
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:NLK-KSR1 (FusionGDB2 ID:59349) |
Fusion Gene Summary for NLK-KSR1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: NLK-KSR1 | Fusion gene ID: 59349 | Hgene | Tgene | Gene symbol | NLK | KSR1 | Gene ID | 51701 | 8844 |
| Gene name | nemo like kinase | kinase suppressor of ras 1 | |
| Synonyms | - | KSR|RSU2 | |
| Cytomap | 17q11.2 | 17q11.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | serine/threonine-protein kinase NLK | kinase suppressor of Ras 1 | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | Q9UBE8 | Q8IVT5 | |
| Ensembl transtripts involved in fusion gene | ENST00000407008, ENST00000582037, ENST00000583517, | ENST00000268763, ENST00000398988, ENST00000578981, ENST00000581975, ENST00000582410, ENST00000319524, ENST00000509603, | |
| Fusion gene scores | * DoF score | 18 X 10 X 9=1620 | 13 X 9 X 7=819 |
| # samples | 20 | 12 | |
| ** MAII score | log2(20/1620*10)=-3.01792190799726 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(12/819*10)=-2.77082904603249 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: NLK [Title/Abstract] AND KSR1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | NLK(26512291)-KSR1(25904518), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | NLK | GO:0050821 | protein stabilization | 25512613 |
| Tgene | KSR1 | GO:0000185 | activation of MAPKKK activity | 29433126 |
 Fusion gene breakpoints across NLK (5'-gene) Fusion gene breakpoints across NLK (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
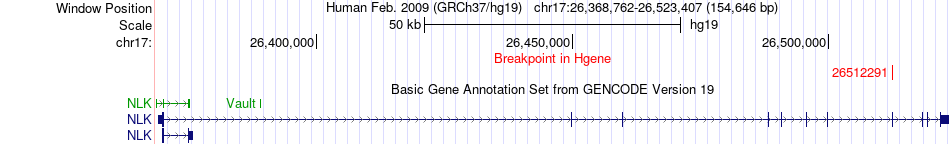 |
 Fusion gene breakpoints across KSR1 (3'-gene) Fusion gene breakpoints across KSR1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
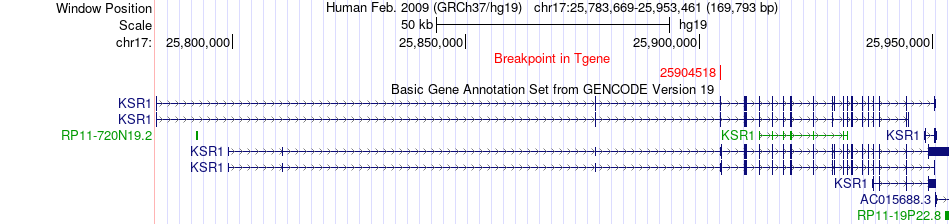 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-BR-4357-01A | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
Top |
Fusion Gene ORF analysis for NLK-KSR1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000407008 | ENST00000268763 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| 5CDS-5UTR | ENST00000407008 | ENST00000398988 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| 5CDS-intron | ENST00000407008 | ENST00000578981 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| 5CDS-intron | ENST00000407008 | ENST00000581975 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| 5CDS-intron | ENST00000407008 | ENST00000582410 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| In-frame | ENST00000407008 | ENST00000319524 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| In-frame | ENST00000407008 | ENST00000509603 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| intron-3CDS | ENST00000582037 | ENST00000319524 