
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:NIPBL-FGF10 (FusionGDB2 ID:59197) |
Fusion Gene Summary for NIPBL-FGF10 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: NIPBL-FGF10 | Fusion gene ID: 59197 | Hgene | Tgene | Gene symbol | NIPBL | FGF10 | Gene ID | 25836 | 2255 |
| Gene name | NIPBL cohesin loading factor | fibroblast growth factor 10 | |
| Synonyms | CDLS|CDLS1|IDN3|IDN3-B|Scc2 | - | |
| Cytomap | 5p13.2 | 5p12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | nipped-B-like proteinNipped-B homologSCC2 homologdelanginsister chromatid cohesion 2 homolog | fibroblast growth factor 10FGF-10keratinocyte growth factor 2produced by fibroblasts of urinary bladder lamina propria | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q6KC79 | O15520 | |
| Ensembl transtripts involved in fusion gene | ENST00000504430, ENST00000282516, ENST00000448238, | ENST00000264664, | |
| Fusion gene scores | * DoF score | 26 X 13 X 16=5408 | 8 X 3 X 6=144 |
| # samples | 43 | 8 | |
| ** MAII score | log2(43/5408*10)=-3.65268658669272 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(8/144*10)=-0.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: NIPBL [Title/Abstract] AND FGF10 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | NIPBL(36962376)-FGF10(44310632), # samples:4 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | NIPBL | GO:0000122 | negative regulation of transcription by RNA polymerase II | 18854353 |
| Hgene | NIPBL | GO:0031065 | positive regulation of histone deacetylation | 18854353 |
| Hgene | NIPBL | GO:0045892 | negative regulation of transcription, DNA-templated | 18854353 |
| Hgene | NIPBL | GO:0071921 | cohesin loading | 22628566 |
| Tgene | FGF10 | GO:0000187 | activation of MAPK activity | 14975937 |
| Tgene | FGF10 | GO:0010634 | positive regulation of epithelial cell migration | 17500053 |
| Tgene | FGF10 | GO:0010838 | positive regulation of keratinocyte proliferation | 17449030 |
| Tgene | FGF10 | GO:0031532 | actin cytoskeleton reorganization | 17449030 |
| Tgene | FGF10 | GO:0032781 | positive regulation of ATPase activity | 12804770 |
| Tgene | FGF10 | GO:0034394 | protein localization to cell surface | 17449030 |
| Tgene | FGF10 | GO:0042060 | wound healing | 11896977|19152659 |
| Tgene | FGF10 | GO:0043410 | positive regulation of MAPK cascade | 12804770 |
| Tgene | FGF10 | GO:0045739 | positive regulation of DNA repair | 14975937 |
| Tgene | FGF10 | GO:0046579 | positive regulation of Ras protein signal transduction | 12804770 |
| Tgene | FGF10 | GO:0048538 | thymus development | 17969154 |
| Tgene | FGF10 | GO:0048754 | branching morphogenesis of an epithelial tube | 17959718 |
| Tgene | FGF10 | GO:0050671 | positive regulation of lymphocyte proliferation | 19152659 |
