
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:NPLOC4-ALYREF (FusionGDB2 ID:59941) |
Fusion Gene Summary for NPLOC4-ALYREF |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: NPLOC4-ALYREF | Fusion gene ID: 59941 | Hgene | Tgene | Gene symbol | NPLOC4 | ALYREF | Gene ID | 55666 | 10189 |
| Gene name | NPL4 homolog, ubiquitin recognition factor | Aly/REF export factor | |
| Synonyms | NPL4 | ALY|ALY/REF|BEF|REF|THOC4 | |
| Cytomap | 17q25.3 | 17q25.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | nuclear protein localization protein 4 homologNPLOC4 ubiquitin recognition factornuclear protein localization 4 homolog | THO complex subunit 4THO complex 4ally of AML-1 and LEF-1bZIP enhancing factorbZIP-enhancing factor BEFtho4transcriptional coactivator Aly/REF | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q8TAT6 | Q86V81 | |
| Ensembl transtripts involved in fusion gene | ENST00000331134, ENST00000374747, ENST00000539314, ENST00000572760, ENST00000573876, ENST00000574344, | ENST00000512673, ENST00000331204, ENST00000505490, | |
| Fusion gene scores | * DoF score | 19 X 17 X 11=3553 | 3 X 3 X 3=27 |
| # samples | 30 | 4 | |
| ** MAII score | log2(30/3553*10)=-3.56600328283534 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/27*10)=0.567040592723894 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: NPLOC4 [Title/Abstract] AND ALYREF [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | NPLOC4(79567367)-ALYREF(79848696), # samples:2 NPLOC4(79589192)-ALYREF(79846021), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | NPLOC4-ALYREF seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. NPLOC4-ALYREF seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | ALYREF | GO:0006405 | RNA export from nucleus | 28418038 |
| Tgene | ALYREF | GO:0046784 | viral mRNA export from host cell nucleus | 18974867 |
 Fusion gene breakpoints across NPLOC4 (5'-gene) Fusion gene breakpoints across NPLOC4 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
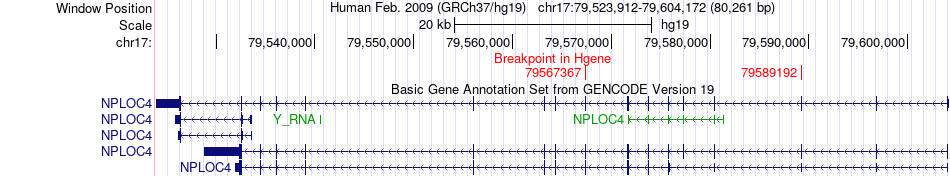 |
 Fusion gene breakpoints across ALYREF (3'-gene) Fusion gene breakpoints across ALYREF (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
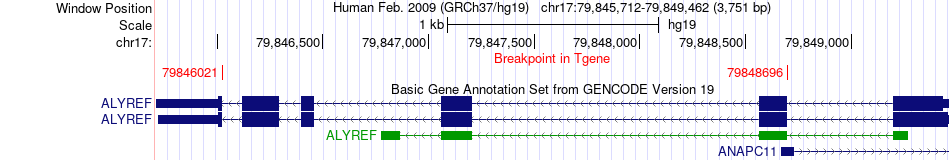 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LIHC | TCGA-FV-A496-01A | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - |
| ChimerDB4 | READ | TCGA-AF-6655-01A | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
| ChimerDB4 | READ | TCGA-AF-6655 | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
Top |
Fusion Gene ORF analysis for NPLOC4-ALYREF |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000331134 | ENST00000512673 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - |
| 5CDS-5UTR | ENST00000374747 | ENST00000512673 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - |
| 5CDS-5UTR | ENST00000539314 | ENST00000512673 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - |
| 5CDS-intron | ENST00000331134 | ENST00000512673 | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