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| intron-3CDS | ENST00000582037 | ENST00000509603 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| intron-3CDS | ENST00000583517 | ENST00000319524 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| intron-3CDS | ENST00000583517 | ENST00000509603 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| intron-5UTR | ENST00000582037 | ENST00000268763 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| intron-5UTR | ENST00000582037 | ENST00000398988 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| intron-5UTR | ENST00000583517 | ENST00000268763 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| intron-5UTR | ENST00000583517 | ENST00000398988 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| intron-intron | ENST00000582037 | ENST00000578981 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| intron-intron | ENST00000582037 | ENST00000581975 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| intron-intron | ENST00000582037 | ENST00000582410 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| intron-intron | ENST00000583517 | ENST00000578981 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| intron-intron | ENST00000583517 | ENST00000581975 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
| intron-intron | ENST00000583517 | ENST00000582410 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000407008 | NLK | chr17 | 26512291 | - | ENST00000509603 | KSR1 | chr17 | 25904518 | + | 4213 | 1954 | 718 | 4212 | 1165 |
| ENST00000407008 | NLK | chr17 | 26512291 | - | ENST00000319524 | KSR1 | chr17 | 25904518 | + | 4653 | 1954 | 718 | 4353 | 1211 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000407008 | ENST00000509603 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + | 0.24458443 | 0.7554156 |
| ENST00000407008 | ENST00000319524 | NLK | chr17 | 26512291 | - | KSR1 | chr17 | 25904518 | + | 0.007872632 | 0.9921274 |
Top |
Fusion Genomic Features for NLK-KSR1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
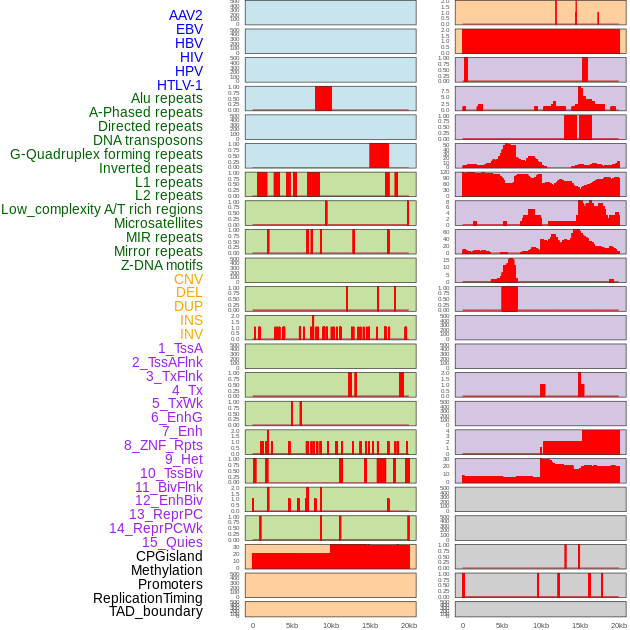 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for NLK-KSR1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:26512291/chr17:25904518) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NLK | KSR1 |
| FUNCTION: Serine/threonine-protein kinase that regulates a number of transcription factors with key roles in cell fate determination. Positive effector of the non-canonical Wnt signaling pathway, acting downstream of WNT5A, MAP3K7/TAK1 and HIPK2. Negative regulator of the canonical Wnt/beta-catenin signaling pathway. Binds to and phosphorylates TCF7L2/TCF4 and LEF1, promoting the dissociation of the TCF7L2/LEF1/beta-catenin complex from DNA, as well as the ubiquitination