| Tgene | FGF10 | GO:0050673 | epithelial cell proliferation | 19224135 |
| Tgene | FGF10 | GO:0050674 | urothelial cell proliferation | 11923311 |
| Tgene | FGF10 | GO:0050677 | positive regulation of urothelial cell proliferation | 11923311 |
| Tgene | FGF10 | GO:0050679 | positive regulation of epithelial cell proliferation | 15690149 |
| Tgene | FGF10 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 17449030 |
| Tgene | FGF10 | GO:0050918 | positive chemotaxis | 9740653|17449030 |
| Tgene | FGF10 | GO:0051549 | positive regulation of keratinocyte migration | 17449030|19152659 |
| Tgene | FGF10 | GO:0060428 | lung epithelium development | 9740653 |
| Tgene | FGF10 | GO:0060447 | bud outgrowth involved in lung branching | 9740653 |
| Tgene | FGF10 | GO:0060595 | fibroblast growth factor receptor signaling pathway involved in mammary gland specification | 11923311 |
| Tgene | FGF10 | GO:0061033 | secretion by lung epithelial cell involved in lung growth | 10541313 |
| Tgene | FGF10 | GO:0070371 | ERK1 and ERK2 cascade | 17959718 |
| Tgene | FGF10 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 14975937 |
| Tgene | FGF10 | GO:0071157 | negative regulation of cell cycle arrest | 17188682 |
| Tgene | FGF10 | GO:0071338 | positive regulation of hair follicle cell proliferation | 16086254 |
| Tgene | FGF10 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 17188682 |
 Fusion gene breakpoints across NIPBL (5'-gene) Fusion gene breakpoints across NIPBL (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
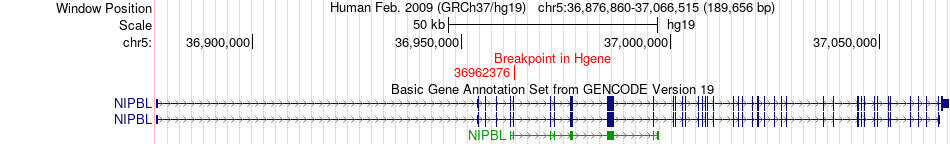 |
 Fusion gene breakpoints across FGF10 (3'-gene) Fusion gene breakpoints across FGF10 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
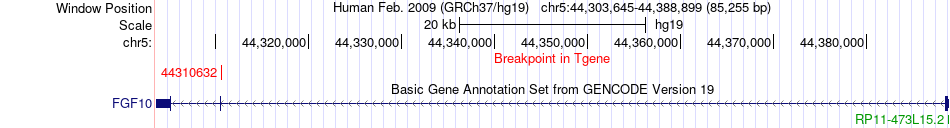 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUSC | TCGA-22-4595-01A | NIPBL | chr5 | 36962376 | - | FGF10 | chr5 | 44310632 | - |
| ChimerDB4 | LUSC | TCGA-22-4595-01A | NIPBL | chr5 | 36962376 | + | FGF10 | chr5 | 44310632 | - |
| ChimerDB4 | LUSC | TCGA-22-4595 | NIPBL | chr5 | 36962376 | + | FGF10 | chr5 | 44310632 | - |
Top |
Fusion Gene ORF analysis for NIPBL-FGF10 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000504430 | ENST00000264664 | NIPBL | chr5 | 36962376 | + | FGF10 | chr5 | 44310632 | - |
| In-frame | ENST00000282516 | ENST00000264664 | NIPBL | chr5 | 36962376 | + | FGF10 | chr5 | 44310632 | - |
| In-frame | ENST00000448238 | ENST00000264664 | NIPBL | chr5 | 36962376 | + | FGF10 | chr5 | 44310632 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000282516 | NIPBL | chr5 | 36962376 | + | ENST00000264664 | FGF10 | chr5 | 44310632 | - | 2862 | 1109 | 499 | 1410 | 303 |
