
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:NPEPPS-CREB3L1 (FusionGDB2 ID:59877) |
Fusion Gene Summary for NPEPPS-CREB3L1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: NPEPPS-CREB3L1 | Fusion gene ID: 59877 | Hgene | Tgene | Gene symbol | NPEPPS | CREB3L1 | Gene ID | 9520 | 90993 |
| Gene name | aminopeptidase puromycin sensitive | cAMP responsive element binding protein 3 like 1 | |
| Synonyms | AAP-S|MP100|PSA | OASIS|OI16 | |
| Cytomap | 17q21.32 | 11p11.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | puromycin-sensitive aminopeptidasecytosol alanyl aminopeptidasemetalloproteinase MP100 | cyclic AMP-responsive element-binding protein 3-like protein 1BBF-2 homologcAMP-responsive element-binding protein 3-like protein 1old astrocyte specifically-induced substance | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | P55786 | Q96BA8 | |
| Ensembl transtripts involved in fusion gene | ENST00000525037, ENST00000322157, ENST00000530173, ENST00000544660, | ENST00000288400, ENST00000529193, ENST00000534616, | |
| Fusion gene scores | * DoF score | 18 X 9 X 9=1458 | 10 X 10 X 4=400 |
| # samples | 19 | 8 | |
| ** MAII score | log2(19/1458*10)=-2.93991939599599 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(8/400*10)=-2.32192809488736 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: NPEPPS [Title/Abstract] AND CREB3L1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | NPEPPS(45623359)-CREB3L1(46341815), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | NPEPPS | GO:0071456 | cellular response to hypoxia | 21056661 |
| Tgene | CREB3L1 | GO:0010629 | negative regulation of gene expression | 27121396 |
| Tgene | CREB3L1 | GO:0032967 | positive regulation of collagen biosynthetic process | 25310401 |
| Tgene | CREB3L1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway | 27121396 |
| Tgene | CREB3L1 | GO:0045892 | negative regulation of transcription, DNA-templated | 27121396 |
 Fusion gene breakpoints across NPEPPS (5'-gene) Fusion gene breakpoints across NPEPPS (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
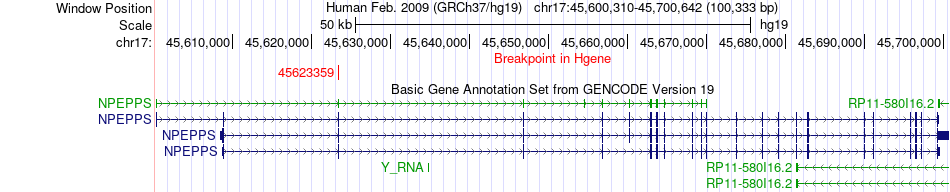 |
 Fusion gene breakpoints across CREB3L1 (3'-gene) Fusion gene breakpoints across CREB3L1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
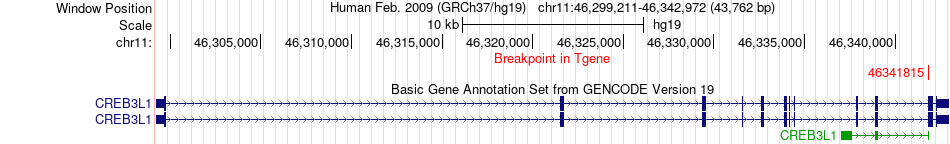 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | UCS | TCGA-NA-A5I1-01A | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + |
| ChimerDB4 | UCS | TCGA-NA-A5I1 | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + |
Top |
Fusion Gene ORF analysis for NPEPPS-CREB3L1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000525037 | ENST00000288400 | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + |
| 3UTR-3CDS | ENST00000525037 | ENST00000529193 | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + |
| 3UTR-3UTR | ENST00000525037 | ENST00000534616 | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + |
