
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:NOVA1-FUS (FusionGDB2 ID:59799) |
Fusion Gene Summary for NOVA1-FUS |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: NOVA1-FUS | Fusion gene ID: 59799 | Hgene | Tgene | Gene symbol | NOVA1 | FUS | Gene ID | 4857 | 2521 |
| Gene name | NOVA alternative splicing regulator 1 | FUS RNA binding protein | |
| Synonyms | Nova-1 | ALS6|ETM4|FUS1|HNRNPP2|POMP75|TLS | |
| Cytomap | 14q12 | 16p11.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | RNA-binding protein Nova-1neuro-oncological ventral antigen 1onconeural ventral antigen 1paraneoplastic Ri antigenventral neuron-specific protein 1 | RNA-binding protein FUS75 kDa DNA-pairing proteinfus-like proteinfused in sarcomafusion gene in myxoid liposarcomaheterogeneous nuclear ribonucleoprotein P2oncogene FUSoncogene TLStranslocated in liposarcoma protein | |
| Modification date | 20200327 | 20200329 | |
| UniProtAcc | . | P35637 | |
| Ensembl transtripts involved in fusion gene | ENST00000344429, ENST00000465357, ENST00000539517, ENST00000547619, ENST00000574031, ENST00000267422, ENST00000551754, | ENST00000474990, ENST00000254108, ENST00000380244, ENST00000568685, | |
| Fusion gene scores | * DoF score | 7 X 5 X 5=175 | 20 X 13 X 10=2600 |
| # samples | 8 | 22 | |
| ** MAII score | log2(8/175*10)=-1.12928301694497 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(22/2600*10)=-3.56293619439116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: NOVA1 [Title/Abstract] AND FUS [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | NOVA1(27064616)-FUS(31199646), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | FUS | GO:0006355 | regulation of transcription, DNA-templated | 26124092 |
| Tgene | FUS | GO:0006357 | regulation of transcription by RNA polymerase II | 25453086 |
| Tgene | FUS | GO:0008380 | RNA splicing | 26124092 |
| Tgene | FUS | GO:0043484 | regulation of RNA splicing | 25453086|27731383 |
| Tgene | FUS | GO:0048255 | mRNA stabilization | 27378374 |
| Tgene | FUS | GO:0051260 | protein homooligomerization | 25453086 |
| Tgene | FUS | GO:1905168 | positive regulation of double-strand break repair via homologous recombination | 10567410 |
 Fusion gene breakpoints across NOVA1 (5'-gene) Fusion gene breakpoints across NOVA1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
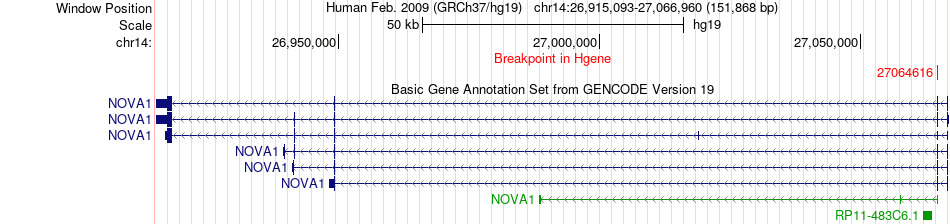 |
 Fusion gene breakpoints across FUS (3'-gene) Fusion gene breakpoints across FUS (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
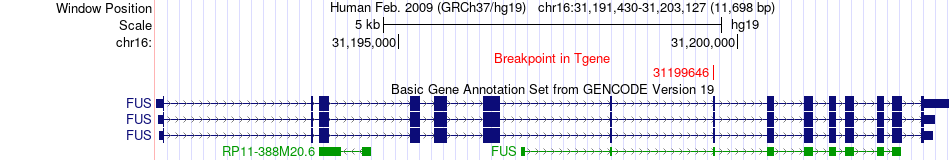 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LGG | TCGA-S9-A7J0 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
Top |
Fusion Gene ORF analysis for NOVA1-FUS |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000344429 | ENST00000474990 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| 5CDS-3UTR | ENST00000465357 | ENST00000474990 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| 5CDS-3UTR | ENST00000539517 | ENST00000474990 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| 5CDS-3UTR | ENST00000547619 | ENST00000474990 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| 5CDS-3UTR | ENST00000574031 | ENST00000474990 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| 5UTR-3CDS | ENST00000267422 | ENST00000254108 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| 5UTR-3CDS | ENST00000267422 | ENST00000380244 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| 5UTR-3CDS | ENST00000267422 | ENST00000568685 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| 5UTR-3CDS | ENST00000551754 | ENST00000254108 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| 5UTR-3CDS | ENST00000551754 | ENST00000380244 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| 5UTR-3CDS | ENST00000551754 | ENST00000568685 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| 5UTR-3UTR | ENST00000267422 | ENST00000474990 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| 5UTR-3UTR | ENST00000551754 | ENST00000474990 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| In-frame | ENST00000344429 | ENST00000254108 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| In-frame | ENST00000344429 | ENST00000380244 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| In-frame | ENST00000344429 | ENST00000568685 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| In-frame | ENST00000465357 | ENST00000254108 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| In-frame | ENST00000465357 | ENST00000380244 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| In-frame | ENST00000465357 | ENST00000568685 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| In-frame | ENST00000539517 | ENST00000254108 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| In-frame | ENST00000539517 | ENST00000380244 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| In-frame | ENST00000539517 | ENST00000568685 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| In-frame | ENST00000547619 | ENST00000254108 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| In-frame | ENST00000547619 | ENST00000380244 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| In-frame | ENST00000547619 | ENST00000568685 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| In-frame | ENST00000574031 | ENST00000254108 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| In-frame | ENST00000574031 | ENST00000380244 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