| ENST00000448238 | NIPBL | chr5 | 36962376 | + | ENST00000264664 | FGF10 | chr5 | 44310632 | - | 2831 | 1078 | 468 | 1379 | 303 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000282516 | ENST00000264664 | NIPBL | chr5 | 36962376 | + | FGF10 | chr5 | 44310632 | - | 0.000523799 | 0.99947625 |
| ENST00000448238 | ENST00000264664 | NIPBL | chr5 | 36962376 | + | FGF10 | chr5 | 44310632 | - | 0.000533962 | 0.999466 |
Top |
Fusion Genomic Features for NIPBL-FGF10 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for NIPBL-FGF10 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:36962376/chr5:44310632) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NIPBL | FGF10 |
| FUNCTION: Plays an important role in the loading of the cohesin complex on to DNA. Forms a heterodimeric complex (also known as cohesin loading complex) with MAU2/SCC4 which mediates the loading of the cohesin complex onto chromatin (PubMed:22628566, PubMed:28914604). Plays a role in cohesin loading at sites of DNA damage. Its recruitment to double-strand breaks (DSBs) sites occurs in a CBX3-, RNF8- and RNF168-dependent manner whereas its recruitment to UV irradiation-induced DNA damage sites occurs in a ATM-, ATR-, RNF8- and RNF168-dependent manner (PubMed:28167679). Along with ZNF609, promotes cortical neuron migration during brain development by regulating the transcription of crucial genes in this process. Preferentially binds promoters containing paused RNA polymerase II. Up-regulates the expression of SEMA3A, NRP1, PLXND1 and GABBR2 genes, among others (By similarity). {ECO:0000250|UniProtKB:Q6KCD5, ECO:0000269|PubMed:22628566, ECO:0000269|PubMed:28167679, ECO:0000269|PubMed:28914604}. | FUNCTION: Plays an important role in the regulation of embryonic development, cell proliferation and cell differentiation. Required for normal branching morphogenesis. May play a role in wound healing. {ECO:0000269|PubMed:16597617}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NIPBL | chr5:36962376 | chr5:44310632 | ENST00000282516 | + | 6 | 47 | 418_462 | 203 | 2805.0 | Compositional bias | Note=Gln-rich |
| Hgene | NIPBL | chr5:36962376 | chr5:44310632 | ENST00000448238 | + | 6 | 46 | 418_462 | 203 | 2698.0 | Compositional bias | Note=Gln-rich |
| Hgene | NIPBL | chr5:36962376 | chr5:44310632 | ENST00000282516 | + | 6 | 47 | 996_1009 | 203 | 2805.0 | Motif | PxVxL motif |
| Hgene | NIPBL | chr5:36962376 | chr5:44310632 | ENST00000448238 | + | 6 | 46 | 996_1009 | 203 | 2698.0 | Motif | PxVxL motif |
| Hgene | NIPBL | chr5:36962376 | chr5:44310632 | ENST00000282516 | + | 6 | 47 | 1767_1805 | 203 | 2805.0 | Repeat | Note=HEAT 1 |
| Hgene | NIPBL | chr5:36962376 | chr5:44310632 | ENST00000282516 | + | 6 | 47 | 1843_1881 | 203 | 2805.0 | Repeat | Note=HEAT 2 |
