTGCGCAAGGACATCCGGCCCGAG TGCCGCGAGGACTTCGTGCTGAGCATCACCGGCAAGAAGGCGCCGGGCTGCGTGCTCTCCAACCCCGACCAGAAGGGCAAGATGCGGCGC ATCGACTGTCTCCGGCAGGCGGACAAGGTGTGGCGGCTGGACCTGGTCATGGTCATCCTGTTCAAGGGCATCCCGCTGGAGAGCACCGAC GGCGAGCGCCTGGTCAAGGCTGCGCAGTGCGGTCACCCGGTCCTGTGCGTGCAGCCGCACCACATTGGCGTGGCCGTCAAGGAGCTGGAC CTCTACCTGGCCTACTTCGTGCGTGAGCGAGATGCAGAGCAAAGCGGCAGTCCCCGGACAGGGATGGGCTCTGACCAGGAGGACAGCAAG CCCATCACGCTGGACACGACCGACTTCCAGGAGAGCTTTGTCACCTCCGGCGTGTTCAGCGTCACTGAGCTCATCCAAGTGTCCCGGACA CCCGTGGTGACTGGAACAGGACCCAACTTCTCCCTGGGGGAGCTGCAGGGGCACCTGGCATACGACCTGAACCCAGCCAGCACTGGCCTC AGAAGAACGCTGCCCAGCACCTCCTCCAGTGGGAGCAAGCGGCACAAATCGGGCTCGATGGAGGAAGACGTGGACACGAGCCCTGGCGGC GATTACTACACTTCGCCCAGCTCGCCCACGAGTAGCAGCCGCAACTGGACGGAGGACATGGAAGGAGCCAGTGGAATCCTGAAGGGCTTC GACCCACTCCTCAACCTTGTGCTGGACGGCACCATTGAGTACATGCGAGACCCTGACGACCAGTACAAGCTCACGGAGGACACCCGGCAG CTGGGCCTCGTGGTGTGCCGGGGCACGTCCGTGGTGCTAATCTGCCCGCAGGACGGCATGGAGGCCATCCCCAACCCCTTCATCCAGCAG CAGGACGCCTAGCCTGGCCGGGGGCGCGGGGGGTGCAGGGCAGGCCCGAGCAGCTCGGTTTCCCGCGGACTTGGCTGCTGCTCCCACCGC >58913_58913_3_NFIC-LSM7_NFIC_chr19_3435205_ENST00000395111_LSM7_chr19_2324195_ENST00000252622_length(amino acids)=381AA_BP=310 MDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHEKRMSKDEERAVKDELLGEKPEVKQKWASRLLAKLRKDIRPECREDFVLSITGK KAPGCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKGIPLESTDGERLVKAAQCGHPVLCVQPHHIGVAVKELDLYLAYFVRERDA EQSGSPRTGMGSDQEDSKPITLDTTDFQESFVTSGVFSVTELIQVSRTPVVTGTGPNFSLGELQGHLAYDLNPASTGLRRTLPSTSSSGS KRHKSGSMEEDVDTSPGGDYYTSPSSPTSSSRNWTEDMEGASGILKGFDPLLNLVLDGTIEYMRDPDDQYKLTEDTRQLGLVVCRGTSVV -------------------------------------------------------------- >58913_58913_4_NFIC-LSM7_NFIC_chr19_3435205_ENST00000443272_LSM7_chr19_2324195_ENST00000252622_length(transcript)=1387nt_BP=1009nt AAGTTCAGCGCGCCCGCTCCGGCCGGCCCTGCGCCTCCCGCCGCGCCCGGGATGTATTCGTCCCCGCTCTGCCTCACCCAGGATGAGTTC CACCCGTTCATCGAGGCCCTGCTGCCTCACGTCCGCGCCTTCGCCTACACCTGGTTCAACCTGCAGGCGCGGAAGCGCAAGTACTTCAAG