| 5CDS-3UTR | ENST00000322157 | ENST00000534616 | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + |
| 5CDS-3UTR | ENST00000530173 | ENST00000534616 | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + |
| 5CDS-3UTR | ENST00000544660 | ENST00000534616 | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + |
| In-frame | ENST00000322157 | ENST00000288400 | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + |
| In-frame | ENST00000322157 | ENST00000529193 | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + |
| In-frame | ENST00000530173 | ENST00000288400 | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + |
| In-frame | ENST00000530173 | ENST00000529193 | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + |
| In-frame | ENST00000544660 | ENST00000288400 | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + |
| In-frame | ENST00000544660 | ENST00000529193 | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000530173 | NPEPPS | chr17 | 45623359 | + | ENST00000529193 | CREB3L1 | chr11 | 46341815 | + | 1336 | 358 | 30 | 659 | 209 |
| ENST00000530173 | NPEPPS | chr17 | 45623359 | + | ENST00000288400 | CREB3L1 | chr11 | 46341815 | + | 1334 | 358 | 30 | 659 | 209 |
| ENST00000322157 | NPEPPS | chr17 | 45623359 | + | ENST00000529193 | CREB3L1 | chr11 | 46341815 | + | 1555 | 577 | 66 | 878 | 270 |
| ENST00000322157 | NPEPPS | chr17 | 45623359 | + | ENST00000288400 | CREB3L1 | chr11 | 46341815 | + | 1553 | 577 | 66 | 878 | 270 |
| ENST00000544660 | NPEPPS | chr17 | 45623359 | + | ENST00000529193 | CREB3L1 | chr11 | 46341815 | + | 1292 | 314 | 13 | 615 | 200 |
| ENST00000544660 | NPEPPS | chr17 | 45623359 | + | ENST00000288400 | CREB3L1 | chr11 | 46341815 | + | 1290 | 314 | 13 | 615 | 200 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000530173 | ENST00000529193 | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + | 0.025895497 | 0.97410446 |
| ENST00000530173 | ENST00000288400 | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + | 0.026064781 | 0.9739352 |
| ENST00000322157 | ENST00000529193 | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + | 0.044976186 | 0.95502377 |
| ENST00000322157 | ENST00000288400 | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + | 0.046533797 | 0.9534662 |
| ENST00000544660 | ENST00000529193 | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + | 0.009365995 | 0.9906341 |
| ENST00000544660 | ENST00000288400 | NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341815 | + | 0.009596954 | 0.99040306 |
Top |
Fusion Genomic Features for NPEPPS-CREB3L1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341814 | + | 2.60E-08 | 1 |
| NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341814 | + | 2.60E-08 | 1 |
| NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341814 | + | 2.60E-08 | 1 |
| NPEPPS | chr17 | 45623359 | + | CREB3L1 | chr11 | 46341814 | + | 2.60E-08 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