| Hgene | NIPBL | chr5:36962376 | chr5:44310632 | ENST00000282516 | + | 6 | 47 | 1945_1984 | 203 | 2805.0 | Repeat | Note=HEAT 3 |
| Hgene | NIPBL | chr5:36962376 | chr5:44310632 | ENST00000282516 | + | 6 | 47 | 2227_2267 | 203 | 2805.0 | Repeat | Note=HEAT 4 |
| Hgene | NIPBL | chr5:36962376 | chr5:44310632 | ENST00000282516 | + | 6 | 47 | 2313_2351 | 203 | 2805.0 | Repeat | Note=HEAT 5 |
| Hgene | NIPBL | chr5:36962376 | chr5:44310632 | ENST00000448238 | + | 6 | 46 | 1767_1805 | 203 | 2698.0 | Repeat | Note=HEAT 1 |
| Hgene | NIPBL | chr5:36962376 | chr5:44310632 | ENST00000448238 | + | 6 | 46 | 1843_1881 | 203 | 2698.0 | Repeat | Note=HEAT 2 |
| Hgene | NIPBL | chr5:36962376 | chr5:44310632 | ENST00000448238 | + | 6 | 46 | 1945_1984 | 203 | 2698.0 | Repeat | Note=HEAT 3 |
| Hgene | NIPBL | chr5:36962376 | chr5:44310632 | ENST00000448238 | + | 6 | 46 | 2227_2267 | 203 | 2698.0 | Repeat | Note=HEAT 4 |
| Hgene | NIPBL | chr5:36962376 | chr5:44310632 | ENST00000448238 | + | 6 | 46 | 2313_2351 | 203 | 2698.0 | Repeat | Note=HEAT 5 |
| Tgene | FGF10 | chr5:36962376 | chr5:44310632 | ENST00000264664 | 0 | 3 | 52_62 | 108 | 209.0 | Compositional bias | Note=Poly-Ser |
Top |
Fusion Gene Sequence for NIPBL-FGF10 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >59197_59197_1_NIPBL-FGF10_NIPBL_chr5_36962376_ENST00000282516_FGF10_chr5_44310632_ENST00000264664_length(transcript)=2862nt_BP=1109nt TCCGGTCGGCATTTTGTTCTGAGAGGGAGAGACGGAACGAGAGAGAGACACACACAGGGCTCCTTCCCCCCGCCCTCCCCCCCCTCCCTC CGTCGGTACCGACTCACCCGACACCACCAAGCCGCAGGGAGGGACGCCCCCGCCGACAGGAGAATTGGTTCCCGGGCCCGCGGCGATGCC CCCCCGGTAGCTCGGGCCCGTGGTCGGGTGTTTGTGAGTGTTTCTATGTGGGAGAAGGAGGAGGAGGAGGAAGAAGAAGCAACGATTTGT CTTCTCGGCTGGTCTCCCCCCGGCTCTACATGTTCCCCGCACTGAGGAGACGGAAGAGGAGCCGTAGCCACCCCCCCTCCCGGCCCGGAT TATAGTCTCTCGCCACAGCGGCCTCGGCCTCCCCTTGGATTCAGACGCCGATTCGCCCAGTGTTTGGGAAATGGGAAGTAATGACAGCTG GCACCTGAACTAAGTACTTTTATAGGCAACACCATTCCAGAAATTCAGGATGAATGGGGATATGCCCCATGTCCCCATTACTACTCTTGC GGGGATTGCTAGTCTCACAGACCTCCTGAACCAGCTGCCTCTTCCATCTCCTTTACCTGCTACAACTACAAAGAGCCTTCTCTTTAATGC ACGAATAGCAGAAGAGGTGAACTGCCTTTTGGCTTGTAGGGATGACAATTTGGTTTCACAGCTTGTCCATAGCCTCAACCAGGTATCAAC AGATCACATAGAGTTGAAAGATAACCTTGGCAGTGATGACCCAGAAGGTGACATACCAGTCTTGTTGCAGGCCGTCCTGGCAAGGAGTCC TAATGTTTTCAGGGAGAAAAGCATGCAGAACAGATATGTACAAAGTGGAATGATGATGTCTCAGTATAAACTTTCTCAGAATTCCATGCA CAGTAGTCCTGCATCTTCCAATTATCAACAAACCACTATCTCACATAGCCCCTCCAGCCGGTTTGTGCCACCACAGACAAGCTCTGGGAA CAGATTTATGCCACAGCAAAATAGCCCAGTGCCTAGTCCATACGCCCCACAAAGCCCTGCAGGATACATGCCATATTCCCATCCTTCAAG TTACACAACACATCCACAGATGCAACAAGGCATCCTGGAGATAACATCAGTAGAAATCGGAGTTGTTGCCGTCAAAGCCATTAACAGCAA CTATTACTTAGCCATGAACAAGAAGGGGAAACTCTATGGCTCAAAAGAATTTAACAATGACTGTAAGCTGAAGGAGAGGATAGAGGAAAA TGGATACAATACCTATGCATCATTTAACTGGCAGCATAATGGGAGGCAAATGTATGTGGCATTGAATGGAAAAGGAGCTCCAAGGAGAGG