AAGCACGAGAAGCGGATGTCGAAGGACGAGGAGCGTGCGGTCAAGGACGAGCTGCTGGGCGAGAAGCCCGAGGTCAAGCAGAAGTGGGCG TCGCGGCTGCTGGCCAAGCTGCGCAAGGACATCCGGCCCGAGTGCCGCGAGGACTTCGTGCTGAGCATCACCGGCAAGAAGGCGCCGGGC TGCGTGCTCTCCAACCCCGACCAGAAGGGCAAGATGCGGCGCATCGACTGTCTCCGGCAGGCGGACAAGGTGTGGCGGCTGGACCTGGTC ATGGTCATCCTGTTCAAGGGCATCCCGCTGGAGAGCACCGACGGCGAGCGCCTGGTCAAGGCTGCGCAGTGCGGTCACCCGGTCCTGTGC GTGCAGCCGCACCACATTGGCGTGGCCGTCAAGGAGCTGGACCTCTACCTGGCCTACTTCGTGCGTGAGCGAGATGCAGAGCAAAGCGGC AGTCCCCGGACAGGGATGGGCTCTGACCAGGAGGACAGCAAGCCCATCACGCTGGACACGACCGACTTCCAGGAGAGCTTTGTCACCTCC GGCGTGTTCAGCGTCACTGAGCTCATCCAAGTGTCCCGGACACCCGTGGTGACTGGAACAGGACCCAACTTCTCCCTGGGGGAGCTGCAG GGGCACCTGGCATACGACCTGAACCCAGCCAGCACTGGCCTCAGAAGAACGCTGCCCAGCACCTCCTCCAGTGGGAGCAAGCGGCACAAA TCGGGCTCGATGGAGGAAGACGTGGACACGAGCCCTGGCGGCGATTACTACACTTCGCCCAGCTCGCCCACGAGTAGCAGCCGCAACTGG ACGGAGGACATGGAAGGAGCCAGTGGAATCCTGAAGGGCTTCGACCCACTCCTCAACCTTGTGCTGGACGGCACCATTGAGTACATGCGA GACCCTGACGACCAGTACAAGCTCACGGAGGACACCCGGCAGCTGGGCCTCGTGGTGTGCCGGGGCACGTCCGTGGTGCTAATCTGCCCG CAGGACGGCATGGAGGCCATCCCCAACCCCTTCATCCAGCAGCAGGACGCCTAGCCTGGCCGGGGGCGCGGGGGGTGCAGGGCAGGCCCG AGCAGCTCGGTTTCCCGCGGACTTGGCTGCTGCTCCCACCGCAGTACCGCCTCCTGGAACGGAAGCATTTTCCTTTTTGTATAGGTTGAA >58913_58913_4_NFIC-LSM7_NFIC_chr19_3435205_ENST00000443272_LSM7_chr19_2324195_ENST00000252622_length(amino acids)=390AA_BP=319 MYSSPLCLTQDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHEKRMSKDEERAVKDELLGEKPEVKQKWASRLLAKLRKDIRPECRE DFVLSITGKKAPGCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKGIPLESTDGERLVKAAQCGHPVLCVQPHHIGVAVKELDLYL AYFVRERDAEQSGSPRTGMGSDQEDSKPITLDTTDFQESFVTSGVFSVTELIQVSRTPVVTGTGPNFSLGELQGHLAYDLNPASTGLRRT LPSTSSSGSKRHKSGSMEEDVDTSPGGDYYTSPSSPTSSSRNWTEDMEGASGILKGFDPLLNLVLDGTIEYMRDPDDQYKLTEDTRQLGL -------------------------------------------------------------- >58913_58913_5_NFIC-LSM7_NFIC_chr19_3435205_ENST00000586919_LSM7_chr19_2324195_ENST00000252622_length(transcript)=1237nt_BP=859nt ATGGATGAGTTCCACCCGTTCATCGAGGCCCTGCTGCCTCACGTCCGCGCCTTCGCCTACACCTGGTTCAACCTGCAGGCGCGGAAGCGC