and subsequent proteolysis of LEF1. Together these effects inhibit the transcriptional activation of canonical Wnt/beta-catenin target genes. Negative regulator of the Notch signaling pathway. Binds to and phosphorylates NOTCH1, thereby preventing the formation of a transcriptionally active ternary complex of NOTCH1, RBPJ/RBPSUH and MAML1. Negative regulator of the MYB family of transcription factors. Phosphorylation of MYB leads to its subsequent proteolysis while phosphorylation of MYBL1 and MYBL2 inhibits their interaction with the coactivator CREBBP. Other transcription factors may also be inhibited by direct phosphorylation of CREBBP itself. Acts downstream of IL6 and MAP3K7/TAK1 to phosphorylate STAT3, which is in turn required for activation of NLK by MAP3K7/TAK1. Upon IL1B stimulus, cooperates with ATF5 to activate the transactivation activity of C/EBP subfamily members. Phosphorylates ATF5 but also stabilizes ATF5 protein levels in a kinase-independent manner (PubMed:25512613). {ECO:0000250|UniProtKB:O54949, ECO:0000269|PubMed:12482967, ECO:0000269|PubMed:14960582, ECO:0000269|PubMed:15004007, ECO:0000269|PubMed:15764709, ECO:0000269|PubMed:20061393, ECO:0000269|PubMed:20118921, ECO:0000269|PubMed:20874444, ECO:0000269|PubMed:21454679, ECO:0000269|PubMed:25512613}. | FUNCTION: Part of a multiprotein signaling complex which promotes phosphorylation of Raf family members and activation of downstream MAP kinases (By similarity). Independently of its kinase activity, acts as MAP2K1/MEK1 and MAP2K2/MEK2-dependent allosteric activator of BRAF; upon binding to MAP2K1/MEK1 or MAP2K2/MEK2, dimerizes with BRAF and promotes BRAF-mediated phosphorylation of MAP2K1/MEK1 and/or MAP2K2/MEK2 (PubMed:29433126). Promotes activation of MAPK1 and/or MAPK3, both in response to EGF and to cAMP (By similarity). Its kinase activity is unsure (By similarity). Some protein kinase activity has been detected in vitro, however the physiological relevance of this activity is unknown (By similarity). {ECO:0000250|UniProtKB:Q61097, ECO:0000269|PubMed:29433126}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NLK | chr17:26512291 | chr17:25904518 | ENST00000407008 | - | 8 | 11 | 106_111 | 412 | 528.0 | Compositional bias | Note=Poly-Ala |
| Hgene | NLK | chr17:26512291 | chr17:25904518 | ENST00000407008 | - | 8 | 11 | 115_119 | 412 | 528.0 | Compositional bias | Note=Poly-Ala |
| Hgene | NLK | chr17:26512291 | chr17:25904518 | ENST00000407008 | - | 8 | 11 | 22_25 | 412 | 528.0 | Compositional bias | Note=Poly-Ala |
| Hgene | NLK | chr17:26512291 | chr17:25904518 | ENST00000407008 | - | 8 | 11 | 27_34 | 412 | 528.0 | Compositional bias | Note=Poly-His |