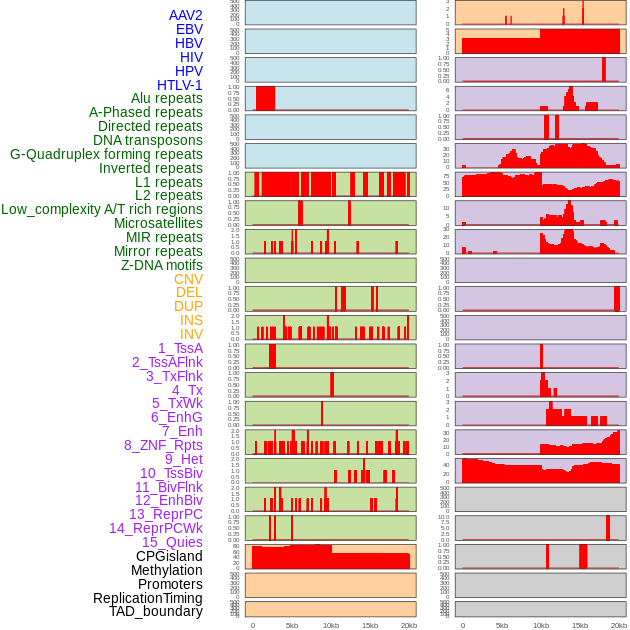 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
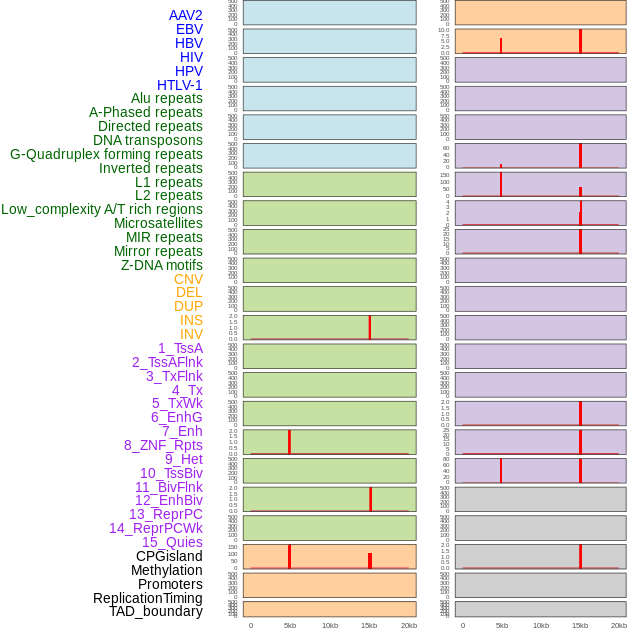 |
Top |
Fusion Protein Features for NPEPPS-CREB3L1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:45623359/chr11:46341815) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NPEPPS | CREB3L1 |
| FUNCTION: Aminopeptidase with broad substrate specificity for several peptides. Involved in proteolytic events essential for cell growth and viability. May act as regulator of neuropeptide activity. Plays a role in the antigen-processing pathway for MHC class I molecules. Involved in the N-terminal trimming of cytotoxic T-cell epitope precursors. Digests the poly-Q peptides found in many cellular proteins. Digests tau from normal brain more efficiently than tau from Alzheimer disease brain. {ECO:0000269|PubMed:10978616, ECO:0000269|PubMed:11062501, ECO:0000269|PubMed:17154549, ECO:0000269|PubMed:17318184, ECO:0000269|PubMed:19917696}. | FUNCTION: Transcription factor involved in unfolded protein response (UPR). Binds the DNA consensus sequence 5'-GTGXGCXGC-3' (PubMed:21767813). In the absence of endoplasmic reticulum (ER) stress, inserted into ER membranes, with N-terminal DNA-binding and transcription activation domains oriented toward the cytosolic face of the membrane. In response to ER stress, transported to the Golgi, where it is cleaved in a site-specific manner by resident proteases S1P/MBTPS1 and S2P/MBTPS2. The released N-terminal cytosolic domain is translocated to the nucleus to effect transcription of specific target genes. Plays a critical role in bone formation through the transcription of COL1A1, and possibly COL1A2, and the secretion of bone matrix proteins. Directly binds to the UPR element (UPRE)-like sequence in an osteoblast-specific COL1A1 promoter region and induces its transcription. Does not regulate COL1A1 in other tissues, such as skin (By similarity). Required to protect astrocytes from ER stress-induced cell death. In astrocytes, binds to the cAMP response element (CRE) of the BiP/HSPA5 promoter and participate in its transcriptional activation (By similarity). Required for TGFB1 to activate genes involved in the assembly of collagen extracellular matrix (PubMed:25310401). {ECO:0000250|UniProtKB:Q9Z125, ECO:0000269|PubMed:12054625, ECO:0000269|PubMed:21767813, ECO:0000269|PubMed:25310401}.; FUNCTION: (Microbial infection) May play a role in limiting virus spread by inhibiting proliferation of virus-infected cells. Upon infection with diverse DNA and RNA viruses, inhibits cell-cycle progression by binding to promoters and activating transcription of genes encoding cell-cycle inhibitors, such as p21/CDKN1A (PubMed:21767813). {ECO:0000269|PubMed:21767813}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000288400 | 9 | 12 | 423_426 | 419 | 520.0 | Motif | S1P recognition | |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000529193 | 9 | 12 | 423_426 | 419 | 520.0 | Motif | S1P recognition |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NPEPPS | chr17:45623359 | chr11:46341815 | ENST00000322157 | + | 2 | 23 | 726_730 | 113 | 920.0 | Motif | Nuclear localization signal |