AAGTACTTCAAGAAGCACGAGAAGCGGATGTCGAAGGACGAGGAGCGTGCGGTCAAGGACGAGCTGCTGGGCGAGAAGCCCGAGGTCAAG CAGAAGTGGGCGTCGCGGCTGCTGGCCAAGCTGCGCAAGGACATCCGGCCCGAGTGCCGCGAGGACTTCGTGCTGAGCATCACCGGCAAG AAGGCGCCGGGCTGCGTGCTCTCCAACCCCGACCAGAAGGGCAAGATGCGGCGCATCGACTGTCTCCGGCAGGCGGACAAGGTGTGGCGG CTGGACCTGGTCATGGTCATCCTGTTCAAGGGCATCCCGCTGGAGAGCACCGACGGCGAGCGCCTGGTCAAGGCTGCGCAGTGCGGTCAC CCGGTCCTGTGCGTGCAGCCGCACCACATTGGCGTGGCCGTCAAGGAGCTGGACCTCTACCTGGCCTACTTCGTGCGTGAGCGAGACACG ACCGACTTCCAGGAGAGCTTTGTCACCTCCGGCGTGTTCAGCGTCACTGAGCTCATCCAAGTGTCCCGGACACCCGTGGTGACTGGAACA GGACCCAACTTCTCCCTGGGGGAGCTGCAGGGGCACCTGGCATACGACCTGAACCCAGCCAGCACTGGCCTCAGAAGAACGCTGCCCAGC ACCTCCTCCAGTGGGAGCAAGCGGCACAAATCGGGCTCGATGGAGGAAGACGTGGACACGAGCCCTGGCGGCGATTACTACACTTCGCCC AGCTCGCCCACGAGTAGCAGCCGCAACTGGACGGAGGACATGGAAGGAGCCAGTGGAATCCTGAAGGGCTTCGACCCACTCCTCAACCTT GTGCTGGACGGCACCATTGAGTACATGCGAGACCCTGACGACCAGTACAAGCTCACGGAGGACACCCGGCAGCTGGGCCTCGTGGTGTGC CGGGGCACGTCCGTGGTGCTAATCTGCCCGCAGGACGGCATGGAGGCCATCCCCAACCCCTTCATCCAGCAGCAGGACGCCTAGCCTGGC CGGGGGCGCGGGGGGTGCAGGGCAGGCCCGAGCAGCTCGGTTTCCCGCGGACTTGGCTGCTGCTCCCACCGCAGTACCGCCTCCTGGAAC >58913_58913_5_NFIC-LSM7_NFIC_chr19_3435205_ENST00000586919_LSM7_chr19_2324195_ENST00000252622_length(amino acids)=357AA_BP=286 MDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHEKRMSKDEERAVKDELLGEKPEVKQKWASRLLAKLRKDIRPECREDFVLSITGK KAPGCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKGIPLESTDGERLVKAAQCGHPVLCVQPHHIGVAVKELDLYLAYFVRERDT TDFQESFVTSGVFSVTELIQVSRTPVVTGTGPNFSLGELQGHLAYDLNPASTGLRRTLPSTSSSGSKRHKSGSMEEDVDTSPGGDYYTSP -------------------------------------------------------------- >58913_58913_6_NFIC-LSM7_NFIC_chr19_3435205_ENST00000589123_LSM7_chr19_2324195_ENST00000252622_length(transcript)=1429nt_BP=1051nt GCTCGCTCCCTCCCCCGCGCGCCCTCCCTCGCCGCCTCCTCCCGCCGCCTGCGGCCCCCCCCTCGCCGGGGACCGAGCGCGCTCGCTCCG GCGCCGGCCTCGCCTCCTCGCAGCAGCGCCATGGATGAGTTCCACCCGTTCATCGAGGCCCTGCTGCCTCACGTCCGCGCCTTCGCCTAC ACCTGGTTCAACCTGCAGGCGCGGAAGCGCAAGTACTTCAAGAAGCACGAGAAGCGGATGTCGAAGGACGAGGAGCGTGCGGTCAAGGAC