| 5CDS-intron | ENST00000374747 | ENST00000512673 | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
| 5UTR-3CDS | ENST00000539314 | ENST00000331204 | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
| 5UTR-3CDS | ENST00000539314 | ENST00000505490 | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
| 5UTR-intron | ENST00000539314 | ENST00000512673 | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
| Frame-shift | ENST00000331134 | ENST00000331204 | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
| Frame-shift | ENST00000331134 | ENST00000505490 | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
| Frame-shift | ENST00000374747 | ENST00000331204 | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
| Frame-shift | ENST00000374747 | ENST00000505490 | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
| Frame-shift | ENST00000539314 | ENST00000331204 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - |
| Frame-shift | ENST00000539314 | ENST00000505490 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - |
| In-frame | ENST00000331134 | ENST00000331204 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - |
| In-frame | ENST00000331134 | ENST00000505490 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - |
| In-frame | ENST00000374747 | ENST00000331204 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - |
| In-frame | ENST00000374747 | ENST00000505490 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - |
| intron-3CDS | ENST00000572760 | ENST00000331204 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - |
| intron-3CDS | ENST00000572760 | ENST00000331204 | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
| intron-3CDS | ENST00000572760 | ENST00000505490 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - |
| intron-3CDS | ENST00000572760 | ENST00000505490 | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
| intron-3CDS | ENST00000573876 | ENST00000331204 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - |
| intron-3CDS | ENST00000573876 | ENST00000331204 | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
| intron-3CDS | ENST00000573876 | ENST00000505490 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - |
| intron-3CDS | ENST00000573876 | ENST00000505490 | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
| intron-3CDS | ENST00000574344 | ENST00000331204 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - |
| intron-3CDS | ENST00000574344 | ENST00000331204 | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
| intron-3CDS | ENST00000574344 | ENST00000505490 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - |
| intron-3CDS | ENST00000574344 | ENST00000505490 | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
| intron-5UTR | ENST00000572760 | ENST00000512673 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - |
| intron-5UTR | ENST00000573876 | ENST00000512673 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - |
| intron-5UTR | ENST00000574344 | ENST00000512673 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - |
| intron-intron | ENST00000572760 | ENST00000512673 | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
| intron-intron | ENST00000573876 | ENST00000512673 | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
| intron-intron | ENST00000574344 | ENST00000512673 | NPLOC4 | chr17 | 79589192 | - | ALYREF | chr17 | 79846021 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000331134 | NPLOC4 | chr17 | 79567367 | - | ENST00000331204 | ALYREF | chr17 | 79848696 | - | 1968 | 1137 | 3 | 1673 | 556 |
| ENST00000331134 | NPLOC4 | chr17 | 79567367 | - | ENST00000505490 | ALYREF | chr17 | 79848696 | - | 1961 | 1137 | 3 | 1673 | 556 |
| ENST00000374747 | NPLOC4 | chr17 | 79567367 | - | ENST00000331204 | ALYREF | chr17 | 79848696 | - | 1882 | 1051 | 1 | 1587 | 528 |
| ENST00000374747 | NPLOC4 | chr17 | 79567367 | - | ENST00000505490 | ALYREF | chr17 | 79848696 | - | 1875 | 1051 | 1 | 1587 | 528 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000331134 | ENST00000331204 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - | 0.004223161 | 0.9957768 |
| ENST00000331134 | ENST00000505490 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - | 0.00394886 | 0.99605113 |
| ENST00000374747 | ENST00000331204 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - | 0.004099949 | 0.9959001 |
| ENST00000374747 | ENST00000505490 | NPLOC4 | chr17 | 79567367 | - | ALYREF | chr17 | 79848696 | - | 0.003849288 | 0.99615073 |
Top |
Fusion Genomic Features for NPLOC4-ALYREF |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
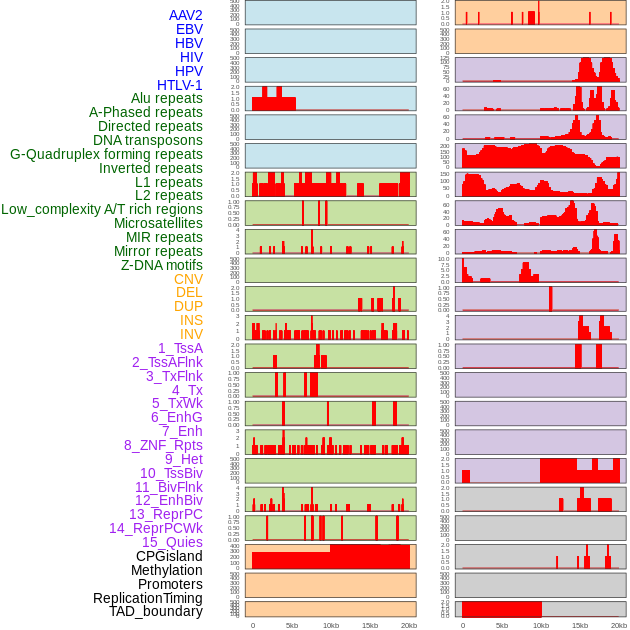 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for NPLOC4-ALYREF |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:79567367/chr17:79848696) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NPLOC4 | ALYREF |
| FUNCTION: The ternary complex containing UFD1, VCP and NPLOC4 binds ubiquitinated proteins and is necessary for the export of misfolded proteins from the ER to the cytoplasm, where they are degraded by the proteasome. The NPLOC4-UFD1-VCP complex regulates spindle disassembly at the end of mitosis and is necessary for the formation of a closed nuclear envelope (By similarity). Acts as a negative regulator of type I interferon production via the complex formed with VCP and UFD1, which binds to DDX58/RIG-I and recruits RNF125 to promote ubiquitination and degradation of DDX58/RIG-I (PubMed:26471729). {ECO:0000250|UniProtKB:Q9ES54, ECO:0000269|PubMed:26471729}. | FUNCTION: Export adapter involved in nuclear export of spliced and unspliced mRNA. Binds mRNA which is thought to be transferred to the NXF1-NXT1 heterodimer for export (TAP/NFX1 pathway) (PubMed:15833825, PubMed:15998806, PubMed:17190602, PubMed:11707413, PubMed:11675789, PubMed:11979277, PubMed:18364396, PubMed:22144908, PubMed:22893130, PubMed:23222130, PubMed:25662211). Component of the TREX complex which is thought to couple mRNA transcription, processing and nuclear export, and specifically associates with spliced mRNA and not with unspliced pre-mRNA (PubMed:15833825, PubMed:15998806, PubMed:17190602). TREX is recruited to spliced mRNAs by a