| Hgene | NPEPPS | chr17:45623359 | chr11:46341815 | ENST00000322157 | + | 2 | 23 | 316_320 | 113 | 920.0 | Region | Substrate binding |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000288400 | 9 | 12 | 146_154 | 419 | 520.0 | Compositional bias | Note=Poly-Ala | |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000529193 | 9 | 12 | 146_154 | 419 | 520.0 | Compositional bias | Note=Poly-Ala | |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000288400 | 9 | 12 | 290_353 | 419 | 520.0 | Domain | bZIP | |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000529193 | 9 | 12 | 290_353 | 419 | 520.0 | Domain | bZIP | |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000288400 | 9 | 12 | 392_395 | 419 | 520.0 | Motif | S2P recognition | |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000529193 | 9 | 12 | 392_395 | 419 | 520.0 | Motif | S2P recognition | |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000288400 | 9 | 12 | 1_60 | 419 | 520.0 | Region | Note=Required for transcriptional activation | |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000288400 | 9 | 12 | 292_321 | 419 | 520.0 | Region | Basic motif | |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000288400 | 9 | 12 | 332_353 | 419 | 520.0 | Region | Leucine-zipper | |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000529193 | 9 | 12 | 1_60 | 419 | 520.0 | Region | Note=Required for transcriptional activation | |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000529193 | 9 | 12 | 292_321 | 419 | 520.0 | Region | Basic motif | |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000529193 | 9 | 12 | 332_353 | 419 | 520.0 | Region | Leucine-zipper | |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000288400 | 9 | 12 | 1_374 | 419 | 520.0 | Topological domain | Cytoplasmic | |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000288400 | 9 | 12 | 396_519 | 419 | 520.0 | Topological domain | Lumenal | |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000529193 | 9 | 12 | 1_374 | 419 | 520.0 | Topological domain | Cytoplasmic | |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000529193 | 9 | 12 | 396_519 | 419 | 520.0 | Topological domain | Lumenal | |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000288400 | 9 | 12 | 375_395 | 419 | 520.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | CREB3L1 | chr17:45623359 | chr11:46341815 | ENST00000529193 | 9 | 12 | 375_395 | 419 | 520.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
Top |