| In-frame | ENST00000574031 | ENST00000568685 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000465357 | NOVA1 | chr14 | 27064616 | - | ENST00000254108 | FUS | chr16 | 31199646 | + | 1480 | 330 | 50 | 1111 | 353 |
| ENST00000465357 | NOVA1 | chr14 | 27064616 | - | ENST00000380244 | FUS | chr16 | 31199646 | + | 1276 | 330 | 50 | 1111 | 353 |
| ENST00000465357 | NOVA1 | chr14 | 27064616 | - | ENST00000568685 | FUS | chr16 | 31199646 | + | 1252 | 330 | 50 | 1114 | 354 |
| ENST00000539517 | NOVA1 | chr14 | 27064616 | - | ENST00000254108 | FUS | chr16 | 31199646 | + | 1748 | 598 | 318 | 1379 | 353 |
| ENST00000539517 | NOVA1 | chr14 | 27064616 | - | ENST00000380244 | FUS | chr16 | 31199646 | + | 1544 | 598 | 318 | 1379 | 353 |
| ENST00000539517 | NOVA1 | chr14 | 27064616 | - | ENST00000568685 | FUS | chr16 | 31199646 | + | 1520 | 598 | 318 | 1382 | 354 |
| ENST00000344429 | NOVA1 | chr14 | 27064616 | - | ENST00000254108 | FUS | chr16 | 31199646 | + | 1434 | 284 | 4 | 1065 | 353 |
| ENST00000344429 | NOVA1 | chr14 | 27064616 | - | ENST00000380244 | FUS | chr16 | 31199646 | + | 1230 | 284 | 4 | 1065 | 353 |
| ENST00000344429 | NOVA1 | chr14 | 27064616 | - | ENST00000568685 | FUS | chr16 | 31199646 | + | 1206 | 284 | 4 | 1068 | 354 |
| ENST00000547619 | NOVA1 | chr14 | 27064616 | - | ENST00000254108 | FUS | chr16 | 31199646 | + | 1489 | 339 | 59 | 1120 | 353 |
| ENST00000547619 | NOVA1 | chr14 | 27064616 | - | ENST00000380244 | FUS | chr16 | 31199646 | + | 1285 | 339 | 59 | 1120 | 353 |
| ENST00000547619 | NOVA1 | chr14 | 27064616 | - | ENST00000568685 | FUS | chr16 | 31199646 | + | 1261 | 339 | 59 | 1123 | 354 |
| ENST00000574031 | NOVA1 | chr14 | 27064616 | - | ENST00000254108 | FUS | chr16 | 31199646 | + | 1454 | 304 | 24 | 1085 | 353 |
| ENST00000574031 | NOVA1 | chr14 | 27064616 | - | ENST00000380244 | FUS | chr16 | 31199646 | + | 1250 | 304 | 24 | 1085 | 353 |
| ENST00000574031 | NOVA1 | chr14 | 27064616 | - | ENST00000568685 | FUS | chr16 | 31199646 | + | 1226 | 304 | 24 | 1088 | 354 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000465357 | ENST00000254108 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + | 0.001605091 | 0.9983949 |
| ENST00000465357 | ENST00000380244 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + | 0.001795554 | 0.99820447 |
| ENST00000465357 | ENST00000568685 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + | 0.001109415 | 0.9988906 |
| ENST00000539517 | ENST00000254108 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + | 0.0017664 | 0.99823356 |
| ENST00000539517 | ENST00000380244 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + | 0.001992889 | 0.9980071 |
| ENST00000539517 | ENST00000568685 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + | 0.001168823 | 0.9988312 |
| ENST00000344429 | ENST00000254108 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + | 0.00158337 | 0.99841666 |
| ENST00000344429 | ENST00000380244 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + | 0.00184189 | 0.99815816 |
| ENST00000344429 | ENST00000568685 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + | 0.001106777 | 0.9988932 |
| ENST00000547619 | ENST00000254108 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + | 0.001556501 | 0.99844354 |
| ENST00000547619 | ENST00000380244 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + | 0.001762231 | 0.9982377 |
| ENST00000547619 | ENST00000568685 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + | 0.001097372 | 0.99890256 |
| ENST00000574031 | ENST00000254108 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + | 0.001649006 | 0.998351 |
| ENST00000574031 | ENST00000380244 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + | 0.001847299 | 0.9981527 |
| ENST00000574031 | ENST00000568685 | NOVA1 | chr14 | 27064616 | - | FUS | chr16 | 31199646 | + | 0.001180158 | 0.99881977 |
Top |
Fusion Genomic Features for NOVA1-FUS |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| NOVA1 | chr14 | 27064615 | - | FUS | chr16 | 31199645 | + | 2.94E-06 | 0.999997 |
| NOVA1 | chr14 | 27064615 | - | FUS | chr16 | 31199645 | + | 2.94E-06 | 0.999997 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
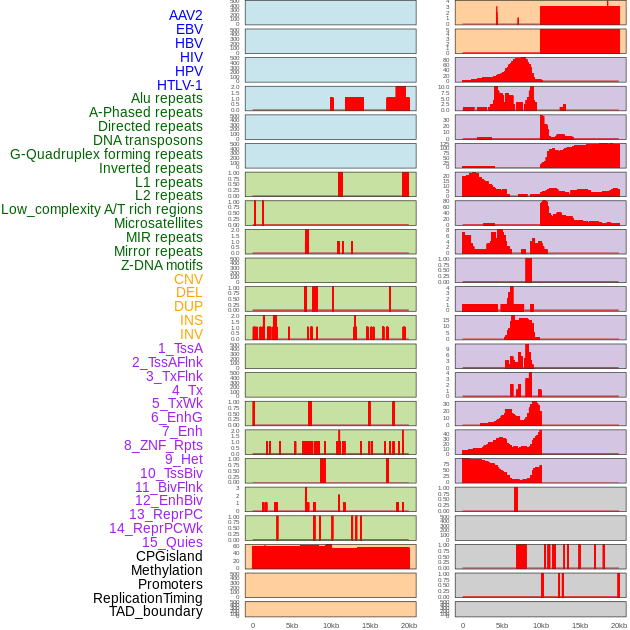 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
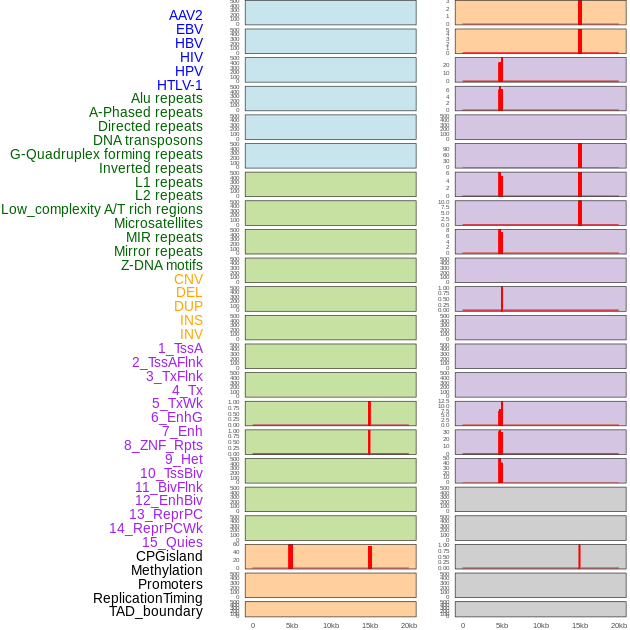 |
Top |
Fusion Protein Features for NOVA1-FUS |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr14:27064616/chr16:31199646) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | FUS |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: DNA/RNA-binding protein that plays a role in various cellular processes such as transcription regulation, RNA splicing, RNA transport, DNA repair and damage response (PubMed:27731383). Binds to nascent pre-mRNAs and acts as a molecular mediator between RNA polymerase II and U1 small nuclear ribonucleoprotein thereby coupling transcription and splicing (PubMed:26124092). Binds also its own pre-mRNA and autoregulates its expression; this autoregulation mechanism is mediated by non-sense-mediated decay (PubMed:24204307). Plays a role in DNA repair mechanisms by promoting D-loop formation and homologous recombination during DNA double-strand break repair (PubMed:10567410). In neuronal cells, plays crucial roles in dendritic spine formation and stability, RNA transport, mRNA stability and synaptic homeostasis (By similarity). {ECO:0000250|UniProtKB:P56959, ECO:0000269|PubMed:10567410, ECO:0000269|PubMed:24204307, ECO:0000269|PubMed:26124092, ECO:0000269|PubMed:27731383}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NOVA1 | chr14:27064616 | chr16:31199646 | ENST00000344429 | - | 2 | 5 | 27_43 | 93 | 182.0 | Motif | Bipartite nuclear localization signal |