ACAGAAAACACGAAGGAAAAACACCTCTGCTCACTTTCTTCCAATGGTGGTACACTCATAGAGGAAGGCAACGTTTGTGGATGCAGTAGA ACCAATGGCTCTTTTGCCAAGAATAGTGGATATTCTTCATGAAGACAGTAGATTGAAAGGCAAAGACACGTTGCAGATGTCTGCTTGCTT AAAAGAAAGCCAGCCTTTGAAGGTTTTTGTATTCACTGCTGACATATGATGTTCTTTTAATTAGTTCTGTGTCATGTCTTATAATCAAGA TATAGGCAGATCGAATGGGATAGAAGTTATTCCCAAGTGAAAAACATTGTGGCTGGGTTTTTTGTTGTTGTTGTCAAGTTTTTGTTTTTA AACCTCTGAGATAGAACTTAAAGGACATAGAACAATCTGTTGAAAGAACGATCTTCGGGAAAGTTATTTATGGAATACGAACTCATATCA AAGACTTCATTGCTCATTCAAGCCTAATGAATCAATGAACAGTAATACATGCAAGCATTTACTGGAAAGCACTTGGGTCATATCATATGC ACAACCAAAGGAGTTCTGGATGTGGTCTCATGGAATAATTGAATAGAATTTAAAAATATAAACATGTTAGTGTGAAACTGTTCTAACAAT ACAAATAGTATGGTATGCTTGTGCATTCTGCCTTCATCCCTTTCTATTTCTTTCTAAGTTATTTATTTAATAGGATGTTAAATATCTTTT GGGGTTTTAAAGAGTATCTCAGCAGCTGTCTTCTGATTTATCTTTTCTTTTTATTCAGCACACCACATGCATGTTCACGACAAAGTGTTT TTAAAACTTGGCGAACACTTCAAAAATAGGAGTTGGGATTAGGGAAGCAGTATGAGTGCCCGTGTGCTATCAGTTGACTTAATTTGCACT TCTGCAGTAATAACCATCAACAATAAATATGGCAATGCTGTGCCATGGCTTGAGTGAGAGATGTCTGCTATCATTTGAAAACATATATTA CTCTCGAGGCTTCCTGTCTCAAGAAATAGACCAGAAGGCCAAATTCTTCTCTTTCAATACATCAGTTTGCCTCCAAGAATATACTAAAAA AAGGAAAATTAATTGCTAAATACATTTAAATAGCCTAGCCTCATTATTTACTCATGATTTCTTGCCAAATGTCATGGCGGTAAAGAGGCT GTCCACATCTCTAAAAACCCTCTGTAAATTCCACATAATGCATCTTTCCCAAAGGAACTATCAAAGAATTTGGTATGAAGCGCAACTCTC CCAGGGGCTTAAACTGAGCAAATCAAATATATACTGGTATATGTGTAACCATATACAAAAACCTGTTCTAGCTGTATGATCTAGTCTTTA CAAAACCAAATAAAACTTGTTTTCTGTAAATTTAAAGAGCTTTACAAAGTTCCATAATGTAACCATATCAAAATTCATTTTGTTAGAGCA >59197_59197_1_NIPBL-FGF10_NIPBL_chr5_36962376_ENST00000282516_FGF10_chr5_44310632_ENST00000264664_length(amino acids)=303AA_BP=203 MNGDMPHVPITTLAGIASLTDLLNQLPLPSPLPATTTKSLLFNARIAEEVNCLLACRDDNLVSQLVHSLNQVSTDHIELKDNLGSDDPEG DIPVLLQAVLARSPNVFREKSMQNRYVQSGMMMSQYKLSQNSMHSSPASSNYQQTTISHSPSSRFVPPQTSSGNRFMPQQNSPVPSPYAP QSPAGYMPYSHPSSYTTHPQMQQGILEITSVEIGVVAVKAINSNYYLAMNKKGKLYGSKEFNNDCKLKERIEENGYNTYASFNWQHNGRQ -------------------------------------------------------------- >59197_59197_2_NIPBL-FGF10_NIPBL_chr5_36962376_ENST00000448238_FGF10_chr5_44310632_ENST00000264664_length(transcript)=2831nt_BP=1078nt ACGGAACGAGAGAGAGACACACACAGGGCTCCTTCCCCCCGCCCTCCCCCCCCTCCCTCCGTCGGTACCGACTCACCCGACACCACCAAG CCGCAGGGAGGGACGCCCCCGCCGACAGGAGAATTGGTTCCCGGGCCCGCGGCGATGCCCCCCCGGTAGCTCGGGCCCGTGGTCGGGTGT TTGTGAGTGTTTCTATGTGGGAGAAGGAGGAGGAGGAGGAAGAAGAAGCAACGATTTGTCTTCTCGGCTGGTCTCCCCCCGGCTCTACAT GTTCCCCGCACTGAGGAGACGGAAGAGGAGCCGTAGCCACCCCCCCTCCCGGCCCGGATTATAGTCTCTCGCCACAGCGGCCTCGGCCTC CCCTTGGATTCAGACGCCGATTCGCCCAGTGTTTGGGAAATGGGAAGTAATGACAGCTGGCACCTGAACTAAGTACTTTTATAGGCAACA CCATTCCAGAAATTCAGGATGAATGGGGATATGCCCCATGTCCCCATTACTACTCTTGCGGGGATTGCTAGTCTCACAGACCTCCTGAAC CAGCTGCCTCTTCCATCTCCTTTACCTGCTACAACTACAAAGAGCCTTCTCTTTAATGCACGAATAGCAGAAGAGGTGAACTGCCTTTTG GCTTGTAGGGATGACAATTTGGTTTCACAGCTTGTCCATAGCCTCAACCAGGTATCAACAGATCACATAGAGTTGAAAGATAACCTTGGC