Fusion Gene Sequence for NPEPPS-CREB3L1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >59877_59877_1_NPEPPS-CREB3L1_NPEPPS_chr17_45623359_ENST00000322157_CREB3L1_chr11_46341815_ENST00000288400_length(transcript)=1553nt_BP=577nt GGGCGCGCCCGGCCGGCGGGGCCGCGCACGCAGCCGCCGCCACCACTTCCCCCTCTCCCTCCCTCCTTGCGGGCCCTCCTCCCCTTCCCT CCCCTCCGCCCCCTTCCCCGTAGGCAGCCCGCCCGCCAGTCCGCCCGCACCGCCTCCTTCCCAGCCCCTAGCGCTCCGGCTGGGTCTCTC CCCCGCCCCCCAGGCTCCCCCGGTCGCTCTCCTCCGGCGGTCGCCCGCGCTCGGTGGATGTGGCTGGCAGCTGCCGCCCCCTCCCTCGCT CGCCGCCTGCTCTTCCTCGGCCCTCCGCCTCCTCCCCTCCTCCTTCTCGTCTTCAGCCGCTCCTCTCGCCGCCGCCTCCACAGCCTGGGC CTCGCCGCGATGCCGGAGAAGAGGCCCTTCGAGCGGCTGCCTGCCGATGTCTCCCCCATCAACTACAGCCTTTGCCTCAAGCCCGACTTG CTGGACTTCACCTTCGAGGGCAAGCTGGAGGCCGCCGCCCAGGTGAGGCAGGCGACTAATCAGATTGTGATGAATTGTGCTGATATTGAT ATTATTACAGCTTCATATGCACCAGAAGGAGATGAAGTGCCCTCCCGAAGCCTCCTATTCTACGATGACGGGGCAGGCTTATGGGAAGAT GGCCGCAGCACCCTGCTGCCCATGGAGCCCCCAGATGGCTGGGAAATCAACCCCGGGGGGCCGGCAGAGCAGCGGCCCCGGGACCACCTG CAGCATGATCACCTGGACAGCACCCACGAGACCACCAAGTACCTGAGTGAGGCCTGGCCTAAAGACGGTGGAAACGGCACCAGCCCCGAC TTCTCCCACTCCAAGGAGTGGTTCCACGACAGGGATCTGGGCCCCAACACCACCATCAAACTCTCCTAGGCCATGCCAAGACCCAGGACA TAGGACGGACCCCTGGTACCCAGAAGAGGAGTTCTTGCTCACTAACCCGGATCCGCCTCGTGCCCCTGCCTCCTGGAGCTTCCCATTCCA GGAGAAAAGGCTCCACTTCCCAGCCCTTCCTTGCCCCTGACATTTGGACTCTTCCCTTGGGCCGACCACTCTGTTCTCATTCTCCTTCCC ACCAACATCCATCCGTCCTTCTCAGACAAACCACTCACTGGGTACCCCACCTCCTCTCTCATATGCCCAACACGACCACTGCCTCCCTGC CCCCACACCTGCACCCAAACAGACACATCAACGCACCCCACTCACAGACACCCCTTACCCCACCCCCACTGTACAGAGACCAAGAACAGA AATTGTTTGTAAATAATGAACCTTATTTTTTATTATTGCCAATCCCCTAAGATATTGTATTTTACAAATCTCCCTCTTCCCTTCGCCCCT CCCTTGTTTTATATTTTATGAAGTTAGTGCGGGCTTTGCTGCTCCCTGGCCCAGGAAAGAGGGACTACCTGACCCTCACCTGGCACCCCC CTGCTGCTGCCCAAGCCGCTGGGCCTTTTTAATTGCCAAACTGCTCTCTTCATCAGCTCAGCACATGCTTTAAGAAAGCAAAACCAAAAA >59877_59877_1_NPEPPS-CREB3L1_NPEPPS_chr17_45623359_ENST00000322157_CREB3L1_chr11_46341815_ENST00000288400_length(amino acids)=270AA_BP=170 MRALLPFPPLRPLPRRQPARQSARTASFPAPSAPAGSLPRPPGSPGRSPPAVARARWMWLAAAAPSLARRLLFLGPPPPPLLLLVFSRSS RRRLHSLGLAAMPEKRPFERLPADVSPINYSLCLKPDLLDFTFEGKLEAAAQVRQATNQIVMNCADIDIITASYAPEGDEVPSRSLLFYD DGAGLWEDGRSTLLPMEPPDGWEINPGGPAEQRPRDHLQHDHLDSTHETTKYLSEAWPKDGGNGTSPDFSHSKEWFHDRDLGPNTTIKLS -------------------------------------------------------------- >59877_59877_2_NPEPPS-CREB3L1_NPEPPS_chr17_45623359_ENST00000322157_CREB3L1_chr11_46341815_ENST00000529193_length(transcript)=1555nt_BP=577nt GGGCGCGCCCGGCCGGCGGGGCCGCGCACGCAGCCGCCGCCACCACTTCCCCCTCTCCCTCCCTCCTTGCGGGCCCTCCTCCCCTTCCCT CCCCTCCGCCCCCTTCCCCGTAGGCAGCCCGCCCGCCAGTCCGCCCGCACCGCCTCCTTCCCAGCCCCTAGCGCTCCGGCTGGGTCTCTC CCCCGCCCCCCAGGCTCCCCCGGTCGCTCTCCTCCGGCGGTCGCCCGCGCTCGGTGGATGTGGCTGGCAGCTGCCGCCCCCTCCCTCGCT CGCCGCCTGCTCTTCCTCGGCCCTCCGCCTCCTCCCCTCCTCCTTCTCGTCTTCAGCCGCTCCTCTCGCCGCCGCCTCCACAGCCTGGGC CTCGCCGCGATGCCGGAGAAGAGGCCCTTCGAGCGGCTGCCTGCCGATGTCTCCCCCATCAACTACAGCCTTTGCCTCAAGCCCGACTTG CTGGACTTCACCTTCGAGGGCAAGCTGGAGGCCGCCGCCCAGGTGAGGCAGGCGACTAATCAGATTGTGATGAATTGTGCTGATATTGAT ATTATTACAGCTTCATATGCACCAGAAGGAGATGAAGTGCCCTCCCGAAGCCTCCTATTCTACGATGACGGGGCAGGCTTATGGGAAGAT