GAGCTGCTGGGCGAGAAGCCCGAGGTCAAGCAGAAGTGGGCGTCGCGGCTGCTGGCCAAGCTGCGCAAGGACATCCGGCCCGAGTGCCGC GAGGACTTCGTGCTGAGCATCACCGGCAAGAAGGCGCCGGGCTGCGTGCTCTCCAACCCCGACCAGAAGGGCAAGATGCGGCGCATCGAC TGTCTCCGGCAGGCGGACAAGGTGTGGCGGCTGGACCTGGTCATGGTCATCCTGTTCAAGGGCATCCCGCTGGAGAGCACCGACGGCGAG CGCCTGGTCAAGGCTGCGCAGTGCGGTCACCCGGTCCTGTGCGTGCAGCCGCACCACATTGGCGTGGCCGTCAAGGAGCTGGACCTCTAC CTGGCCTACTTCGTGCGTGAGCGAGATGCAGAGCAAAGCGGCAGTCCCCGGACAGGGATGGGCTCTGACCAGGAGGACAGCAAGCCCATC ACGCTGGACACGACCGACTTCCAGGAGAGCTTTGTCACCTCCGGCGTGTTCAGCGTCACTGAGCTCATCCAAGTGTCCCGGACACCCGTG GTGACTGGAACAGGACCCAACTTCTCCCTGGGGGAGCTGCAGGGGCACCTGGCATACGACCTGAACCCAGCCAGCACTGGCCTCAGAAGA ACGCTGCCCAGCACCTCCTCCAGTGGGAGCAAGCGGCACAAATCGGGCTCGATGGAGGAAGACGTGGACACGAGCCCTGGCGGCGATTAC TACACTTCGCCCAGCTCGCCCACGAGTAGCAGCCGCAACTGGACGGAGGACATGGAAGGAGCCAGTGGAATCCTGAAGGGCTTCGACCCA CTCCTCAACCTTGTGCTGGACGGCACCATTGAGTACATGCGAGACCCTGACGACCAGTACAAGCTCACGGAGGACACCCGGCAGCTGGGC CTCGTGGTGTGCCGGGGCACGTCCGTGGTGCTAATCTGCCCGCAGGACGGCATGGAGGCCATCCCCAACCCCTTCATCCAGCAGCAGGAC GCCTAGCCTGGCCGGGGGCGCGGGGGGTGCAGGGCAGGCCCGAGCAGCTCGGTTTCCCGCGGACTTGGCTGCTGCTCCCACCGCAGTACC >58913_58913_6_NFIC-LSM7_NFIC_chr19_3435205_ENST00000589123_LSM7_chr19_2324195_ENST00000252622_length(amino acids)=405AA_BP=334 MRPPPRRGPSALAPAPASPPRSSAMDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHEKRMSKDEERAVKDELLGEKPEVKQKWASR LLAKLRKDIRPECREDFVLSITGKKAPGCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKGIPLESTDGERLVKAAQCGHPVLCVQ PHHIGVAVKELDLYLAYFVRERDAEQSGSPRTGMGSDQEDSKPITLDTTDFQESFVTSGVFSVTELIQVSRTPVVTGTGPNFSLGELQGH LAYDLNPASTGLRRTLPSTSSSGSKRHKSGSMEEDVDTSPGGDYYTSPSSPTSSSRNWTEDMEGASGILKGFDPLLNLVLDGTIEYMRDP -------------------------------------------------------------- >58913_58913_7_NFIC-LSM7_NFIC_chr19_3435205_ENST00000590282_LSM7_chr19_2324195_ENST00000252622_length(transcript)=1390nt_BP=1012nt AGTAAGTTCAGCGCGCCCGCTCCGGCCGGCCCTGCGCCTCCCGCCGCGCCCGGGATGTATTCGTCCCCGCTCTGCCTCACCCAGGATGAG TTCCACCCGTTCATCGAGGCCCTGCTGCCTCACGTCCGCGCCTTCGCCTACACCTGGTTCAACCTGCAGGCGCGGAAGCGCAAGTACTTC