| Hgene | NLK | chr17:26512291 | chr17:25904518 | ENST00000407008 | - | 8 | 11 | 42_48 | 412 | 528.0 | Compositional bias | Note=Poly-His |
| Hgene | NLK | chr17:26512291 | chr17:25904518 | ENST00000407008 | - | 8 | 11 | 71_83 | 412 | 528.0 | Compositional bias | Note=Poly-Ala |
| Hgene | NLK | chr17:26512291 | chr17:25904518 | ENST00000407008 | - | 8 | 11 | 298_300 | 412 | 528.0 | Motif | Note=TQE |
| Hgene | NLK | chr17:26512291 | chr17:25904518 | ENST00000407008 | - | 8 | 11 | 144_152 | 412 | 528.0 | Nucleotide binding | ATP |
| Tgene | KSR1 | chr17:26512291 | chr17:25904518 | ENST00000268763 | 2 | 22 | 17_21 | 0 | 787.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | KSR1 | chr17:26512291 | chr17:25904518 | ENST00000268763 | 2 | 22 | 289_292 | 0 | 787.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | KSR1 | chr17:26512291 | chr17:25904518 | ENST00000268763 | 2 | 22 | 613_883 | 0 | 787.0 | Domain | Protein kinase | |
| Tgene | KSR1 | chr17:26512291 | chr17:25904518 | ENST00000268763 | 2 | 22 | 619_627 | 0 | 787.0 | Nucleotide binding | ATP | |
| Tgene | KSR1 | chr17:26512291 | chr17:25904518 | ENST00000268763 | 2 | 22 | 1_172 | 0 | 787.0 | Region | Mediates association with membranes | |
| Tgene | KSR1 | chr17:26512291 | chr17:25904518 | ENST00000268763 | 2 | 22 | 347_391 | 0 | 787.0 | Zinc finger | Phorbol-ester/DAG-type |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NLK | chr17:26512291 | chr17:25904518 | ENST00000407008 | - | 8 | 11 | 138_427 | 412 | 528.0 | Domain | Protein kinase |
| Hgene | NLK | chr17:26512291 | chr17:25904518 | ENST00000407008 | - | 8 | 11 | 428_527 | 412 | 528.0 | Region | Required for homodimerization and kinase activation and localization to the nucleus |
Top |
Fusion Gene Sequence for NLK-KSR1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >59349_59349_1_NLK-KSR1_NLK_chr17_26512291_ENST00000407008_KSR1_chr17_25904518_ENST00000319524_length(transcript)=4653nt_BP=1954nt CCCCGGCTCTCTTTCTCCTCTCTGTCCTGCTGTCGCCTCCTCTCCTCACTCCAGCCTCCTTCCCTGGCTTATTCTGCCCAGGAAAGTTTG GAACCCCCGCCCCCCCTCCCCAATCAGGTCAACCTGACTAGGAGTAGGGGGAGATGTAGGAAGGCCTGAGAGTTCGGGGGGGGGGAAGCT GGAAGCTGGTTTCCCAGAAAATTTCCCCCCATCTTCTCCATTTAGGGTGGTGGGAATTTTTTTTAATCCTGTTTTTGAGGAGGTGGTAGG TGTTTAGAAAAGGATGTGGGAATGGAAGGGGTAGATGAGTCAACCCTCTTTCAAGATTCCAAGGGTGGTGTGTTACCTGATCCTTTTCGT GTGTGGAGTTCAGGGGGTCGGGAGGTTTGGCCTTTATTCCCACATTCAAAGTTGGAACCTCCCCCAAATTAAGAATGGAGTTTTAGTACC CACTTGTGGCAGGAATCCTGGGGGGAAGGGGTCCTTTTTTCCTTTTTCTTTTCTTTTTTCCCTTTTTTTTCCTTTTGGCAACCCACACCC TTCCACACACGCTCACCCCAAAATTAAACACCAAGATCCTCTAACTTGTTTGGATTGACTGATGAAGACATAAAGCTCTATGTTTTTTGA GGTGGAGTGAGTGGTTTTTCTTCATTTTTAAATGGCCAAATGACAGCTTGACCCAGTTTGCTTTCCAATCAAAGGGCATTTATTTTGAAT GTCTCTTTGTGGCGCAAGAGCCAACGCAAAAATGATGGCGGCTTACAATGGCGGTACATCTGCAGCAGCAGCAGGTCACCACCACCACCA