| Hgene | NOVA1 | chr14:27064616 | chr16:31199646 | ENST00000465357 | - | 2 | 4 | 27_43 | 93 | 484.0 | Motif | Bipartite nuclear localization signal |
| Hgene | NOVA1 | chr14:27064616 | chr16:31199646 | ENST00000539517 | - | 2 | 5 | 27_43 | 93 | 508.0 | Motif | Bipartite nuclear localization signal |
| Tgene | FUS | chr14:27064616 | chr16:31199646 | ENST00000254108 | 6 | 15 | 371_526 | 266 | 527.0 | Compositional bias | Note=Arg/Gly-rich | |
| Tgene | FUS | chr14:27064616 | chr16:31199646 | ENST00000380244 | 6 | 15 | 371_526 | 265 | 526.0 | Compositional bias | Note=Arg/Gly-rich | |
| Tgene | FUS | chr14:27064616 | chr16:31199646 | ENST00000254108 | 6 | 15 | 285_371 | 266 | 527.0 | Domain | RRM | |
| Tgene | FUS | chr14:27064616 | chr16:31199646 | ENST00000380244 | 6 | 15 | 285_371 | 265 | 526.0 | Domain | RRM | |
| Tgene | FUS | chr14:27064616 | chr16:31199646 | ENST00000254108 | 6 | 15 | 422_453 | 266 | 527.0 | Zinc finger | RanBP2-type | |
| Tgene | FUS | chr14:27064616 | chr16:31199646 | ENST00000380244 | 6 | 15 | 422_453 | 265 | 526.0 | Zinc finger | RanBP2-type |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NOVA1 | chr14:27064616 | chr16:31199646 | ENST00000344429 | - | 2 | 5 | 273_409 | 93 | 182.0 | Compositional bias | Note=Ala-rich |
| Hgene | NOVA1 | chr14:27064616 | chr16:31199646 | ENST00000465357 | - | 2 | 4 | 273_409 | 93 | 484.0 | Compositional bias | Note=Ala-rich |
| Hgene | NOVA1 | chr14:27064616 | chr16:31199646 | ENST00000539517 | - | 2 | 5 | 273_409 | 93 | 508.0 | Compositional bias | Note=Ala-rich |
| Hgene | NOVA1 | chr14:27064616 | chr16:31199646 | ENST00000344429 | - | 2 | 5 | 171_237 | 93 | 182.0 | Domain | KH 2 |
| Hgene | NOVA1 | chr14:27064616 | chr16:31199646 | ENST00000344429 | - | 2 | 5 | 421_488 | 93 | 182.0 | Domain | KH 3 |
| Hgene | NOVA1 | chr14:27064616 | chr16:31199646 | ENST00000344429 | - | 2 | 5 | 49_116 | 93 | 182.0 | Domain | KH 1 |
| Hgene | NOVA1 | chr14:27064616 | chr16:31199646 | ENST00000465357 | - | 2 | 4 | 171_237 | 93 | 484.0 | Domain | KH 2 |
| Hgene | NOVA1 | chr14:27064616 | chr16:31199646 | ENST00000465357 | - | 2 | 4 | 421_488 | 93 | 484.0 | Domain | KH 3 |
| Hgene | NOVA1 | chr14:27064616 | chr16:31199646 | ENST00000465357 | - | 2 | 4 | 49_116 | 93 | 484.0 | Domain | KH 1 |
| Hgene | NOVA1 | chr14:27064616 | chr16:31199646 | ENST00000539517 | - | 2 | 5 | 171_237 | 93 | 508.0 | Domain | KH 2 |
| Hgene | NOVA1 | chr14:27064616 | chr16:31199646 | ENST00000539517 | - | 2 | 5 | 421_488 | 93 | 508.0 | Domain | KH 3 |
| Hgene | NOVA1 | chr14:27064616 | chr16:31199646 | ENST00000539517 | - | 2 | 5 | 49_116 | 93 | 508.0 | Domain | KH 1 |
| Tgene | FUS | chr14:27064616 | chr16:31199646 | ENST00000254108 | 6 | 15 | 166_267 | 266 | 527.0 | Compositional bias | Note=Gly-rich | |
| Tgene | FUS | chr14:27064616 | chr16:31199646 | ENST00000254108 | 6 | 15 | 1_165 | 266 | 527.0 | Compositional bias | Note=Gln/Gly/Ser/Tyr-rich | |
| Tgene | FUS | chr14:27064616 | chr16:31199646 | ENST00000380244 | 6 | 15 | 166_267 | 265 | 526.0 | Compositional bias | Note=Gly-rich | |
| Tgene | FUS | chr14:27064616 | chr16:31199646 | ENST00000380244 | 6 | 15 | 1_165 | 265 | 526.0 | Compositional bias | Note=Gln/Gly/Ser/Tyr-rich |
Top |
Fusion Gene Sequence for NOVA1-FUS |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >59799_59799_1_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000344429_FUS_chr16_31199646_ENST00000254108_length(transcript)=1434nt_BP=284nt AAACATGATGGCGGCAGCTCCCATCCAGCAGAACGGGACCCACACTGGGGTTCCCATAGACCTGGACCCGCCGGACTCGCGGAAAAGGCC GCTGGAAGCCCCCCCTGAAGCCGGCAGCACCAAGAGGACCAATACGGGCGAAGACGGCCAGTATTTTCTAAAGGTTCTCATACCTAGTTA TGCTGCTGGATCTATAATTGGGAAGGGAGGACAGACAATTGTTCAGTTGCAAAAAGAAACTGGAGCCACCATCAAGCTGTCTAAGTCCAA AGATTTTTACCCAGGCCCTCGGGACCAAGGATCACGTCATGACTCCGAACAGGATAATTCAGACAACAACACCATCTTTGTGCAAGGCCT GGGTGAGAATGTTACAATTGAGTCTGTGGCTGATTACTTCAAGCAGATTGGTATTATTAAGACAAACAAGAAAACGGGACAGCCCATGAT TAATTTGTACACAGACAGGGAAACTGGCAAGCTGAAGGGAGAGGCAACGGTCTCTTTTGATGACCCACCTTCAGCTAAAGCAGCTATTGA CTGGTTTGATGGTAAAGAATTCTCCGGAAATCCTATCAAGGTCTCATTTGCTACTCGCCGGGCAGACTTTAATCGGGGTGGTGGCAATGG TCGTGGAGGCCGAGGGCGAGGAGGACCCATGGGCCGTGGAGGCTATGGAGGTGGTGGCAGTGGTGGTGGTGGCCGAGGAGGATTTCCCAG TGGAGGTGGTGGCGGTGGAGGACAGCAGCGAGCTGGTGACTGGAAGTGTCCTAATCCCACCTGTGAGAATATGAACTTCTCTTGGAGGAA TGAATGCAACCAGTGTAAGGCCCCTAAACCAGATGGCCCAGGAGGGGGACCAGGTGGCTCTCACATGGGGGGTAACTACGGGGATGATCG TCGTGGTGGCAGAGGAGGCTATGATCGAGGCGGCTACCGGGGCCGCGGCGGGGACCGTGGAGGCTTCCGAGGGGGCCGGGGTGGTGGGGA CAGAGGTGGCTTTGGCCCTGGCAAGATGGATTCCAGGGGTGAGCACAGACAGGATCGCAGGGAGAGGCCGTATTAATTAGCCTGGCTCCC CAGGTTCTGGAACAGCTTTTTGTCCTGTACCCAGTGTTACCCTCGTTATTTTGTAACCTTCCAATTCCTGATCACCCAAGGGTTTTTTTG TGTCGGACTATGTAATTGTAACTATACCTCTGGTTCCCATTAAAAGTGACCATTTTAGTTAAATTTTGTTCCTCTTCCCCCTTTTCACTT TCCTGGAAGATCGATGTCCCGATCAGGAAGGTAGAGAGTTTTCCTGTTCAGATTACCCTGCCCAGCAGGAACTGGAATACAGTGTTCGGG >59799_59799_1_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000344429_FUS_chr16_31199646_ENST00000254108_length(amino acids)=353AA_BP=209 MMAAAPIQQNGTHTGVPIDLDPPDSRKRPLEAPPEAGSTKRTNTGEDGQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKD FYPGPRDQGSRHDSEQDNSDNNTIFVQGLGENVTIESVADYFKQIGIIKTNKKTGQPMINLYTDRETGKLKGEATVSFDDPPSAKAAIDW FDGKEFSGNPIKVSFATRRADFNRGGGNGRGGRGRGGPMGRGGYGGGGSGGGGRGGFPSGGGGGGGQQRAGDWKCPNPTCENMNFSWRNE -------------------------------------------------------------- >59799_59799_2_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000344429_FUS_chr16_31199646_ENST00000380244_length(transcript)=1230nt_BP=284nt