AGTGATGACCCAGAAGGTGACATACCAGTCTTGTTGCAGGCCGTCCTGGCAAGGAGTCCTAATGTTTTCAGGGAGAAAAGCATGCAGAAC AGATATGTACAAAGTGGAATGATGATGTCTCAGTATAAACTTTCTCAGAATTCCATGCACAGTAGTCCTGCATCTTCCAATTATCAACAA ACCACTATCTCACATAGCCCCTCCAGCCGGTTTGTGCCACCACAGACAAGCTCTGGGAACAGATTTATGCCACAGCAAAATAGCCCAGTG CCTAGTCCATACGCCCCACAAAGCCCTGCAGGATACATGCCATATTCCCATCCTTCAAGTTACACAACACATCCACAGATGCAACAAGGC ATCCTGGAGATAACATCAGTAGAAATCGGAGTTGTTGCCGTCAAAGCCATTAACAGCAACTATTACTTAGCCATGAACAAGAAGGGGAAA CTCTATGGCTCAAAAGAATTTAACAATGACTGTAAGCTGAAGGAGAGGATAGAGGAAAATGGATACAATACCTATGCATCATTTAACTGG CAGCATAATGGGAGGCAAATGTATGTGGCATTGAATGGAAAAGGAGCTCCAAGGAGAGGACAGAAAACACGAAGGAAAAACACCTCTGCT CACTTTCTTCCAATGGTGGTACACTCATAGAGGAAGGCAACGTTTGTGGATGCAGTAGAACCAATGGCTCTTTTGCCAAGAATAGTGGAT ATTCTTCATGAAGACAGTAGATTGAAAGGCAAAGACACGTTGCAGATGTCTGCTTGCTTAAAAGAAAGCCAGCCTTTGAAGGTTTTTGTA TTCACTGCTGACATATGATGTTCTTTTAATTAGTTCTGTGTCATGTCTTATAATCAAGATATAGGCAGATCGAATGGGATAGAAGTTATT CCCAAGTGAAAAACATTGTGGCTGGGTTTTTTGTTGTTGTTGTCAAGTTTTTGTTTTTAAACCTCTGAGATAGAACTTAAAGGACATAGA ACAATCTGTTGAAAGAACGATCTTCGGGAAAGTTATTTATGGAATACGAACTCATATCAAAGACTTCATTGCTCATTCAAGCCTAATGAA TCAATGAACAGTAATACATGCAAGCATTTACTGGAAAGCACTTGGGTCATATCATATGCACAACCAAAGGAGTTCTGGATGTGGTCTCAT GGAATAATTGAATAGAATTTAAAAATATAAACATGTTAGTGTGAAACTGTTCTAACAATACAAATAGTATGGTATGCTTGTGCATTCTGC CTTCATCCCTTTCTATTTCTTTCTAAGTTATTTATTTAATAGGATGTTAAATATCTTTTGGGGTTTTAAAGAGTATCTCAGCAGCTGTCT TCTGATTTATCTTTTCTTTTTATTCAGCACACCACATGCATGTTCACGACAAAGTGTTTTTAAAACTTGGCGAACACTTCAAAAATAGGA GTTGGGATTAGGGAAGCAGTATGAGTGCCCGTGTGCTATCAGTTGACTTAATTTGCACTTCTGCAGTAATAACCATCAACAATAAATATG GCAATGCTGTGCCATGGCTTGAGTGAGAGATGTCTGCTATCATTTGAAAACATATATTACTCTCGAGGCTTCCTGTCTCAAGAAATAGAC CAGAAGGCCAAATTCTTCTCTTTCAATACATCAGTTTGCCTCCAAGAATATACTAAAAAAAGGAAAATTAATTGCTAAATACATTTAAAT AGCCTAGCCTCATTATTTACTCATGATTTCTTGCCAAATGTCATGGCGGTAAAGAGGCTGTCCACATCTCTAAAAACCCTCTGTAAATTC CACATAATGCATCTTTCCCAAAGGAACTATCAAAGAATTTGGTATGAAGCGCAACTCTCCCAGGGGCTTAAACTGAGCAAATCAAATATA TACTGGTATATGTGTAACCATATACAAAAACCTGTTCTAGCTGTATGATCTAGTCTTTACAAAACCAAATAAAACTTGTTTTCTGTAAAT TTAAAGAGCTTTACAAAGTTCCATAATGTAACCATATCAAAATTCATTTTGTTAGAGCAGGTATAGAAAAGAGTACATAAGAGTTTACCA >59197_59197_2_NIPBL-FGF10_NIPBL_chr5_36962376_ENST00000448238_FGF10_chr5_44310632_ENST00000264664_length(amino acids)=303AA_BP=203 MNGDMPHVPITTLAGIASLTDLLNQLPLPSPLPATTTKSLLFNARIAEEVNCLLACRDDNLVSQLVHSLNQVSTDHIELKDNLGSDDPEG DIPVLLQAVLARSPNVFREKSMQNRYVQSGMMMSQYKLSQNSMHSSPASSNYQQTTISHSPSSRFVPPQTSSGNRFMPQQNSPVPSPYAP QSPAGYMPYSHPSSYTTHPQMQQGILEITSVEIGVVAVKAINSNYYLAMNKKGKLYGSKEFNNDCKLKERIEENGYNTYASFNWQHNGRQ -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for NIPBL-FGF10 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for NIPBL-FGF10 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for NIPBL-FGF10 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