GGCCGCAGCACCCTGCTGCCCATGGAGCCCCCAGATGGCTGGGAAATCAACCCCGGGGGGCCGGCAGAGCAGCGGCCCCGGGACCACCTG CAGCATGATCACCTGGACAGCACCCACGAGACCACCAAGTACCTGAGTGAGGCCTGGCCTAAAGACGGTGGAAACGGCACCAGCCCCGAC TTCTCCCACTCCAAGGAGTGGTTCCACGACAGGGATCTGGGCCCCAACACCACCATCAAACTCTCCTAGGCCATGCCAAGACCCAGGACA TAGGACGGACCCCTGGTACCCAGAAGAGGAGTTCTTGCTCACTAACCCGGATCCGCCTCGTGCCCCTGCCTCCTGGAGCTTCCCATTCCA GGAGAAAAGGCTCCACTTCCCAGCCCTTCCTTGCCCCTGACATTTGGACTCTTCCCTTGGGCCGACCACTCTGTTCTCATTCTCCTTCCC ACCAACATCCATCCGTCCTTCTCAGACAAACCACTCACTGGGTACCCCACCTCCTCTCTCATATGCCCAACACGACCACTGCCTCCCTGC CCCCACACCTGCACCCAAACAGACACATCAACGCACCCCACTCACAGACACCCCTTACCCCACCCCCACTGTACAGAGACCAAGAACAGA AATTGTTTGTAAATAATGAACCTTATTTTTTATTATTGCCAATCCCCTAAGATATTGTATTTTACAAATCTCCCTCTTCCCTTCGCCCCT CCCTTGTTTTATATTTTATGAAGTTAGTGCGGGCTTTGCTGCTCCCTGGCCCAGGAAAGAGGGACTACCTGACCCTCACCTGGCACCCCC CTGCTGCTGCCCAAGCCGCTGGGCCTTTTTAATTGCCAAACTGCTCTCTTCATCAGCTCAGCACATGCTTTAAGAAAGCAAAACCAAAAA >59877_59877_2_NPEPPS-CREB3L1_NPEPPS_chr17_45623359_ENST00000322157_CREB3L1_chr11_46341815_ENST00000529193_length(amino acids)=270AA_BP=170 MRALLPFPPLRPLPRRQPARQSARTASFPAPSAPAGSLPRPPGSPGRSPPAVARARWMWLAAAAPSLARRLLFLGPPPPPLLLLVFSRSS RRRLHSLGLAAMPEKRPFERLPADVSPINYSLCLKPDLLDFTFEGKLEAAAQVRQATNQIVMNCADIDIITASYAPEGDEVPSRSLLFYD DGAGLWEDGRSTLLPMEPPDGWEINPGGPAEQRPRDHLQHDHLDSTHETTKYLSEAWPKDGGNGTSPDFSHSKEWFHDRDLGPNTTIKLS -------------------------------------------------------------- >59877_59877_3_NPEPPS-CREB3L1_NPEPPS_chr17_45623359_ENST00000530173_CREB3L1_chr11_46341815_ENST00000288400_length(transcript)=1334nt_BP=358nt AAGTTACGGAAGAGTCAAGGCTGGGACCTCATGGCATCATTTTCCATGGATTGCTCTCCAAGTTTTTGTGTTCCTGGCCTTTGGAACCCC TTCATCATATTCCGCAGCCGCTCCTCTCGCCGCCGCCTCCACAGCCTGGGCCTCGCCGCGATGCCGGAGAAGAGGCCCTTCGAGCGGCTG CCTGCCGATGTCTCCCCCATCAACTACAGCCTTTGCCTCAAGCCCGACTTGCTGGACTTCACCTTCGAGGGCAAGCTGGAGGCCGCCGCC CAGGTGAGGCAGGCGACTAATCAGATTGTGATGAATTGTGCTGATATTGATATTATTACAGCTTCATATGCACCAGAAGGAGATGAAGTG CCCTCCCGAAGCCTCCTATTCTACGATGACGGGGCAGGCTTATGGGAAGATGGCCGCAGCACCCTGCTGCCCATGGAGCCCCCAGATGGC TGGGAAATCAACCCCGGGGGGCCGGCAGAGCAGCGGCCCCGGGACCACCTGCAGCATGATCACCTGGACAGCACCCACGAGACCACCAAG TACCTGAGTGAGGCCTGGCCTAAAGACGGTGGAAACGGCACCAGCCCCGACTTCTCCCACTCCAAGGAGTGGTTCCACGACAGGGATCTG GGCCCCAACACCACCATCAAACTCTCCTAGGCCATGCCAAGACCCAGGACATAGGACGGACCCCTGGTACCCAGAAGAGGAGTTCTTGCT CACTAACCCGGATCCGCCTCGTGCCCCTGCCTCCTGGAGCTTCCCATTCCAGGAGAAAAGGCTCCACTTCCCAGCCCTTCCTTGCCCCTG ACATTTGGACTCTTCCCTTGGGCCGACCACTCTGTTCTCATTCTCCTTCCCACCAACATCCATCCGTCCTTCTCAGACAAACCACTCACT GGGTACCCCACCTCCTCTCTCATATGCCCAACACGACCACTGCCTCCCTGCCCCCACACCTGCACCCAAACAGACACATCAACGCACCCC ACTCACAGACACCCCTTACCCCACCCCCACTGTACAGAGACCAAGAACAGAAATTGTTTGTAAATAATGAACCTTATTTTTTATTATTGC CAATCCCCTAAGATATTGTATTTTACAAATCTCCCTCTTCCCTTCGCCCCTCCCTTGTTTTATATTTTATGAAGTTAGTGCGGGCTTTGC TGCTCCCTGGCCCAGGAAAGAGGGACTACCTGACCCTCACCTGGCACCCCCCTGCTGCTGCCCAAGCCGCTGGGCCTTTTTAATTGCCAA >59877_59877_3_NPEPPS-CREB3L1_NPEPPS_chr17_45623359_ENST00000530173_CREB3L1_chr11_46341815_ENST00000288400_length(amino acids)=209AA_BP=109 