transcription-independent mechanism, binds to mRNA upstream of the exon-junction complex (EJC) and is recruited in a splicing- and cap-dependent manner to a region near the 5' end of the mRNA where it functions in mRNA export to the cytoplasm (PubMed:15833825, PubMed:15998806, PubMed:17190602). TREX recruitment occurs via an interaction between ALYREF/THOC4 and the cap-binding protein NCBP1 (PubMed:15833825, PubMed:15998806, PubMed:17190602). The TREX complex is essential for the export of Kaposi's sarcoma-associated herpesvirus (KSHV) intronless mRNAs and infectious virus production; ALYREF/THOC4 mediates the recruitment of the TREX complex to the intronless viral mRNA (PubMed:18974867). Required for TREX complex assembly and for linking DDX39B to the cap-binding complex (CBC) (PubMed:15998806, PubMed:17984224). In conjunction with THOC5 functions in NXF1-NXT1 mediated nuclear export of HSP70 mRNA; both proteins enhance the RNA binding activity of NXF1 and are required for NXF1 localization to the nuclear rim (PubMed:19165146). Involved in the nuclear export of intronless mRNA; proposed to be recruited to intronless mRNA by ATP-bound DDX39B. Involved in transcription elongation and genome stability (PubMed:12438613, PubMed:17984224). Involved in mRNA export of C5-methylcytosine (m5C)-containing mRNAs: specifically recognizes and binds m5C mRNAs and mediates their nucleo-cytoplasmic shuttling (PubMed:28418038). {ECO:0000269|PubMed:11675789, ECO:0000269|PubMed:11707413, ECO:0000269|PubMed:11979277, ECO:0000269|PubMed:12438613, ECO:0000269|PubMed:15833825, ECO:0000269|PubMed:15998806, ECO:0000269|PubMed:17190602, ECO:0000269|PubMed:17984224, ECO:0000269|PubMed:18364396, ECO:0000269|PubMed:18974867, ECO:0000269|PubMed:19165146, ECO:0000269|PubMed:22144908, ECO:0000269|PubMed:22893130, ECO:0000269|PubMed:23222130, ECO:0000269|PubMed:25662211, ECO:0000269|PubMed:28418038}.; FUNCTION: Acts as chaperone and promotes the dimerization of transcription factors containing basic leucine zipper (bZIP) domains and thereby promotes transcriptional activation. {ECO:0000269|PubMed:10488337}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ALYREF | chr17:79567367 | chr17:79848696 | ENST00000331204 | 0 | 6 | 106_183 | 79 | 258.0 | Domain | RRM |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NPLOC4 | chr17:79567367 | chr17:79848696 | ENST00000331134 | - | 9 | 17 | 226_363 | 307 | 609.0 | Domain | MPN |
| Hgene | NPLOC4 | chr17:79567367 | chr17:79848696 | ENST00000374747 | - | 9 | 16 | 226_363 | 307 | 618.0 | Domain | MPN |
| Hgene | NPLOC4 | chr17:79567367 | chr17:79848696 | ENST00000331134 | - | 9 | 17 | 580_608 | 307 | 609.0 | Zinc finger | RanBP2-type |
| Hgene | NPLOC4 | chr17:79567367 | chr17:79848696 | ENST00000374747 | - | 9 | 16 | 580_608 | 307 | 618.0 | Zinc finger | RanBP2-type |
| Tgene | ALYREF | chr17:79567367 | chr17:79848696 | ENST00000331204 | 0 | 6 | 21_231 | 79 | 258.0 | Compositional bias | Note=Ala/Arg/Gly-rich |
Top |