TCACCACCACCTTCCACACCTCCCTCCTCCTCACCTGCACCACCACCACCACCCTCAACACCATCTTCATCCGGGGTCGGCTGCCGCTGT ACACCCTGTACAGCAGCACACCTCTTCGGCAGCTGCGGCAGCCGCAGCAGCGGCTGCAGCTGCAGCCATGTTAAACCCTGGGCAACAACA GCCATATTTCCCATCACCGGCACCGGGGCAGGCTCCTGGACCAGCTGCAGCAGCCCCAGCTCAGGTACAGGCTGCCGCAGCTGCTACAGT TAAGGCGCACCATCATCAGCACTCGCATCATCCACAGCAGCAGCTGGATATTGAGCCGGATAGACCTATTGGATATGGAGCCTTTGGTGT TGTCTGGTCAGTAACAGATCCAAGAGATGGAAAGAGAGTAGCGCTCAAAAAGATGCCCAACGTCTTCCAGAATCTGGTCTCTTGCAAAAG GGTCTTCCGGGAATTGAAGATGTTGTGTTTTTTTAAGCATGATAATGTACTCTCTGCCCTTGACATACTCCAACCTCCACACATTGACTA TTTTGAAGAAATATATGTTGTCACAGAATTGATGCAGAGTGACCTACATAAAATTATCGTCTCTCCTCAACCACTCAGCTCAGATCATGT CAAAGTTTTTCTTTATCAGATTTTGCGAGGTTTGAAATATCTCCATTCAGCTGGCATTTTACATCGAGACATTAAGCCAGGGAATCTCCT TGTGAACAGCAACTGTGTTCTAAAGATTTGTGATTTTGGATTGGCCAGAGTGGAAGAATTAGATGAATCCCGTCATATGACTCAGGAAGT TGTTACTCAGTATTATCGGGCTCCAGAAATCCTGATGGGCAGCCGTCATTACAGCAATGCTATTGACATCTGGTCTGTGGGATGTATCTT TGCAGAACTACTAGGACGAAGAATATTGTTTCAGGCACAGAGTCCCATTCAGCAGTTGGATTTGATCACGGATCTGTTGGGCACACCATC ACTGGAAGCAATGAGGACAGCTTGTGAAGGCGCTAAGGCACATATACTCAGGGGTCCTCATAAACAGCCATCTCTTCCTGTACTCTATAC CCTGTCTAGCCAGGCTACACATGAAGCTGTTCATCTCCTTTGCAGGATGTTGGTCTTTGATCCAGAGATCCCCCGAGACCTCACGCTGGA TGCCCTGCTGGAGATGAATGAGGCCAAGGTGAAGGAGACGCTGCGGCGCTGTGGGGCCAGCGGGGATGAGTGTGGCCGTCTGCAGTATGC CCTCACCTGCCTGCGGAAGGTGACAGGCCTGGGAGGGGAGCACAAGGAGGACTCCAGTTGGAGTTCATTGGATGCGCGGCGGGAAAGTGG CTCAGGGCCTTCCACGGACACCCTCTCAGCAGCCAGCCTGCCCTGGCCCCCAGGGAGCTCCCAGCTGGGCAGAGCAGGCAACAGCGCCCA GGGCCCACGCTCCATCTCCGTGTCAGCTCTGCCCGCCTCAGACTCCCCCACCCCCAGCTTCAGTGAGGGCCTCTCAGACACCTGTATTCC CCTGCACGCCAGCGGCCGGCTGACCCCCCGTGCCCTGCACAGCTTCATCACCCCGCCCACCACACCCCAGCTGCGACGGCACACCAAGCT GAAGCCACCACGGACGCCCCCCCCACCCAGCCGCAAGGTCTTCCAGCTGCTGCCCAGCTTCCCCACACTCACCCGGAGCAAGTCCCATGA GTCTCAGCTGGGGAACCGCATTGATGACGTCTCCTCGATGAGGTTTGATCTCTCGCATGGATCCCCACAGATGGTACGGAGGGATATCGG GCTGTCGGTGACGCACAGGTTCTCCACCAAGTCCTGGCTGTCGCAGGTCTGCCACGTGTGCCAGAAGAGCATGATATTTGGAGTGAAGTG CAAGCATTGCAGGTTGAAGTGTCACAACAAATGTACCAAAGAAGCCCCTGCCTGTAGAATATCCTTCCTGCCACTAACTCGGCTTCGGAG GACAGAATCTGTCCCCTCGGACATCAACAACCCGGTGGACAGAGCAGCCGAACCCCATTTTGGAACCCTCCCCAAAGCACTGACAAAGAA GGAGCACCCTCCGGCCATGAATCACCTGGACTCCAGCAGCAACCCTTCCTCCACCACCTCCTCCACACCCTCCTCACCGGCGCCCTTCCC GACATCATCCAACCCATCCAGCGCCACCACGCCCCCCAACCCCTCACCTGGCCAGCGGGACAGCAGGTTCAACTTCCCAGCTGCCTACTT CATTCATCATAGACAGCAGTTTATCTTTCCAGTGCCATCTGCTGGCCATTGCTGGAAATGCCTCCTTATTGCAGAAAGTTTAAAGGAAAA CGCTTTCAACATTTCAGCCTTTGCACACGCAGCCCCGCTCCCTGAAGCTGCCGACGGTACCCGGCTCGATGACCAGCCGAAAGCAGATGT GTTGGAAGCTCACGAAGCGGAGGCTGAGGAGCCAGAGGCTGGCAAGTCAGAGGCAGAAGACGATGAGGACGAGGTGGACGACTTGCCGAG CTCTCGCCGGCCCTGGCGGGGCCCCATCTCTCGCAAGGCCAGCCAGACCAGCGTGTACCTGCAGGAGTGGGACATCCCCTTCGAGCAGGT AGAGCTGGGCGAGCCCATCGGGCAGGGCCGCTGGGGCCGGGTGCACCGCGGCCGCTGGCATGGCGAGGTGGCCATTCGCCTGCTGGAGAT