MASFSMDCSPSFCVPGLWNPFIIFRSRSSRRRLHSLGLAAMPEKRPFERLPADVSPINYSLCLKPDLLDFTFEGKLEAAAQVRQATNQIV MNCADIDIITASYAPEGDEVPSRSLLFYDDGAGLWEDGRSTLLPMEPPDGWEINPGGPAEQRPRDHLQHDHLDSTHETTKYLSEAWPKDG -------------------------------------------------------------- >59877_59877_4_NPEPPS-CREB3L1_NPEPPS_chr17_45623359_ENST00000530173_CREB3L1_chr11_46341815_ENST00000529193_length(transcript)=1336nt_BP=358nt AAGTTACGGAAGAGTCAAGGCTGGGACCTCATGGCATCATTTTCCATGGATTGCTCTCCAAGTTTTTGTGTTCCTGGCCTTTGGAACCCC TTCATCATATTCCGCAGCCGCTCCTCTCGCCGCCGCCTCCACAGCCTGGGCCTCGCCGCGATGCCGGAGAAGAGGCCCTTCGAGCGGCTG CCTGCCGATGTCTCCCCCATCAACTACAGCCTTTGCCTCAAGCCCGACTTGCTGGACTTCACCTTCGAGGGCAAGCTGGAGGCCGCCGCC CAGGTGAGGCAGGCGACTAATCAGATTGTGATGAATTGTGCTGATATTGATATTATTACAGCTTCATATGCACCAGAAGGAGATGAAGTG CCCTCCCGAAGCCTCCTATTCTACGATGACGGGGCAGGCTTATGGGAAGATGGCCGCAGCACCCTGCTGCCCATGGAGCCCCCAGATGGC TGGGAAATCAACCCCGGGGGGCCGGCAGAGCAGCGGCCCCGGGACCACCTGCAGCATGATCACCTGGACAGCACCCACGAGACCACCAAG TACCTGAGTGAGGCCTGGCCTAAAGACGGTGGAAACGGCACCAGCCCCGACTTCTCCCACTCCAAGGAGTGGTTCCACGACAGGGATCTG GGCCCCAACACCACCATCAAACTCTCCTAGGCCATGCCAAGACCCAGGACATAGGACGGACCCCTGGTACCCAGAAGAGGAGTTCTTGCT CACTAACCCGGATCCGCCTCGTGCCCCTGCCTCCTGGAGCTTCCCATTCCAGGAGAAAAGGCTCCACTTCCCAGCCCTTCCTTGCCCCTG ACATTTGGACTCTTCCCTTGGGCCGACCACTCTGTTCTCATTCTCCTTCCCACCAACATCCATCCGTCCTTCTCAGACAAACCACTCACT GGGTACCCCACCTCCTCTCTCATATGCCCAACACGACCACTGCCTCCCTGCCCCCACACCTGCACCCAAACAGACACATCAACGCACCCC ACTCACAGACACCCCTTACCCCACCCCCACTGTACAGAGACCAAGAACAGAAATTGTTTGTAAATAATGAACCTTATTTTTTATTATTGC CAATCCCCTAAGATATTGTATTTTACAAATCTCCCTCTTCCCTTCGCCCCTCCCTTGTTTTATATTTTATGAAGTTAGTGCGGGCTTTGC TGCTCCCTGGCCCAGGAAAGAGGGACTACCTGACCCTCACCTGGCACCCCCCTGCTGCTGCCCAAGCCGCTGGGCCTTTTTAATTGCCAA >59877_59877_4_NPEPPS-CREB3L1_NPEPPS_chr17_45623359_ENST00000530173_CREB3L1_chr11_46341815_ENST00000529193_length(amino acids)=209AA_BP=109 MASFSMDCSPSFCVPGLWNPFIIFRSRSSRRRLHSLGLAAMPEKRPFERLPADVSPINYSLCLKPDLLDFTFEGKLEAAAQVRQATNQIV MNCADIDIITASYAPEGDEVPSRSLLFYDDGAGLWEDGRSTLLPMEPPDGWEINPGGPAEQRPRDHLQHDHLDSTHETTKYLSEAWPKDG -------------------------------------------------------------- >59877_59877_5_NPEPPS-CREB3L1_NPEPPS_chr17_45623359_ENST00000544660_CREB3L1_chr11_46341815_ENST00000288400_length(transcript)=1290nt_BP=314nt CCTCGCTCGCCGCCTGCTCTTCCTCGGCCCTCCGCCTCCTCCCCTCCTCCTTCTCGTCTTCAGCCGCTCCTCTCGCCGCCGCCTCCACAG CCTGGGCCTCGCCGCGATGCCGGAGAAGAGGCCCTTCGAGCGGCTGCCTGCCGATGTCTCCCCCATCAACTACAGCCTTTGCCTCAAGCC CGACTTGCTGGACTTCACCTTCGAGGGCAAGCTGGAGGCCGCCGCCCAGGTGAGGCAGGCGACTAATCAGATTGTGATGAATTGTGCTGA TATTGATATTATTACAGCTTCATATGCACCAGAAGGAGATGAAGTGCCCTCCCGAAGCCTCCTATTCTACGATGACGGGGCAGGCTTATG GGAAGATGGCCGCAGCACCCTGCTGCCCATGGAGCCCCCAGATGGCTGGGAAATCAACCCCGGGGGGCCGGCAGAGCAGCGGCCCCGGGA CCACCTGCAGCATGATCACCTGGACAGCACCCACGAGACCACCAAGTACCTGAGTGAGGCCTGGCCTAAAGACGGTGGAAACGGCACCAG CCCCGACTTCTCCCACTCCAAGGAGTGGTTCCACGACAGGGATCTGGGCCCCAACACCACCATCAAACTCTCCTAGGCCATGCCAAGACC CAGGACATAGGACGGACCCCTGGTACCCAGAAGAGGAGTTCTTGCTCACTAACCCGGATCCGCCTCGTGCCCCTGCCTCCTGGAGCTTCC