AAGAAGCACGAGAAGCGGATGTCGAAGGACGAGGAGCGTGCGGTCAAGGACGAGCTGCTGGGCGAGAAGCCCGAGGTCAAGCAGAAGTGG GCGTCGCGGCTGCTGGCCAAGCTGCGCAAGGACATCCGGCCCGAGTGCCGCGAGGACTTCGTGCTGAGCATCACCGGCAAGAAGGCGCCG GGCTGCGTGCTCTCCAACCCCGACCAGAAGGGCAAGATGCGGCGCATCGACTGTCTCCGGCAGGCGGACAAGGTGTGGCGGCTGGACCTG GTCATGGTCATCCTGTTCAAGGGCATCCCGCTGGAGAGCACCGACGGCGAGCGCCTGGTCAAGGCTGCGCAGTGCGGTCACCCGGTCCTG TGCGTGCAGCCGCACCACATTGGCGTGGCCGTCAAGGAGCTGGACCTCTACCTGGCCTACTTCGTGCGTGAGCGAGATGCAGAGCAAAGC GGCAGTCCCCGGACAGGGATGGGCTCTGACCAGGAGGACAGCAAGCCCATCACGCTGGACACGACCGACTTCCAGGAGAGCTTTGTCACC TCCGGCGTGTTCAGCGTCACTGAGCTCATCCAAGTGTCCCGGACACCCGTGGTGACTGGAACAGGACCCAACTTCTCCCTGGGGGAGCTG CAGGGGCACCTGGCATACGACCTGAACCCAGCCAGCACTGGCCTCAGAAGAACGCTGCCCAGCACCTCCTCCAGTGGGAGCAAGCGGCAC AAATCGGGCTCGATGGAGGAAGACGTGGACACGAGCCCTGGCGGCGATTACTACACTTCGCCCAGCTCGCCCACGAGTAGCAGCCGCAAC TGGACGGAGGACATGGAAGGAGCCAGTGGAATCCTGAAGGGCTTCGACCCACTCCTCAACCTTGTGCTGGACGGCACCATTGAGTACATG CGAGACCCTGACGACCAGTACAAGCTCACGGAGGACACCCGGCAGCTGGGCCTCGTGGTGTGCCGGGGCACGTCCGTGGTGCTAATCTGC CCGCAGGACGGCATGGAGGCCATCCCCAACCCCTTCATCCAGCAGCAGGACGCCTAGCCTGGCCGGGGGCGCGGGGGGTGCAGGGCAGGC CCGAGCAGCTCGGTTTCCCGCGGACTTGGCTGCTGCTCCCACCGCAGTACCGCCTCCTGGAACGGAAGCATTTTCCTTTTTGTATAGGTT >58913_58913_7_NFIC-LSM7_NFIC_chr19_3435205_ENST00000590282_LSM7_chr19_2324195_ENST00000252622_length(amino acids)=390AA_BP=319 MYSSPLCLTQDEFHPFIEALLPHVRAFAYTWFNLQARKRKYFKKHEKRMSKDEERAVKDELLGEKPEVKQKWASRLLAKLRKDIRPECRE DFVLSITGKKAPGCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKGIPLESTDGERLVKAAQCGHPVLCVQPHHIGVAVKELDLYL AYFVRERDAEQSGSPRTGMGSDQEDSKPITLDTTDFQESFVTSGVFSVTELIQVSRTPVVTGTGPNFSLGELQGHLAYDLNPASTGLRRT LPSTSSSGSKRHKSGSMEEDVDTSPGGDYYTSPSSPTSSSRNWTEDMEGASGILKGFDPLLNLVLDGTIEYMRDPDDQYKLTEDTRQLGL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for NFIC-LSM7 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for NFIC-LSM7 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for NFIC-LSM7 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