GGACGGCCACAACCAGGACCACCTGAAGCTCTTCAAGAAAGAGGTGATGAACTACCGGCAGACGCGGCATGAGAACGTGGTGCTCTTCAT GGGGGCCTGCATGAACCCGCCCCACCTGGCCATTATCACCAGCTTCTGCAAGGGGCGGACGTTGCACTCGTTTGTGAGGGACCCCAAGAC GTCTCTGGACATCAACAAGACGAGGCAAATCGCTCAGGAGATCATCAAGGGCATGGGATATCTTCATGCCAAGGGCATCGTACACAAAGA TCTCAAATCTAAGAACGTCTTCTATGACAACGGCAAGGTGGTCATCACAGACTTCGGGCTGTTTGGGATCTCAGGCGTGGTCCGAGAGGG ACGGCGTGAGAACCAGCTAAAGCTGTCCCACGACTGGCTGTGCTATCTGGCCCCTGAGATTGTACGCGAGATGACCCCCGGGAAGGACGA GGATCAGCTGCCATTCTCCAAAGCTGCTGATGTCTATGCATTTGGGACTGTTTGGTATGAGCTGCAAGCAAGAGACTGGCCCTTGAAGAA CCAGGCTGCAGAGGCATCCATCTGGCAGATTGGAAGCGGGGAAGGAATGAAGCGTGTCCTGACTTCTGTCAGCTTGGGGAAGGAAGTCAG TGAGATCCTGTCGGCCTGCTGGGCTTTCGACCTGCAGGAGAGACCCAGCTTCAGCCTGCTGATGGACATGCTGGAGAAACTTCCCAAGCT GAACCGGCGGCTCTCCCACCCTGGACACTTCTGGAAGTCAGCTGACATTAACAGCAGCAAAGTTGTACCCCGGTTTGAAAGGTTTGGCTT GGGCGTCCTGGAGTCCAGTAATCCAAAGATGTAGCCAGCCATATGGTTTTTCGCTGCTGATCTCTTTCTTTTTAAAATGTGTTTCTGAAA CATCCCAACAACCACCACGACAAAAAAACACTGCCTGCCCAGCGCTGCAAACCAGGAGCACACGTCCTAGATTCAGACTGTTGGCCATAA ACCCCACTCGGGAGATGGAGCTGCACCTGCTATTTCTTAAAATGACACCACCAACAACCAAACCTGTCATGACAGACAGCAAATGTTTAC >59349_59349_1_NLK-KSR1_NLK_chr17_26512291_ENST00000407008_KSR1_chr17_25904518_ENST00000319524_length(amino acids)=1211AA_BP=412 MSLCGARANAKMMAAYNGGTSAAAAGHHHHHHHHLPHLPPPHLHHHHHPQHHLHPGSAAAVHPVQQHTSSAAAAAAAAAAAAAMLNPGQQ QPYFPSPAPGQAPGPAAAAPAQVQAAAAATVKAHHHQHSHHPQQQLDIEPDRPIGYGAFGVVWSVTDPRDGKRVALKKMPNVFQNLVSCK RVFRELKMLCFFKHDNVLSALDILQPPHIDYFEEIYVVTELMQSDLHKIIVSPQPLSSDHVKVFLYQILRGLKYLHSAGILHRDIKPGNL LVNSNCVLKICDFGLARVEELDESRHMTQEVVTQYYRAPEILMGSRHYSNAIDIWSVGCIFAELLGRRILFQAQSPIQQLDLITDLLGTP SLEAMRTACEGAKAHILRGPHKQPSLPVLYTLSSQATHEAVHLLCRMLVFDPEIPRDLTLDALLEMNEAKVKETLRRCGASGDECGRLQY ALTCLRKVTGLGGEHKEDSSWSSLDARRESGSGPSTDTLSAASLPWPPGSSQLGRAGNSAQGPRSISVSALPASDSPTPSFSEGLSDTCI PLHASGRLTPRALHSFITPPTTPQLRRHTKLKPPRTPPPPSRKVFQLLPSFPTLTRSKSHESQLGNRIDDVSSMRFDLSHGSPQMVRRDI GLSVTHRFSTKSWLSQVCHVCQKSMIFGVKCKHCRLKCHNKCTKEAPACRISFLPLTRLRRTESVPSDINNPVDRAAEPHFGTLPKALTK KEHPPAMNHLDSSSNPSSTTSSTPSSPAPFPTSSNPSSATTPPNPSPGQRDSRFNFPAAYFIHHRQQFIFPVPSAGHCWKCLLIAESLKE NAFNISAFAHAAPLPEAADGTRLDDQPKADVLEAHEAEAEEPEAGKSEAEDDEDEVDDLPSSRRPWRGPISRKASQTSVYLQEWDIPFEQ VELGEPIGQGRWGRVHRGRWHGEVAIRLLEMDGHNQDHLKLFKKEVMNYRQTRHENVVLFMGACMNPPHLAIITSFCKGRTLHSFVRDPK TSLDINKTRQIAQEIIKGMGYLHAKGIVHKDLKSKNVFYDNGKVVITDFGLFGISGVVREGRRENQLKLSHDWLCYLAPEIVREMTPGKD EDQLPFSKAADVYAFGTVWYELQARDWPLKNQAAEASIWQIGSGEGMKRVLTSVSLGKEVSEILSACWAFDLQERPSFSLLMDMLEKLPK -------------------------------------------------------------- >59349_59349_2_NLK-KSR1_NLK_chr17_26512291_ENST00000407008_KSR1_chr17_25904518_ENST00000509603_length(transcript)=4213nt_BP=1954nt CCCCGGCTCTCTTTCTCCTCTCTGTCCTGCTGTCGCCTCCTCTCCTCACTCCAGCCTCCTTCCCTGGCTTATTCTGCCCAGGAAAGTTTG GAACCCCCGCCCCCCCTCCCCAATCAGGTCAACCTGACTAGGAGTAGGGGGAGATGTAGGAAGGCCTGAGAGTTCGGGGGGGGGGAAGCT