CATTCCAGGAGAAAAGGCTCCACTTCCCAGCCCTTCCTTGCCCCTGACATTTGGACTCTTCCCTTGGGCCGACCACTCTGTTCTCATTCT CCTTCCCACCAACATCCATCCGTCCTTCTCAGACAAACCACTCACTGGGTACCCCACCTCCTCTCTCATATGCCCAACACGACCACTGCC TCCCTGCCCCCACACCTGCACCCAAACAGACACATCAACGCACCCCACTCACAGACACCCCTTACCCCACCCCCACTGTACAGAGACCAA GAACAGAAATTGTTTGTAAATAATGAACCTTATTTTTTATTATTGCCAATCCCCTAAGATATTGTATTTTACAAATCTCCCTCTTCCCTT CGCCCCTCCCTTGTTTTATATTTTATGAAGTTAGTGCGGGCTTTGCTGCTCCCTGGCCCAGGAAAGAGGGACTACCTGACCCTCACCTGG CACCCCCCTGCTGCTGCCCAAGCCGCTGGGCCTTTTTAATTGCCAAACTGCTCTCTTCATCAGCTCAGCACATGCTTTAAGAAAGCAAAA >59877_59877_5_NPEPPS-CREB3L1_NPEPPS_chr17_45623359_ENST00000544660_CREB3L1_chr11_46341815_ENST00000288400_length(amino acids)=200AA_BP=100 MLFLGPPPPPLLLLVFSRSSRRRLHSLGLAAMPEKRPFERLPADVSPINYSLCLKPDLLDFTFEGKLEAAAQVRQATNQIVMNCADIDII TASYAPEGDEVPSRSLLFYDDGAGLWEDGRSTLLPMEPPDGWEINPGGPAEQRPRDHLQHDHLDSTHETTKYLSEAWPKDGGNGTSPDFS -------------------------------------------------------------- >59877_59877_6_NPEPPS-CREB3L1_NPEPPS_chr17_45623359_ENST00000544660_CREB3L1_chr11_46341815_ENST00000529193_length(transcript)=1292nt_BP=314nt CCTCGCTCGCCGCCTGCTCTTCCTCGGCCCTCCGCCTCCTCCCCTCCTCCTTCTCGTCTTCAGCCGCTCCTCTCGCCGCCGCCTCCACAG CCTGGGCCTCGCCGCGATGCCGGAGAAGAGGCCCTTCGAGCGGCTGCCTGCCGATGTCTCCCCCATCAACTACAGCCTTTGCCTCAAGCC CGACTTGCTGGACTTCACCTTCGAGGGCAAGCTGGAGGCCGCCGCCCAGGTGAGGCAGGCGACTAATCAGATTGTGATGAATTGTGCTGA TATTGATATTATTACAGCTTCATATGCACCAGAAGGAGATGAAGTGCCCTCCCGAAGCCTCCTATTCTACGATGACGGGGCAGGCTTATG GGAAGATGGCCGCAGCACCCTGCTGCCCATGGAGCCCCCAGATGGCTGGGAAATCAACCCCGGGGGGCCGGCAGAGCAGCGGCCCCGGGA CCACCTGCAGCATGATCACCTGGACAGCACCCACGAGACCACCAAGTACCTGAGTGAGGCCTGGCCTAAAGACGGTGGAAACGGCACCAG CCCCGACTTCTCCCACTCCAAGGAGTGGTTCCACGACAGGGATCTGGGCCCCAACACCACCATCAAACTCTCCTAGGCCATGCCAAGACC CAGGACATAGGACGGACCCCTGGTACCCAGAAGAGGAGTTCTTGCTCACTAACCCGGATCCGCCTCGTGCCCCTGCCTCCTGGAGCTTCC CATTCCAGGAGAAAAGGCTCCACTTCCCAGCCCTTCCTTGCCCCTGACATTTGGACTCTTCCCTTGGGCCGACCACTCTGTTCTCATTCT CCTTCCCACCAACATCCATCCGTCCTTCTCAGACAAACCACTCACTGGGTACCCCACCTCCTCTCTCATATGCCCAACACGACCACTGCC TCCCTGCCCCCACACCTGCACCCAAACAGACACATCAACGCACCCCACTCACAGACACCCCTTACCCCACCCCCACTGTACAGAGACCAA GAACAGAAATTGTTTGTAAATAATGAACCTTATTTTTTATTATTGCCAATCCCCTAAGATATTGTATTTTACAAATCTCCCTCTTCCCTT CGCCCCTCCCTTGTTTTATATTTTATGAAGTTAGTGCGGGCTTTGCTGCTCCCTGGCCCAGGAAAGAGGGACTACCTGACCCTCACCTGG CACCCCCCTGCTGCTGCCCAAGCCGCTGGGCCTTTTTAATTGCCAAACTGCTCTCTTCATCAGCTCAGCACATGCTTTAAGAAAGCAAAA >59877_59877_6_NPEPPS-CREB3L1_NPEPPS_chr17_45623359_ENST00000544660_CREB3L1_chr11_46341815_ENST00000529193_length(amino acids)=200AA_BP=100 MLFLGPPPPPLLLLVFSRSSRRRLHSLGLAAMPEKRPFERLPADVSPINYSLCLKPDLLDFTFEGKLEAAAQVRQATNQIVMNCADIDII TASYAPEGDEVPSRSLLFYDDGAGLWEDGRSTLLPMEPPDGWEINPGGPAEQRPRDHLQHDHLDSTHETTKYLSEAWPKDGGNGTSPDFS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for NPEPPS-CREB3L1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for NPEPPS-CREB3L1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for NPEPPS-CREB3L1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