AAACATGATGGCGGCAGCTCCCATCCAGCAGAACGGGACCCACACTGGGGTTCCCATAGACCTGGACCCGCCGGACTCGCGGAAAAGGCC GCTGGAAGCCCCCCCTGAAGCCGGCAGCACCAAGAGGACCAATACGGGCGAAGACGGCCAGTATTTTCTAAAGGTTCTCATACCTAGTTA TGCTGCTGGATCTATAATTGGGAAGGGAGGACAGACAATTGTTCAGTTGCAAAAAGAAACTGGAGCCACCATCAAGCTGTCTAAGTCCAA AGATTTTTACCCAGGCCCTCGGGACCAAGGATCACGTCATGACTCCGAACAGGATAATTCAGACAACAACACCATCTTTGTGCAAGGCCT GGGTGAGAATGTTACAATTGAGTCTGTGGCTGATTACTTCAAGCAGATTGGTATTATTAAGACAAACAAGAAAACGGGACAGCCCATGAT TAATTTGTACACAGACAGGGAAACTGGCAAGCTGAAGGGAGAGGCAACGGTCTCTTTTGATGACCCACCTTCAGCTAAAGCAGCTATTGA CTGGTTTGATGGTAAAGAATTCTCCGGAAATCCTATCAAGGTCTCATTTGCTACTCGCCGGGCAGACTTTAATCGGGGTGGTGGCAATGG TCGTGGAGGCCGAGGGCGAGGAGGACCCATGGGCCGTGGAGGCTATGGAGGTGGTGGCAGTGGTGGTGGTGGCCGAGGAGGATTTCCCAG TGGAGGTGGTGGCGGTGGAGGACAGCAGCGAGCTGGTGACTGGAAGTGTCCTAATCCCACCTGTGAGAATATGAACTTCTCTTGGAGGAA TGAATGCAACCAGTGTAAGGCCCCTAAACCAGATGGCCCAGGAGGGGGACCAGGTGGCTCTCACATGGGGGGTAACTACGGGGATGATCG TCGTGGTGGCAGAGGAGGCTATGATCGAGGCGGCTACCGGGGCCGCGGCGGGGACCGTGGAGGCTTCCGAGGGGGCCGGGGTGGTGGGGA CAGAGGTGGCTTTGGCCCTGGCAAGATGGATTCCAGGGGTGAGCACAGACAGGATCGCAGGGAGAGGCCGTATTAATTAGCCTGGCTCCC CAGGTTCTGGAACAGCTTTTTGTCCTGTACCCAGTGTTACCCTCGTTATTTTGTAACCTTCCAATTCCTGATCACCCAAGGGTTTTTTTG >59799_59799_2_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000344429_FUS_chr16_31199646_ENST00000380244_length(amino acids)=353AA_BP=209 MMAAAPIQQNGTHTGVPIDLDPPDSRKRPLEAPPEAGSTKRTNTGEDGQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKD FYPGPRDQGSRHDSEQDNSDNNTIFVQGLGENVTIESVADYFKQIGIIKTNKKTGQPMINLYTDRETGKLKGEATVSFDDPPSAKAAIDW FDGKEFSGNPIKVSFATRRADFNRGGGNGRGGRGRGGPMGRGGYGGGGSGGGGRGGFPSGGGGGGGQQRAGDWKCPNPTCENMNFSWRNE -------------------------------------------------------------- >59799_59799_3_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000344429_FUS_chr16_31199646_ENST00000568685_length(transcript)=1206nt_BP=284nt AAACATGATGGCGGCAGCTCCCATCCAGCAGAACGGGACCCACACTGGGGTTCCCATAGACCTGGACCCGCCGGACTCGCGGAAAAGGCC GCTGGAAGCCCCCCCTGAAGCCGGCAGCACCAAGAGGACCAATACGGGCGAAGACGGCCAGTATTTTCTAAAGGTTCTCATACCTAGTTA TGCTGCTGGATCTATAATTGGGAAGGGAGGACAGACAATTGTTCAGTTGCAAAAAGAAACTGGAGCCACCATCAAGCTGTCTAAGTCCAA AGATTTTTACCCAGGCCCTCGGGACCAAGGATCACGTCATGACTCCGCAGAACAGGATAATTCAGACAACAACACCATCTTTGTGCAAGG CCTGGGTGAGAATGTTACAATTGAGTCTGTGGCTGATTACTTCAAGCAGATTGGTATTATTAAGACAAACAAGAAAACGGGACAGCCCAT GATTAATTTGTACACAGACAGGGAAACTGGCAAGCTGAAGGGAGAGGCAACGGTCTCTTTTGATGACCCACCTTCAGCTAAAGCAGCTAT TGACTGGTTTGATGGTAAAGAATTCTCCGGAAATCCTATCAAGGTCTCATTTGCTACTCGCCGGGCAGACTTTAATCGGGGTGGTGGCAA TGGTCGTGGAGGCCGAGGGCGAGGAGGACCCATGGGCCGTGGAGGCTATGGAGGTGGTGGCAGTGGTGGTGGTGGCCGAGGAGGATTTCC CAGTGGAGGTGGTGGCGGTGGAGGACAGCAGCGAGCTGGTGACTGGAAGTGTCCTAATCCCACCTGTGAGAATATGAACTTCTCTTGGAG GAATGAATGCAACCAGTGTAAGGCCCCTAAACCAGATGGCCCAGGAGGGGGACCAGGTGGCTCTCACATGGGGGGTAACTACGGGGATGA TCGTCGTGGTGGCAGAGGAGGCTATGATCGAGGCGGCTACCGGGGCCGCGGCGGGGACCGTGGAGGCTTCCGAGGGGGCCGGGGTGGTGG GGACAGAGGTGGCTTTGGCCCTGGCAAGATGGATTCCAGGGGTGAGCACAGACAGGATCGCAGGGAGAGGCCGTATTAATTAGCCTGGCT CCCCAGGTTCTGGAACAGCTTTTTGTCCTGTACCCAGTGTTACCCTCGTTATTTTGTAACCTTCCAATTCCTGATCACCCAAGGGTTTTT >59799_59799_3_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000344429_FUS_chr16_31199646_ENST00000568685_length(amino acids)=354AA_BP=210 MMAAAPIQQNGTHTGVPIDLDPPDSRKRPLEAPPEAGSTKRTNTGEDGQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKD FYPGPRDQGSRHDSAEQDNSDNNTIFVQGLGENVTIESVADYFKQIGIIKTNKKTGQPMINLYTDRETGKLKGEATVSFDDPPSAKAAID WFDGKEFSGNPIKVSFATRRADFNRGGGNGRGGRGRGGPMGRGGYGGGGSGGGGRGGFPSGGGGGGGQQRAGDWKCPNPTCENMNFSWRN -------------------------------------------------------------- >59799_59799_4_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000465357_FUS_chr16_31199646_ENST00000254108_length(transcript)=1480nt_BP=330nt AAAACAAAAGGGAGAACCTTCTCCCGGTAGCAGCGGCAGGAACTGCAAACATGATGGCGGCAGCTCCCATCCAGCAGAACGGGACCCACA CTGGGGTTCCCATAGACCTGGACCCGCCGGACTCGCGGAAAAGGCCGCTGGAAGCCCCCCCTGAAGCCGGCAGCACCAAGAGGACCAATA CGGGCGAAGACGGCCAGTATTTTCTAAAGGTTCTCATACCTAGTTATGCTGCTGGATCTATAATTGGGAAGGGAGGACAGACAATTGTTC AGTTGCAAAAAGAAACTGGAGCCACCATCAAGCTGTCTAAGTCCAAAGATTTTTACCCAGGCCCTCGGGACCAAGGATCACGTCATGACT CCGAACAGGATAATTCAGACAACAACACCATCTTTGTGCAAGGCCTGGGTGAGAATGTTACAATTGAGTCTGTGGCTGATTACTTCAAGC AGATTGGTATTATTAAGACAAACAAGAAAACGGGACAGCCCATGATTAATTTGTACACAGACAGGGAAACTGGCAAGCTGAAGGGAGAGG CAACGGTCTCTTTTGATGACCCACCTTCAGCTAAAGCAGCTATTGACTGGTTTGATGGTAAAGAATTCTCCGGAAATCCTATCAAGGTCT CATTTGCTACTCGCCGGGCAGACTTTAATCGGGGTGGTGGCAATGGTCGTGGAGGCCGAGGGCGAGGAGGACCCATGGGCCGTGGAGGCT ATGGAGGTGGTGGCAGTGGTGGTGGTGGCCGAGGAGGATTTCCCAGTGGAGGTGGTGGCGGTGGAGGACAGCAGCGAGCTGGTGACTGGA AGTGTCCTAATCCCACCTGTGAGAATATGAACTTCTCTTGGAGGAATGAATGCAACCAGTGTAAGGCCCCTAAACCAGATGGCCCAGGAG GGGGACCAGGTGGCTCTCACATGGGGGGTAACTACGGGGATGATCGTCGTGGTGGCAGAGGAGGCTATGATCGAGGCGGCTACCGGGGCC GCGGCGGGGACCGTGGAGGCTTCCGAGGGGGCCGGGGTGGTGGGGACAGAGGTGGCTTTGGCCCTGGCAAGATGGATTCCAGGGGTGAGC ACAGACAGGATCGCAGGGAGAGGCCGTATTAATTAGCCTGGCTCCCCAGGTTCTGGAACAGCTTTTTGTCCTGTACCCAGTGTTACCCTC GTTATTTTGTAACCTTCCAATTCCTGATCACCCAAGGGTTTTTTTGTGTCGGACTATGTAATTGTAACTATACCTCTGGTTCCCATTAAA AGTGACCATTTTAGTTAAATTTTGTTCCTCTTCCCCCTTTTCACTTTCCTGGAAGATCGATGTCCCGATCAGGAAGGTAGAGAGTTTTCC TGTTCAGATTACCCTGCCCAGCAGGAACTGGAATACAGTGTTCGGGGAGAAGGCCAAATGATATCCTTGAGAGCAGAGATTAAACTTTTC >59799_59799_4_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000465357_FUS_chr16_31199646_ENST00000254108_length(amino acids)=353AA_BP=209 MMAAAPIQQNGTHTGVPIDLDPPDSRKRPLEAPPEAGSTKRTNTGEDGQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKD FYPGPRDQGSRHDSEQDNSDNNTIFVQGLGENVTIESVADYFKQIGIIKTNKKTGQPMINLYTDRETGKLKGEATVSFDDPPSAKAAIDW FDGKEFSGNPIKVSFATRRADFNRGGGNGRGGRGRGGPMGRGGYGGGGSGGGGRGGFPSGGGGGGGQQRAGDWKCPNPTCENMNFSWRNE -------------------------------------------------------------- >59799_59799_5_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000465357_FUS_chr16_31199646_ENST00000380244_length(transcript)=1276nt_BP=330nt AAAACAAAAGGGAGAACCTTCTCCCGGTAGCAGCGGCAGGAACTGCAAACATGATGGCGGCAGCTCCCATCCAGCAGAACGGGACCCACA CTGGGGTTCCCATAGACCTGGACCCGCCGGACTCGCGGAAAAGGCCGCTGGAAGCCCCCCCTGAAGCCGGCAGCACCAAGAGGACCAATA CGGGCGAAGACGGCCAGTATTTTCTAAAGGTTCTCATACCTAGTTATGCTGCTGGATCTATAATTGGGAAGGGAGGACAGACAATTGTTC AGTTGCAAAAAGAAACTGGAGCCACCATCAAGCTGTCTAAGTCCAAAGATTTTTACCCAGGCCCTCGGGACCAAGGATCACGTCATGACT CCGAACAGGATAATTCAGACAACAACACCATCTTTGTGCAAGGCCTGGGTGAGAATGTTACAATTGAGTCTGTGGCTGATTACTTCAAGC AGATTGGTATTATTAAGACAAACAAGAAAACGGGACAGCCCATGATTAATTTGTACACAGACAGGGAAACTGGCAAGCTGAAGGGAGAGG CAACGGTCTCTTTTGATGACCCACCTTCAGCTAAAGCAGCTATTGACTGGTTTGATGGTAAAGAATTCTCCGGAAATCCTATCAAGGTCT CATTTGCTACTCGCCGGGCAGACTTTAATCGGGGTGGTGGCAATGGTCGTGGAGGCCGAGGGCGAGGAGGACCCATGGGCCGTGGAGGCT ATGGAGGTGGTGGCAGTGGTGGTGGTGGCCGAGGAGGATTTCCCAGTGGAGGTGGTGGCGGTGGAGGACAGCAGCGAGCTGGTGACTGGA AGTGTCCTAATCCCACCTGTGAGAATATGAACTTCTCTTGGAGGAATGAATGCAACCAGTGTAAGGCCCCTAAACCAGATGGCCCAGGAG GGGGACCAGGTGGCTCTCACATGGGGGGTAACTACGGGGATGATCGTCGTGGTGGCAGAGGAGGCTATGATCGAGGCGGCTACCGGGGCC GCGGCGGGGACCGTGGAGGCTTCCGAGGGGGCCGGGGTGGTGGGGACAGAGGTGGCTTTGGCCCTGGCAAGATGGATTCCAGGGGTGAGC ACAGACAGGATCGCAGGGAGAGGCCGTATTAATTAGCCTGGCTCCCCAGGTTCTGGAACAGCTTTTTGTCCTGTACCCAGTGTTACCCTC GTTATTTTGTAACCTTCCAATTCCTGATCACCCAAGGGTTTTTTTGTGTCGGACTATGTAATTGTAACTATACCTCTGGTTCCCATTAAA >59799_59799_5_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000465357_FUS_chr16_31199646_ENST00000380244_length(amino acids)=353AA_BP=209 MMAAAPIQQNGTHTGVPIDLDPPDSRKRPLEAPPEAGSTKRTNTGEDGQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKD FYPGPRDQGSRHDSEQDNSDNNTIFVQGLGENVTIESVADYFKQIGIIKTNKKTGQPMINLYTDRETGKLKGEATVSFDDPPSAKAAIDW FDGKEFSGNPIKVSFATRRADFNRGGGNGRGGRGRGGPMGRGGYGGGGSGGGGRGGFPSGGGGGGGQQRAGDWKCPNPTCENMNFSWRNE -------------------------------------------------------------- >59799_59799_6_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000465357_FUS_chr16_31199646_ENST00000568685_length(transcript)=1252nt_BP=330nt AAAACAAAAGGGAGAACCTTCTCCCGGTAGCAGCGGCAGGAACTGCAAACATGATGGCGGCAGCTCCCATCCAGCAGAACGGGACCCACA CTGGGGTTCCCATAGACCTGGACCCGCCGGACTCGCGGAAAAGGCCGCTGGAAGCCCCCCCTGAAGCCGGCAGCACCAAGAGGACCAATA CGGGCGAAGACGGCCAGTATTTTCTAAAGGTTCTCATACCTAGTTATGCTGCTGGATCTATAATTGGGAAGGGAGGACAGACAATTGTTC AGTTGCAAAAAGAAACTGGAGCCACCATCAAGCTGTCTAAGTCCAAAGATTTTTACCCAGGCCCTCGGGACCAAGGATCACGTCATGACT CCGCAGAACAGGATAATTCAGACAACAACACCATCTTTGTGCAAGGCCTGGGTGAGAATGTTACAATTGAGTCTGTGGCTGATTACTTCA AGCAGATTGGTATTATTAAGACAAACAAGAAAACGGGACAGCCCATGATTAATTTGTACACAGACAGGGAAACTGGCAAGCTGAAGGGAG AGGCAACGGTCTCTTTTGATGACCCACCTTCAGCTAAAGCAGCTATTGACTGGTTTGATGGTAAAGAATTCTCCGGAAATCCTATCAAGG TCTCATTTGCTACTCGCCGGGCAGACTTTAATCGGGGTGGTGGCAATGGTCGTGGAGGCCGAGGGCGAGGAGGACCCATGGGCCGTGGAG GCTATGGAGGTGGTGGCAGTGGTGGTGGTGGCCGAGGAGGATTTCCCAGTGGAGGTGGTGGCGGTGGAGGACAGCAGCGAGCTGGTGACT GGAAGTGTCCTAATCCCACCTGTGAGAATATGAACTTCTCTTGGAGGAATGAATGCAACCAGTGTAAGGCCCCTAAACCAGATGGCCCAG GAGGGGGACCAGGTGGCTCTCACATGGGGGGTAACTACGGGGATGATCGTCGTGGTGGCAGAGGAGGCTATGATCGAGGCGGCTACCGGG GCCGCGGCGGGGACCGTGGAGGCTTCCGAGGGGGCCGGGGTGGTGGGGACAGAGGTGGCTTTGGCCCTGGCAAGATGGATTCCAGGGGTG AGCACAGACAGGATCGCAGGGAGAGGCCGTATTAATTAGCCTGGCTCCCCAGGTTCTGGAACAGCTTTTTGTCCTGTACCCAGTGTTACC >59799_59799_6_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000465357_FUS_chr16_31199646_ENST00000568685_length(amino acids)=354AA_BP=210 MMAAAPIQQNGTHTGVPIDLDPPDSRKRPLEAPPEAGSTKRTNTGEDGQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKD FYPGPRDQGSRHDSAEQDNSDNNTIFVQGLGENVTIESVADYFKQIGIIKTNKKTGQPMINLYTDRETGKLKGEATVSFDDPPSAKAAID WFDGKEFSGNPIKVSFATRRADFNRGGGNGRGGRGRGGPMGRGGYGGGGSGGGGRGGFPSGGGGGGGQQRAGDWKCPNPTCENMNFSWRN -------------------------------------------------------------- >59799_59799_7_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000539517_FUS_chr16_31199646_ENST00000254108_length(transcript)=1748nt_BP=598nt CTCTCCCTTCTCCACTCTCTCCCCCTGTCTCCTTTCTTCTTCTTCTTTCACCCTCCGTCTCTCACACCCCCTCCATTCCCCTGTCTCCTT TCTGACACTGCACTGCAGCTGCTCCTCAGCCCTGCCCCCTCCCCAGTGAGAACAAACCAGCAACATTGCTTTTTTTCCTAAAGAGATTTA TATTGATCCGATTAAAAAAAAAAAACCTTAAGAAACCCCAAACGCAAAAAAAAAAAAAAAAAAAAAAGAAAAAAGAAAAGAAAAAGCCAA AACAAAAGGGAGAACCTTCTCCCGGTAGCAGCGGCAGGAACTGCAAACATGATGGCGGCAGCTCCCATCCAGCAGAACGGGACCCACACT GGGGTTCCCATAGACCTGGACCCGCCGGACTCGCGGAAAAGGCCGCTGGAAGCCCCCCCTGAAGCCGGCAGCACCAAGAGGACCAATACG GGCGAAGACGGCCAGTATTTTCTAAAGGTTCTCATACCTAGTTATGCTGCTGGATCTATAATTGGGAAGGGAGGACAGACAATTGTTCAG TTGCAAAAAGAAACTGGAGCCACCATCAAGCTGTCTAAGTCCAAAGATTTTTACCCAGGCCCTCGGGACCAAGGATCACGTCATGACTCC GAACAGGATAATTCAGACAACAACACCATCTTTGTGCAAGGCCTGGGTGAGAATGTTACAATTGAGTCTGTGGCTGATTACTTCAAGCAG ATTGGTATTATTAAGACAAACAAGAAAACGGGACAGCCCATGATTAATTTGTACACAGACAGGGAAACTGGCAAGCTGAAGGGAGAGGCA ACGGTCTCTTTTGATGACCCACCTTCAGCTAAAGCAGCTATTGACTGGTTTGATGGTAAAGAATTCTCCGGAAATCCTATCAAGGTCTCA TTTGCTACTCGCCGGGCAGACTTTAATCGGGGTGGTGGCAATGGTCGTGGAGGCCGAGGGCGAGGAGGACCCATGGGCCGTGGAGGCTAT GGAGGTGGTGGCAGTGGTGGTGGTGGCCGAGGAGGATTTCCCAGTGGAGGTGGTGGCGGTGGAGGACAGCAGCGAGCTGGTGACTGGAAG TGTCCTAATCCCACCTGTGAGAATATGAACTTCTCTTGGAGGAATGAATGCAACCAGTGTAAGGCCCCTAAACCAGATGGCCCAGGAGGG GGACCAGGTGGCTCTCACATGGGGGGTAACTACGGGGATGATCGTCGTGGTGGCAGAGGAGGCTATGATCGAGGCGGCTACCGGGGCCGC GGCGGGGACCGTGGAGGCTTCCGAGGGGGCCGGGGTGGTGGGGACAGAGGTGGCTTTGGCCCTGGCAAGATGGATTCCAGGGGTGAGCAC AGACAGGATCGCAGGGAGAGGCCGTATTAATTAGCCTGGCTCCCCAGGTTCTGGAACAGCTTTTTGTCCTGTACCCAGTGTTACCCTCGT TATTTTGTAACCTTCCAATTCCTGATCACCCAAGGGTTTTTTTGTGTCGGACTATGTAATTGTAACTATACCTCTGGTTCCCATTAAAAG TGACCATTTTAGTTAAATTTTGTTCCTCTTCCCCCTTTTCACTTTCCTGGAAGATCGATGTCCCGATCAGGAAGGTAGAGAGTTTTCCTG