GGAAGCTGGTTTCCCAGAAAATTTCCCCCCATCTTCTCCATTTAGGGTGGTGGGAATTTTTTTTAATCCTGTTTTTGAGGAGGTGGTAGG TGTTTAGAAAAGGATGTGGGAATGGAAGGGGTAGATGAGTCAACCCTCTTTCAAGATTCCAAGGGTGGTGTGTTACCTGATCCTTTTCGT GTGTGGAGTTCAGGGGGTCGGGAGGTTTGGCCTTTATTCCCACATTCAAAGTTGGAACCTCCCCCAAATTAAGAATGGAGTTTTAGTACC CACTTGTGGCAGGAATCCTGGGGGGAAGGGGTCCTTTTTTCCTTTTTCTTTTCTTTTTTCCCTTTTTTTTCCTTTTGGCAACCCACACCC TTCCACACACGCTCACCCCAAAATTAAACACCAAGATCCTCTAACTTGTTTGGATTGACTGATGAAGACATAAAGCTCTATGTTTTTTGA GGTGGAGTGAGTGGTTTTTCTTCATTTTTAAATGGCCAAATGACAGCTTGACCCAGTTTGCTTTCCAATCAAAGGGCATTTATTTTGAAT GTCTCTTTGTGGCGCAAGAGCCAACGCAAAAATGATGGCGGCTTACAATGGCGGTACATCTGCAGCAGCAGCAGGTCACCACCACCACCA TCACCACCACCTTCCACACCTCCCTCCTCCTCACCTGCACCACCACCACCACCCTCAACACCATCTTCATCCGGGGTCGGCTGCCGCTGT ACACCCTGTACAGCAGCACACCTCTTCGGCAGCTGCGGCAGCCGCAGCAGCGGCTGCAGCTGCAGCCATGTTAAACCCTGGGCAACAACA GCCATATTTCCCATCACCGGCACCGGGGCAGGCTCCTGGACCAGCTGCAGCAGCCCCAGCTCAGGTACAGGCTGCCGCAGCTGCTACAGT TAAGGCGCACCATCATCAGCACTCGCATCATCCACAGCAGCAGCTGGATATTGAGCCGGATAGACCTATTGGATATGGAGCCTTTGGTGT TGTCTGGTCAGTAACAGATCCAAGAGATGGAAAGAGAGTAGCGCTCAAAAAGATGCCCAACGTCTTCCAGAATCTGGTCTCTTGCAAAAG GGTCTTCCGGGAATTGAAGATGTTGTGTTTTTTTAAGCATGATAATGTACTCTCTGCCCTTGACATACTCCAACCTCCACACATTGACTA TTTTGAAGAAATATATGTTGTCACAGAATTGATGCAGAGTGACCTACATAAAATTATCGTCTCTCCTCAACCACTCAGCTCAGATCATGT CAAAGTTTTTCTTTATCAGATTTTGCGAGGTTTGAAATATCTCCATTCAGCTGGCATTTTACATCGAGACATTAAGCCAGGGAATCTCCT TGTGAACAGCAACTGTGTTCTAAAGATTTGTGATTTTGGATTGGCCAGAGTGGAAGAATTAGATGAATCCCGTCATATGACTCAGGAAGT TGTTACTCAGTATTATCGGGCTCCAGAAATCCTGATGGGCAGCCGTCATTACAGCAATGCTATTGACATCTGGTCTGTGGGATGTATCTT TGCAGAACTACTAGGACGAAGAATATTGTTTCAGGCACAGAGTCCCATTCAGCAGTTGGATTTGATCACGGATCTGTTGGGCACACCATC ACTGGAAGCAATGAGGACAGCTTGTGAAGGCGCTAAGGCACATATACTCAGGGGTCCTCATAAACAGCCATCTCTTCCTGTACTCTATAC CCTGTCTAGCCAGGCTACACATGAAGCTGTTCATCTCCTTTGCAGGATGTTGGTCTTTGATCCAGAGATCCCCCGAGACCTCACGCTGGA TGCCCTGCTGGAGATGAATGAGGCCAAGGTGAAGGAGACGCTGCGGCGCTGTGGGGCCAGCGGGGATGAGTGTGGCCGTCTGCAGTATGC CCTCACCTGCCTGCGGAAGGTGACAGGCCTGGGAGGGGAGCACAAGGAGGACTCCAGTTGGAGTTCATTGGATGCGCGGCGGGAAAGTGG CTCAGGGCCTTCCACGGACACCCTCTCAGCAGCCAGCCTGCCCTGGCCCCCAGGGAGCTCCCAGCTGGGCAGAGCAGGCAACAGCGCCCA GGGCCCACGCTCCATCTCCGTGTCAGCTCTGCCCGCCTCAGACTCCCCCACCCCCAGCTTCAGTGAGGGCCTCTCAGACACCTGTATTCC CCTGCACGCCAGCGGCCGGCTGACCCCCCGTGCCCTGCACAGCTTCATCACCCCGCCCACCACACCCCAGCTGCGACGGCACACCAAGCT GAAGCCACCACGGACGCCCCCCCCACCCAGCCGCAAGGTCTTCCAGCTGCTGCCCAGCTTCCCCACACTCACCCGGAGCAAGTCCCATGA GTCTCAGCTGGGGAACCGCATTGATGACGTCTCCTCGATGAGGTTTGATCTCTCGCATGGATCCCCACAGATGGTACGGAGGGATATCGG GCTGTCGGTGACGCACAGGTTCTCCACCAAGTCCTGGCTGTCGCAGGTCTGCCACGTGTGCCAGAAGAGCATGATATTTGGAGTGAAGTG CAAGCATTGCAGGTTGAAGTGTCACAACAAATGTACCAAAGAAGCCCCTGCCTGTAGAATATCCTTCCTGCCACTAACTCGGCTTCGGAG GACAGAATCTGTCCCCTCGGACATCAACAACCCGGTGGACAGAGCAGCCGAACCCCATTTTGGAACCCTCCCCAAAGCACTGACAAAGAA