TTCAGATTACCCTGCCCAGCAGGAACTGGAATACAGTGTTCGGGGAGAAGGCCAAATGATATCCTTGAGAGCAGAGATTAAACTTTTCTG >59799_59799_7_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000539517_FUS_chr16_31199646_ENST00000254108_length(amino acids)=353AA_BP=209 MMAAAPIQQNGTHTGVPIDLDPPDSRKRPLEAPPEAGSTKRTNTGEDGQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKD FYPGPRDQGSRHDSEQDNSDNNTIFVQGLGENVTIESVADYFKQIGIIKTNKKTGQPMINLYTDRETGKLKGEATVSFDDPPSAKAAIDW FDGKEFSGNPIKVSFATRRADFNRGGGNGRGGRGRGGPMGRGGYGGGGSGGGGRGGFPSGGGGGGGQQRAGDWKCPNPTCENMNFSWRNE -------------------------------------------------------------- >59799_59799_8_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000539517_FUS_chr16_31199646_ENST00000380244_length(transcript)=1544nt_BP=598nt CTCTCCCTTCTCCACTCTCTCCCCCTGTCTCCTTTCTTCTTCTTCTTTCACCCTCCGTCTCTCACACCCCCTCCATTCCCCTGTCTCCTT TCTGACACTGCACTGCAGCTGCTCCTCAGCCCTGCCCCCTCCCCAGTGAGAACAAACCAGCAACATTGCTTTTTTTCCTAAAGAGATTTA TATTGATCCGATTAAAAAAAAAAAACCTTAAGAAACCCCAAACGCAAAAAAAAAAAAAAAAAAAAAAGAAAAAAGAAAAGAAAAAGCCAA AACAAAAGGGAGAACCTTCTCCCGGTAGCAGCGGCAGGAACTGCAAACATGATGGCGGCAGCTCCCATCCAGCAGAACGGGACCCACACT GGGGTTCCCATAGACCTGGACCCGCCGGACTCGCGGAAAAGGCCGCTGGAAGCCCCCCCTGAAGCCGGCAGCACCAAGAGGACCAATACG GGCGAAGACGGCCAGTATTTTCTAAAGGTTCTCATACCTAGTTATGCTGCTGGATCTATAATTGGGAAGGGAGGACAGACAATTGTTCAG TTGCAAAAAGAAACTGGAGCCACCATCAAGCTGTCTAAGTCCAAAGATTTTTACCCAGGCCCTCGGGACCAAGGATCACGTCATGACTCC GAACAGGATAATTCAGACAACAACACCATCTTTGTGCAAGGCCTGGGTGAGAATGTTACAATTGAGTCTGTGGCTGATTACTTCAAGCAG ATTGGTATTATTAAGACAAACAAGAAAACGGGACAGCCCATGATTAATTTGTACACAGACAGGGAAACTGGCAAGCTGAAGGGAGAGGCA ACGGTCTCTTTTGATGACCCACCTTCAGCTAAAGCAGCTATTGACTGGTTTGATGGTAAAGAATTCTCCGGAAATCCTATCAAGGTCTCA TTTGCTACTCGCCGGGCAGACTTTAATCGGGGTGGTGGCAATGGTCGTGGAGGCCGAGGGCGAGGAGGACCCATGGGCCGTGGAGGCTAT GGAGGTGGTGGCAGTGGTGGTGGTGGCCGAGGAGGATTTCCCAGTGGAGGTGGTGGCGGTGGAGGACAGCAGCGAGCTGGTGACTGGAAG TGTCCTAATCCCACCTGTGAGAATATGAACTTCTCTTGGAGGAATGAATGCAACCAGTGTAAGGCCCCTAAACCAGATGGCCCAGGAGGG GGACCAGGTGGCTCTCACATGGGGGGTAACTACGGGGATGATCGTCGTGGTGGCAGAGGAGGCTATGATCGAGGCGGCTACCGGGGCCGC GGCGGGGACCGTGGAGGCTTCCGAGGGGGCCGGGGTGGTGGGGACAGAGGTGGCTTTGGCCCTGGCAAGATGGATTCCAGGGGTGAGCAC AGACAGGATCGCAGGGAGAGGCCGTATTAATTAGCCTGGCTCCCCAGGTTCTGGAACAGCTTTTTGTCCTGTACCCAGTGTTACCCTCGT TATTTTGTAACCTTCCAATTCCTGATCACCCAAGGGTTTTTTTGTGTCGGACTATGTAATTGTAACTATACCTCTGGTTCCCATTAAAAG >59799_59799_8_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000539517_FUS_chr16_31199646_ENST00000380244_length(amino acids)=353AA_BP=209 MMAAAPIQQNGTHTGVPIDLDPPDSRKRPLEAPPEAGSTKRTNTGEDGQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKD FYPGPRDQGSRHDSEQDNSDNNTIFVQGLGENVTIESVADYFKQIGIIKTNKKTGQPMINLYTDRETGKLKGEATVSFDDPPSAKAAIDW FDGKEFSGNPIKVSFATRRADFNRGGGNGRGGRGRGGPMGRGGYGGGGSGGGGRGGFPSGGGGGGGQQRAGDWKCPNPTCENMNFSWRNE -------------------------------------------------------------- >59799_59799_9_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000539517_FUS_chr16_31199646_ENST00000568685_length(transcript)=1520nt_BP=598nt CTCTCCCTTCTCCACTCTCTCCCCCTGTCTCCTTTCTTCTTCTTCTTTCACCCTCCGTCTCTCACACCCCCTCCATTCCCCTGTCTCCTT TCTGACACTGCACTGCAGCTGCTCCTCAGCCCTGCCCCCTCCCCAGTGAGAACAAACCAGCAACATTGCTTTTTTTCCTAAAGAGATTTA TATTGATCCGATTAAAAAAAAAAAACCTTAAGAAACCCCAAACGCAAAAAAAAAAAAAAAAAAAAAAGAAAAAAGAAAAGAAAAAGCCAA AACAAAAGGGAGAACCTTCTCCCGGTAGCAGCGGCAGGAACTGCAAACATGATGGCGGCAGCTCCCATCCAGCAGAACGGGACCCACACT GGGGTTCCCATAGACCTGGACCCGCCGGACTCGCGGAAAAGGCCGCTGGAAGCCCCCCCTGAAGCCGGCAGCACCAAGAGGACCAATACG GGCGAAGACGGCCAGTATTTTCTAAAGGTTCTCATACCTAGTTATGCTGCTGGATCTATAATTGGGAAGGGAGGACAGACAATTGTTCAG TTGCAAAAAGAAACTGGAGCCACCATCAAGCTGTCTAAGTCCAAAGATTTTTACCCAGGCCCTCGGGACCAAGGATCACGTCATGACTCC GCAGAACAGGATAATTCAGACAACAACACCATCTTTGTGCAAGGCCTGGGTGAGAATGTTACAATTGAGTCTGTGGCTGATTACTTCAAG CAGATTGGTATTATTAAGACAAACAAGAAAACGGGACAGCCCATGATTAATTTGTACACAGACAGGGAAACTGGCAAGCTGAAGGGAGAG GCAACGGTCTCTTTTGATGACCCACCTTCAGCTAAAGCAGCTATTGACTGGTTTGATGGTAAAGAATTCTCCGGAAATCCTATCAAGGTC TCATTTGCTACTCGCCGGGCAGACTTTAATCGGGGTGGTGGCAATGGTCGTGGAGGCCGAGGGCGAGGAGGACCCATGGGCCGTGGAGGC TATGGAGGTGGTGGCAGTGGTGGTGGTGGCCGAGGAGGATTTCCCAGTGGAGGTGGTGGCGGTGGAGGACAGCAGCGAGCTGGTGACTGG AAGTGTCCTAATCCCACCTGTGAGAATATGAACTTCTCTTGGAGGAATGAATGCAACCAGTGTAAGGCCCCTAAACCAGATGGCCCAGGA GGGGGACCAGGTGGCTCTCACATGGGGGGTAACTACGGGGATGATCGTCGTGGTGGCAGAGGAGGCTATGATCGAGGCGGCTACCGGGGC CGCGGCGGGGACCGTGGAGGCTTCCGAGGGGGCCGGGGTGGTGGGGACAGAGGTGGCTTTGGCCCTGGCAAGATGGATTCCAGGGGTGAG CACAGACAGGATCGCAGGGAGAGGCCGTATTAATTAGCCTGGCTCCCCAGGTTCTGGAACAGCTTTTTGTCCTGTACCCAGTGTTACCCT >59799_59799_9_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000539517_FUS_chr16_31199646_ENST00000568685_length(amino acids)=354AA_BP=210 MMAAAPIQQNGTHTGVPIDLDPPDSRKRPLEAPPEAGSTKRTNTGEDGQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKD FYPGPRDQGSRHDSAEQDNSDNNTIFVQGLGENVTIESVADYFKQIGIIKTNKKTGQPMINLYTDRETGKLKGEATVSFDDPPSAKAAID WFDGKEFSGNPIKVSFATRRADFNRGGGNGRGGRGRGGPMGRGGYGGGGSGGGGRGGFPSGGGGGGGQQRAGDWKCPNPTCENMNFSWRN -------------------------------------------------------------- >59799_59799_10_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000547619_FUS_chr16_31199646_ENST00000254108_length(transcript)=1489nt_BP=339nt GAAAAAGCCAAAACAAAAGGGAGAACCTTCTCCCGGTAGCAGCGGCAGGAACTGCAAACATGATGGCGGCAGCTCCCATCCAGCAGAACG GGACCCACACTGGGGTTCCCATAGACCTGGACCCGCCGGACTCGCGGAAAAGGCCGCTGGAAGCCCCCCCTGAAGCCGGCAGCACCAAGA GGACCAATACGGGCGAAGACGGCCAGTATTTTCTAAAGGTTCTCATACCTAGTTATGCTGCTGGATCTATAATTGGGAAGGGAGGACAGA CAATTGTTCAGTTGCAAAAAGAAACTGGAGCCACCATCAAGCTGTCTAAGTCCAAAGATTTTTACCCAGGCCCTCGGGACCAAGGATCAC GTCATGACTCCGAACAGGATAATTCAGACAACAACACCATCTTTGTGCAAGGCCTGGGTGAGAATGTTACAATTGAGTCTGTGGCTGATT ACTTCAAGCAGATTGGTATTATTAAGACAAACAAGAAAACGGGACAGCCCATGATTAATTTGTACACAGACAGGGAAACTGGCAAGCTGA AGGGAGAGGCAACGGTCTCTTTTGATGACCCACCTTCAGCTAAAGCAGCTATTGACTGGTTTGATGGTAAAGAATTCTCCGGAAATCCTA TCAAGGTCTCATTTGCTACTCGCCGGGCAGACTTTAATCGGGGTGGTGGCAATGGTCGTGGAGGCCGAGGGCGAGGAGGACCCATGGGCC GTGGAGGCTATGGAGGTGGTGGCAGTGGTGGTGGTGGCCGAGGAGGATTTCCCAGTGGAGGTGGTGGCGGTGGAGGACAGCAGCGAGCTG GTGACTGGAAGTGTCCTAATCCCACCTGTGAGAATATGAACTTCTCTTGGAGGAATGAATGCAACCAGTGTAAGGCCCCTAAACCAGATG GCCCAGGAGGGGGACCAGGTGGCTCTCACATGGGGGGTAACTACGGGGATGATCGTCGTGGTGGCAGAGGAGGCTATGATCGAGGCGGCT ACCGGGGCCGCGGCGGGGACCGTGGAGGCTTCCGAGGGGGCCGGGGTGGTGGGGACAGAGGTGGCTTTGGCCCTGGCAAGATGGATTCCA GGGGTGAGCACAGACAGGATCGCAGGGAGAGGCCGTATTAATTAGCCTGGCTCCCCAGGTTCTGGAACAGCTTTTTGTCCTGTACCCAGT GTTACCCTCGTTATTTTGTAACCTTCCAATTCCTGATCACCCAAGGGTTTTTTTGTGTCGGACTATGTAATTGTAACTATACCTCTGGTT CCCATTAAAAGTGACCATTTTAGTTAAATTTTGTTCCTCTTCCCCCTTTTCACTTTCCTGGAAGATCGATGTCCCGATCAGGAAGGTAGA GAGTTTTCCTGTTCAGATTACCCTGCCCAGCAGGAACTGGAATACAGTGTTCGGGGAGAAGGCCAAATGATATCCTTGAGAGCAGAGATT >59799_59799_10_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000547619_FUS_chr16_31199646_ENST00000254108_length(amino acids)=353AA_BP=209 MMAAAPIQQNGTHTGVPIDLDPPDSRKRPLEAPPEAGSTKRTNTGEDGQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKD FYPGPRDQGSRHDSEQDNSDNNTIFVQGLGENVTIESVADYFKQIGIIKTNKKTGQPMINLYTDRETGKLKGEATVSFDDPPSAKAAIDW FDGKEFSGNPIKVSFATRRADFNRGGGNGRGGRGRGGPMGRGGYGGGGSGGGGRGGFPSGGGGGGGQQRAGDWKCPNPTCENMNFSWRNE -------------------------------------------------------------- >59799_59799_11_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000547619_FUS_chr16_31199646_ENST00000380244_length(transcript)=1285nt_BP=339nt GAAAAAGCCAAAACAAAAGGGAGAACCTTCTCCCGGTAGCAGCGGCAGGAACTGCAAACATGATGGCGGCAGCTCCCATCCAGCAGAACG GGACCCACACTGGGGTTCCCATAGACCTGGACCCGCCGGACTCGCGGAAAAGGCCGCTGGAAGCCCCCCCTGAAGCCGGCAGCACCAAGA GGACCAATACGGGCGAAGACGGCCAGTATTTTCTAAAGGTTCTCATACCTAGTTATGCTGCTGGATCTATAATTGGGAAGGGAGGACAGA CAATTGTTCAGTTGCAAAAAGAAACTGGAGCCACCATCAAGCTGTCTAAGTCCAAAGATTTTTACCCAGGCCCTCGGGACCAAGGATCAC GTCATGACTCCGAACAGGATAATTCAGACAACAACACCATCTTTGTGCAAGGCCTGGGTGAGAATGTTACAATTGAGTCTGTGGCTGATT ACTTCAAGCAGATTGGTATTATTAAGACAAACAAGAAAACGGGACAGCCCATGATTAATTTGTACACAGACAGGGAAACTGGCAAGCTGA AGGGAGAGGCAACGGTCTCTTTTGATGACCCACCTTCAGCTAAAGCAGCTATTGACTGGTTTGATGGTAAAGAATTCTCCGGAAATCCTA TCAAGGTCTCATTTGCTACTCGCCGGGCAGACTTTAATCGGGGTGGTGGCAATGGTCGTGGAGGCCGAGGGCGAGGAGGACCCATGGGCC GTGGAGGCTATGGAGGTGGTGGCAGTGGTGGTGGTGGCCGAGGAGGATTTCCCAGTGGAGGTGGTGGCGGTGGAGGACAGCAGCGAGCTG GTGACTGGAAGTGTCCTAATCCCACCTGTGAGAATATGAACTTCTCTTGGAGGAATGAATGCAACCAGTGTAAGGCCCCTAAACCAGATG GCCCAGGAGGGGGACCAGGTGGCTCTCACATGGGGGGTAACTACGGGGATGATCGTCGTGGTGGCAGAGGAGGCTATGATCGAGGCGGCT ACCGGGGCCGCGGCGGGGACCGTGGAGGCTTCCGAGGGGGCCGGGGTGGTGGGGACAGAGGTGGCTTTGGCCCTGGCAAGATGGATTCCA GGGGTGAGCACAGACAGGATCGCAGGGAGAGGCCGTATTAATTAGCCTGGCTCCCCAGGTTCTGGAACAGCTTTTTGTCCTGTACCCAGT GTTACCCTCGTTATTTTGTAACCTTCCAATTCCTGATCACCCAAGGGTTTTTTTGTGTCGGACTATGTAATTGTAACTATACCTCTGGTT >59799_59799_11_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000547619_FUS_chr16_31199646_ENST00000380244_length(amino acids)=353AA_BP=209 MMAAAPIQQNGTHTGVPIDLDPPDSRKRPLEAPPEAGSTKRTNTGEDGQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKD FYPGPRDQGSRHDSEQDNSDNNTIFVQGLGENVTIESVADYFKQIGIIKTNKKTGQPMINLYTDRETGKLKGEATVSFDDPPSAKAAIDW FDGKEFSGNPIKVSFATRRADFNRGGGNGRGGRGRGGPMGRGGYGGGGSGGGGRGGFPSGGGGGGGQQRAGDWKCPNPTCENMNFSWRNE -------------------------------------------------------------- >59799_59799_12_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000547619_FUS_chr16_31199646_ENST00000568685_length(transcript)=1261nt_BP=339nt GAAAAAGCCAAAACAAAAGGGAGAACCTTCTCCCGGTAGCAGCGGCAGGAACTGCAAACATGATGGCGGCAGCTCCCATCCAGCAGAACG GGACCCACACTGGGGTTCCCATAGACCTGGACCCGCCGGACTCGCGGAAAAGGCCGCTGGAAGCCCCCCCTGAAGCCGGCAGCACCAAGA GGACCAATACGGGCGAAGACGGCCAGTATTTTCTAAAGGTTCTCATACCTAGTTATGCTGCTGGATCTATAATTGGGAAGGGAGGACAGA CAATTGTTCAGTTGCAAAAAGAAACTGGAGCCACCATCAAGCTGTCTAAGTCCAAAGATTTTTACCCAGGCCCTCGGGACCAAGGATCAC GTCATGACTCCGCAGAACAGGATAATTCAGACAACAACACCATCTTTGTGCAAGGCCTGGGTGAGAATGTTACAATTGAGTCTGTGGCTG ATTACTTCAAGCAGATTGGTATTATTAAGACAAACAAGAAAACGGGACAGCCCATGATTAATTTGTACACAGACAGGGAAACTGGCAAGC TGAAGGGAGAGGCAACGGTCTCTTTTGATGACCCACCTTCAGCTAAAGCAGCTATTGACTGGTTTGATGGTAAAGAATTCTCCGGAAATC CTATCAAGGTCTCATTTGCTACTCGCCGGGCAGACTTTAATCGGGGTGGTGGCAATGGTCGTGGAGGCCGAGGGCGAGGAGGACCCATGG GCCGTGGAGGCTATGGAGGTGGTGGCAGTGGTGGTGGTGGCCGAGGAGGATTTCCCAGTGGAGGTGGTGGCGGTGGAGGACAGCAGCGAG CTGGTGACTGGAAGTGTCCTAATCCCACCTGTGAGAATATGAACTTCTCTTGGAGGAATGAATGCAACCAGTGTAAGGCCCCTAAACCAG ATGGCCCAGGAGGGGGACCAGGTGGCTCTCACATGGGGGGTAACTACGGGGATGATCGTCGTGGTGGCAGAGGAGGCTATGATCGAGGCG GCTACCGGGGCCGCGGCGGGGACCGTGGAGGCTTCCGAGGGGGCCGGGGTGGTGGGGACAGAGGTGGCTTTGGCCCTGGCAAGATGGATT CCAGGGGTGAGCACAGACAGGATCGCAGGGAGAGGCCGTATTAATTAGCCTGGCTCCCCAGGTTCTGGAACAGCTTTTTGTCCTGTACCC AGTGTTACCCTCGTTATTTTGTAACCTTCCAATTCCTGATCACCCAAGGGTTTTTTTGTGTCGGACTATGTAATTGTAACTATACCTCTG >59799_59799_12_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000547619_FUS_chr16_31199646_ENST00000568685_length(amino acids)=354AA_BP=210 MMAAAPIQQNGTHTGVPIDLDPPDSRKRPLEAPPEAGSTKRTNTGEDGQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKD FYPGPRDQGSRHDSAEQDNSDNNTIFVQGLGENVTIESVADYFKQIGIIKTNKKTGQPMINLYTDRETGKLKGEATVSFDDPPSAKAAID WFDGKEFSGNPIKVSFATRRADFNRGGGNGRGGRGRGGPMGRGGYGGGGSGGGGRGGFPSGGGGGGGQQRAGDWKCPNPTCENMNFSWRN -------------------------------------------------------------- >59799_59799_13_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000574031_FUS_chr16_31199646_ENST00000254108_length(transcript)=1454nt_BP=304nt GTAGCAGCGGCAGGAACTGCAAACATGATGGCGGCAGCTCCCATCCAGCAGAACGGGACCCACACTGGGGTTCCCATAGACCTGGACCCG CCGGACTCGCGGAAAAGGCCGCTGGAAGCCCCCCCTGAAGCCGGCAGCACCAAGAGGACCAATACGGGCGAAGACGGCCAGTATTTTCTA