GGAGCACCCTCCGGCCATGAATCACCTGGACTCCAGCAGCAACCCTTCCTCCACCACCTCCTCCACACCCTCCTCACCGGCGCCCTTCCC GACATCATCCAACCCATCCAGCGCCACCACGCCCCCCAACCCCTCACCTGGCCAGCGGGACAGCAGGTTCAACTTCCCAGCTGCCTACTT CATTCATCATAGACAGCAGTTTATCTTTCCAGACATTTCAGCCTTTGCACACGCAGCCCCGCTCCCTGAAGCTGCCGACGGTACCCGGCT CGATGACCAGCCGAAAGCAGATGTGTTGGAAGCTCACGAAGCGGAGGCTGAGGAGCCAGAGGCTGGCAAGTCAGAGGCAGAAGACGATGA GGACGAGGTGGACGACTTGCCGAGCTCTCGCCGGCCCTGGCGGGGCCCCATCTCTCGCAAGGCCAGCCAGACCAGCGTGTACCTGCAGGA GTGGGACATCCCCTTCGAGCAGGTAGAGCTGGGCGAGCCCATCGGGCAGGGCCGCTGGGGCCGGGTGCACCGCGGCCGCTGGCATGGCGA GGTGGCCATTCGCCTGCTGGAGATGGACGGCCACAACCAGGACCACCTGAAGCTCTTCAAGAAAGAGGTGATGAACTACCGGCAGACGCG GCATGAGAACGTGGTGCTCTTCATGGGGGCCTGCATGAACCCGCCCCACCTGGCCATTATCACCAGCTTCTGCAAGGGGCGGACGTTGCA CTCGTTTGTGAGGGACCCCAAGACGTCTCTGGACATCAACAAGACGAGGCAAATCGCTCAGGAGATCATCAAGGGCATGGGATATCTTCA TGCCAAGGGCATCGTACACAAAGATCTCAAATCTAAGAACGTCTTCTATGACAACGGCAAGGTGGTCATCACAGACTTCGGGCTGTTTGG GATCTCAGGCGTGGTCCGAGAGGGACGGCGTGAGAACCAGCTAAAGCTGTCCCACGACTGGCTGTGCTATCTGGCCCCTGAGATTGTACG CGAGATGACCCCCGGGAAGGACGAGGATCAGCTGCCATTCTCCAAAGCTGCTGATGTCTATGCATTTGGGACTGTTTGGTATGAGCTGCA AGCAAGAGACTGGCCCTTGAAGAACCAGGCTGCAGAGGCATCCATCTGGCAGATTGGAAGCGGGGAAGGAATGAAGCGTGTCCTGACTTC TGTCAGCTTGGGGAAGGAAGTCAGTGAGATCCTGTCGGCCTGCTGGGCTTTCGACCTGCAGGAGAGACCCAGCTTCAGCCTGCTGATGGA >59349_59349_2_NLK-KSR1_NLK_chr17_26512291_ENST00000407008_KSR1_chr17_25904518_ENST00000509603_length(amino acids)=1165AA_BP=412 MSLCGARANAKMMAAYNGGTSAAAAGHHHHHHHHLPHLPPPHLHHHHHPQHHLHPGSAAAVHPVQQHTSSAAAAAAAAAAAAAMLNPGQQ QPYFPSPAPGQAPGPAAAAPAQVQAAAAATVKAHHHQHSHHPQQQLDIEPDRPIGYGAFGVVWSVTDPRDGKRVALKKMPNVFQNLVSCK RVFRELKMLCFFKHDNVLSALDILQPPHIDYFEEIYVVTELMQSDLHKIIVSPQPLSSDHVKVFLYQILRGLKYLHSAGILHRDIKPGNL LVNSNCVLKICDFGLARVEELDESRHMTQEVVTQYYRAPEILMGSRHYSNAIDIWSVGCIFAELLGRRILFQAQSPIQQLDLITDLLGTP SLEAMRTACEGAKAHILRGPHKQPSLPVLYTLSSQATHEAVHLLCRMLVFDPEIPRDLTLDALLEMNEAKVKETLRRCGASGDECGRLQY ALTCLRKVTGLGGEHKEDSSWSSLDARRESGSGPSTDTLSAASLPWPPGSSQLGRAGNSAQGPRSISVSALPASDSPTPSFSEGLSDTCI PLHASGRLTPRALHSFITPPTTPQLRRHTKLKPPRTPPPPSRKVFQLLPSFPTLTRSKSHESQLGNRIDDVSSMRFDLSHGSPQMVRRDI GLSVTHRFSTKSWLSQVCHVCQKSMIFGVKCKHCRLKCHNKCTKEAPACRISFLPLTRLRRTESVPSDINNPVDRAAEPHFGTLPKALTK KEHPPAMNHLDSSSNPSSTTSSTPSSPAPFPTSSNPSSATTPPNPSPGQRDSRFNFPAAYFIHHRQQFIFPDISAFAHAAPLPEAADGTR LDDQPKADVLEAHEAEAEEPEAGKSEAEDDEDEVDDLPSSRRPWRGPISRKASQTSVYLQEWDIPFEQVELGEPIGQGRWGRVHRGRWHG EVAIRLLEMDGHNQDHLKLFKKEVMNYRQTRHENVVLFMGACMNPPHLAIITSFCKGRTLHSFVRDPKTSLDINKTRQIAQEIIKGMGYL HAKGIVHKDLKSKNVFYDNGKVVITDFGLFGISGVVREGRRENQLKLSHDWLCYLAPEIVREMTPGKDEDQLPFSKAADVYAFGTVWYEL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for NLK-KSR1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for NLK-KSR1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for NLK-KSR1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