AAGGTTCTCATACCTAGTTATGCTGCTGGATCTATAATTGGGAAGGGAGGACAGACAATTGTTCAGTTGCAAAAAGAAACTGGAGCCACC ATCAAGCTGTCTAAGTCCAAAGATTTTTACCCAGGCCCTCGGGACCAAGGATCACGTCATGACTCCGAACAGGATAATTCAGACAACAAC ACCATCTTTGTGCAAGGCCTGGGTGAGAATGTTACAATTGAGTCTGTGGCTGATTACTTCAAGCAGATTGGTATTATTAAGACAAACAAG AAAACGGGACAGCCCATGATTAATTTGTACACAGACAGGGAAACTGGCAAGCTGAAGGGAGAGGCAACGGTCTCTTTTGATGACCCACCT TCAGCTAAAGCAGCTATTGACTGGTTTGATGGTAAAGAATTCTCCGGAAATCCTATCAAGGTCTCATTTGCTACTCGCCGGGCAGACTTT AATCGGGGTGGTGGCAATGGTCGTGGAGGCCGAGGGCGAGGAGGACCCATGGGCCGTGGAGGCTATGGAGGTGGTGGCAGTGGTGGTGGT GGCCGAGGAGGATTTCCCAGTGGAGGTGGTGGCGGTGGAGGACAGCAGCGAGCTGGTGACTGGAAGTGTCCTAATCCCACCTGTGAGAAT ATGAACTTCTCTTGGAGGAATGAATGCAACCAGTGTAAGGCCCCTAAACCAGATGGCCCAGGAGGGGGACCAGGTGGCTCTCACATGGGG GGTAACTACGGGGATGATCGTCGTGGTGGCAGAGGAGGCTATGATCGAGGCGGCTACCGGGGCCGCGGCGGGGACCGTGGAGGCTTCCGA GGGGGCCGGGGTGGTGGGGACAGAGGTGGCTTTGGCCCTGGCAAGATGGATTCCAGGGGTGAGCACAGACAGGATCGCAGGGAGAGGCCG TATTAATTAGCCTGGCTCCCCAGGTTCTGGAACAGCTTTTTGTCCTGTACCCAGTGTTACCCTCGTTATTTTGTAACCTTCCAATTCCTG ATCACCCAAGGGTTTTTTTGTGTCGGACTATGTAATTGTAACTATACCTCTGGTTCCCATTAAAAGTGACCATTTTAGTTAAATTTTGTT CCTCTTCCCCCTTTTCACTTTCCTGGAAGATCGATGTCCCGATCAGGAAGGTAGAGAGTTTTCCTGTTCAGATTACCCTGCCCAGCAGGA ACTGGAATACAGTGTTCGGGGAGAAGGCCAAATGATATCCTTGAGAGCAGAGATTAAACTTTTCTGTCATGGGGAAAGTTGGTGTATAAA >59799_59799_13_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000574031_FUS_chr16_31199646_ENST00000254108_length(amino acids)=353AA_BP=209 MMAAAPIQQNGTHTGVPIDLDPPDSRKRPLEAPPEAGSTKRTNTGEDGQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKD FYPGPRDQGSRHDSEQDNSDNNTIFVQGLGENVTIESVADYFKQIGIIKTNKKTGQPMINLYTDRETGKLKGEATVSFDDPPSAKAAIDW FDGKEFSGNPIKVSFATRRADFNRGGGNGRGGRGRGGPMGRGGYGGGGSGGGGRGGFPSGGGGGGGQQRAGDWKCPNPTCENMNFSWRNE -------------------------------------------------------------- >59799_59799_14_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000574031_FUS_chr16_31199646_ENST00000380244_length(transcript)=1250nt_BP=304nt GTAGCAGCGGCAGGAACTGCAAACATGATGGCGGCAGCTCCCATCCAGCAGAACGGGACCCACACTGGGGTTCCCATAGACCTGGACCCG CCGGACTCGCGGAAAAGGCCGCTGGAAGCCCCCCCTGAAGCCGGCAGCACCAAGAGGACCAATACGGGCGAAGACGGCCAGTATTTTCTA AAGGTTCTCATACCTAGTTATGCTGCTGGATCTATAATTGGGAAGGGAGGACAGACAATTGTTCAGTTGCAAAAAGAAACTGGAGCCACC ATCAAGCTGTCTAAGTCCAAAGATTTTTACCCAGGCCCTCGGGACCAAGGATCACGTCATGACTCCGAACAGGATAATTCAGACAACAAC ACCATCTTTGTGCAAGGCCTGGGTGAGAATGTTACAATTGAGTCTGTGGCTGATTACTTCAAGCAGATTGGTATTATTAAGACAAACAAG AAAACGGGACAGCCCATGATTAATTTGTACACAGACAGGGAAACTGGCAAGCTGAAGGGAGAGGCAACGGTCTCTTTTGATGACCCACCT TCAGCTAAAGCAGCTATTGACTGGTTTGATGGTAAAGAATTCTCCGGAAATCCTATCAAGGTCTCATTTGCTACTCGCCGGGCAGACTTT AATCGGGGTGGTGGCAATGGTCGTGGAGGCCGAGGGCGAGGAGGACCCATGGGCCGTGGAGGCTATGGAGGTGGTGGCAGTGGTGGTGGT GGCCGAGGAGGATTTCCCAGTGGAGGTGGTGGCGGTGGAGGACAGCAGCGAGCTGGTGACTGGAAGTGTCCTAATCCCACCTGTGAGAAT ATGAACTTCTCTTGGAGGAATGAATGCAACCAGTGTAAGGCCCCTAAACCAGATGGCCCAGGAGGGGGACCAGGTGGCTCTCACATGGGG GGTAACTACGGGGATGATCGTCGTGGTGGCAGAGGAGGCTATGATCGAGGCGGCTACCGGGGCCGCGGCGGGGACCGTGGAGGCTTCCGA GGGGGCCGGGGTGGTGGGGACAGAGGTGGCTTTGGCCCTGGCAAGATGGATTCCAGGGGTGAGCACAGACAGGATCGCAGGGAGAGGCCG TATTAATTAGCCTGGCTCCCCAGGTTCTGGAACAGCTTTTTGTCCTGTACCCAGTGTTACCCTCGTTATTTTGTAACCTTCCAATTCCTG >59799_59799_14_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000574031_FUS_chr16_31199646_ENST00000380244_length(amino acids)=353AA_BP=209 MMAAAPIQQNGTHTGVPIDLDPPDSRKRPLEAPPEAGSTKRTNTGEDGQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKD FYPGPRDQGSRHDSEQDNSDNNTIFVQGLGENVTIESVADYFKQIGIIKTNKKTGQPMINLYTDRETGKLKGEATVSFDDPPSAKAAIDW FDGKEFSGNPIKVSFATRRADFNRGGGNGRGGRGRGGPMGRGGYGGGGSGGGGRGGFPSGGGGGGGQQRAGDWKCPNPTCENMNFSWRNE -------------------------------------------------------------- >59799_59799_15_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000574031_FUS_chr16_31199646_ENST00000568685_length(transcript)=1226nt_BP=304nt GTAGCAGCGGCAGGAACTGCAAACATGATGGCGGCAGCTCCCATCCAGCAGAACGGGACCCACACTGGGGTTCCCATAGACCTGGACCCG CCGGACTCGCGGAAAAGGCCGCTGGAAGCCCCCCCTGAAGCCGGCAGCACCAAGAGGACCAATACGGGCGAAGACGGCCAGTATTTTCTA AAGGTTCTCATACCTAGTTATGCTGCTGGATCTATAATTGGGAAGGGAGGACAGACAATTGTTCAGTTGCAAAAAGAAACTGGAGCCACC ATCAAGCTGTCTAAGTCCAAAGATTTTTACCCAGGCCCTCGGGACCAAGGATCACGTCATGACTCCGCAGAACAGGATAATTCAGACAAC AACACCATCTTTGTGCAAGGCCTGGGTGAGAATGTTACAATTGAGTCTGTGGCTGATTACTTCAAGCAGATTGGTATTATTAAGACAAAC AAGAAAACGGGACAGCCCATGATTAATTTGTACACAGACAGGGAAACTGGCAAGCTGAAGGGAGAGGCAACGGTCTCTTTTGATGACCCA CCTTCAGCTAAAGCAGCTATTGACTGGTTTGATGGTAAAGAATTCTCCGGAAATCCTATCAAGGTCTCATTTGCTACTCGCCGGGCAGAC TTTAATCGGGGTGGTGGCAATGGTCGTGGAGGCCGAGGGCGAGGAGGACCCATGGGCCGTGGAGGCTATGGAGGTGGTGGCAGTGGTGGT GGTGGCCGAGGAGGATTTCCCAGTGGAGGTGGTGGCGGTGGAGGACAGCAGCGAGCTGGTGACTGGAAGTGTCCTAATCCCACCTGTGAG AATATGAACTTCTCTTGGAGGAATGAATGCAACCAGTGTAAGGCCCCTAAACCAGATGGCCCAGGAGGGGGACCAGGTGGCTCTCACATG GGGGGTAACTACGGGGATGATCGTCGTGGTGGCAGAGGAGGCTATGATCGAGGCGGCTACCGGGGCCGCGGCGGGGACCGTGGAGGCTTC CGAGGGGGCCGGGGTGGTGGGGACAGAGGTGGCTTTGGCCCTGGCAAGATGGATTCCAGGGGTGAGCACAGACAGGATCGCAGGGAGAGG CCGTATTAATTAGCCTGGCTCCCCAGGTTCTGGAACAGCTTTTTGTCCTGTACCCAGTGTTACCCTCGTTATTTTGTAACCTTCCAATTC >59799_59799_15_NOVA1-FUS_NOVA1_chr14_27064616_ENST00000574031_FUS_chr16_31199646_ENST00000568685_length(amino acids)=354AA_BP=210 MMAAAPIQQNGTHTGVPIDLDPPDSRKRPLEAPPEAGSTKRTNTGEDGQYFLKVLIPSYAAGSIIGKGGQTIVQLQKETGATIKLSKSKD FYPGPRDQGSRHDSAEQDNSDNNTIFVQGLGENVTIESVADYFKQIGIIKTNKKTGQPMINLYTDRETGKLKGEATVSFDDPPSAKAAID WFDGKEFSGNPIKVSFATRRADFNRGGGNGRGGRGRGGPMGRGGYGGGGSGGGGRGGFPSGGGGGGGQQRAGDWKCPNPTCENMNFSWRN -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for NOVA1-FUS |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for NOVA1-FUS |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for NOVA1-FUS |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