Fusion Gene Sequence for NPLOC4-ALYREF |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >59941_59941_1_NPLOC4-ALYREF_NPLOC4_chr17_79567367_ENST00000331134_ALYREF_chr17_79848696_ENST00000331204_length(transcript)=1968nt_BP=1137nt AACCTGGTTTCCCGGCCTCGCGCCCGAGGGGAGCGTATTGAGCGAGGCACCGGGACGTCGGCGGCTGGCGGAGCCGGGCGGGCGAAGCTG GAGCGGCGGTGGCGGCGGCGTAGGGAGGCCGGGGCCGGGGCTGAGGCCGCGGCCGGGTCTGCGAGGCCCTTGGGGCGGCAGGCGGCGGCG GCCCGGGGCTCGAGCCCGGAGGCAGGAGCAGCCGCCATGGCCGAGAGCATCATAATTCGTGTCCAGTCCCCGGATGGAGTGAAGCGGATC ACAGCAACAAAGAGAGAAACAGCAGCAACATTTTTGAAAAAGGTTGCAAAGGAGTTTGGCTTCCAAAATAATGGCTTCTCGGTTTACATC AATAGAAACAAGACCGGAGAGATAACAGCCTCCTCCAACAAATCCCTCAACTTGCTAAAAATCAAGCATGGCGATTTGTTGTTCCTGTTT CCCTCGAGCCTTGCTGGGCCCTCATCTGAAATGGAGACGTCAGTTCCACCGGGCTTCAAAGTCTTTGGCGCTCCCAACGTGGTGGAGGAT GAGATTGATCAGTACCTCAGCAAACAGGACGGGAAGATTTACAGAAGCCGAGACCCACAGCTATGCCGCCACGGCCCTTTGGGGAAATGC GTGCACTGCGTCCCTCTAGAGCCATTCGATGAGGACTATCTAAACCATCTCGAGCCTCCCGTGAAGCACATGTCCTTCCACGCCTACATC CGGAAGCTGACTGGAGGGGCTGACAAGGGGAAGTTTGTTGCCCTGGAGAACATCAGCTGCAAGATTAAGTCAGGGTGCGAGGGGCACCTC CCGTGGCCGAATGGCATCTGTACTAAGTGCCAGCCGAGCGCCATCACGCTGAACAGACAGAAGTACAGGCATGTGGACAATATCATGTTT GAGAATCACACCGTCGCTGACCGCTTTCTTGACTTCTGGAGAAAGACAGGGAACCAGCATTTTGGGTACTTATACGGACGGTACACGGAG CACAAAGACATTCCCCTTGGCATCAGGGCTGAAGTGGCTGCGATTTATGAGCCACCTCAGATTGGTACACAGAACAGCTTGGAGCTTCTT GAGGATCCAAAAGCTGAAGTGGTCGATGAAATTGCTGCCAAACTTGGCCTGCGGAAGCCAAAACAACTTCCCGACAAGTGGCAGCACGAT CTTTTCGACAGTGGCTTCGGCGGTGGTGCCGGCGTGGAGACAGGTGGGAAACTGCTGGTGTCCAATCTGGATTTTGGAGTCTCAGACGCC GATATTCAGGAACTCTTTGCTGAATTTGGAACGCTGAAGAAGGCGGCTGTGCACTATGATCGCTCTGGTCGCAGCTTAGGAACAGCAGAC GTGCACTTTGAGCGGAAGGCAGATGCCCTGAAGGCCATGAAGCAGTACAACGGCGTCCCTCTGGATGGCCGCCCCATGAACATTCAGCTT GTCACGTCACAGATTGACGCACAGCGGAGGCCTGCACAGAGCGTAAACAGAGGTGGCATGACTAGAAACCGTGGCGCTGGAGGTTTTGGT GGTGGTGGAGGCACCCGGAGAGGCACCCGCGGAGGCGCCCGTGGAAGAGGCAGAGGTGCCGGCAGGAATTCAAAGCAGCAGCTTTCGGCA GAGGAGCTGGATGCCCAGCTGGACGCCTATAATGCGAGAATGGACACCAGTTAAACAGACCAGCAAATCCGCGTGCGGAACAGGACCCAG GCGTCTCCTCTTGCTCCCTGGTTGGGGGGCGGTGGCTGGGGCTGTGCGGCCAATGATGGATTTGTTTCTTTTATGTTTTAAAATAGGATT TAAAAACTCATGTAAAGGTTTTTTTTTTTTCTTTTTTTTTTTTTTTAATTCTGAAACAGACCTGTTTTGTACCGAGTTATTTTTGGGATA >59941_59941_1_NPLOC4-ALYREF_NPLOC4_chr17_79567367_ENST00000331134_ALYREF_chr17_79848696_ENST00000331204_length(amino acids)=556AA_BP=20 MVSRPRARGERIERGTGTSAAGGAGRAKLERRWRRRREAGAGAEAAAGSARPLGRQAAAARGSSPEAGAAAMAESIIIRVQSPDGVKRIT ATKRETAATFLKKVAKEFGFQNNGFSVYINRNKTGEITASSNKSLNLLKIKHGDLLFLFPSSLAGPSSEMETSVPPGFKVFGAPNVVEDE IDQYLSKQDGKIYRSRDPQLCRHGPLGKCVHCVPLEPFDEDYLNHLEPPVKHMSFHAYIRKLTGGADKGKFVALENISCKIKSGCEGHLP WPNGICTKCQPSAITLNRQKYRHVDNIMFENHTVADRFLDFWRKTGNQHFGYLYGRYTEHKDIPLGIRAEVAAIYEPPQIGTQNSLELLE DPKAEVVDEIAAKLGLRKPKQLPDKWQHDLFDSGFGGGAGVETGGKLLVSNLDFGVSDADIQELFAEFGTLKKAAVHYDRSGRSLGTADV HFERKADALKAMKQYNGVPLDGRPMNIQLVTSQIDAQRRPAQSVNRGGMTRNRGAGGFGGGGGTRRGTRGGARGRGRGAGRNSKQQLSAE -------------------------------------------------------------- >59941_59941_2_NPLOC4-ALYREF_NPLOC4_chr17_79567367_ENST00000331134_ALYREF_chr17_79848696_ENST00000505490_length(transcript)=1961nt_BP=1137nt AACCTGGTTTCCCGGCCTCGCGCCCGAGGGGAGCGTATTGAGCGAGGCACCGGGACGTCGGCGGCTGGCGGAGCCGGGCGGGCGAAGCTG GAGCGGCGGTGGCGGCGGCGTAGGGAGGCCGGGGCCGGGGCTGAGGCCGCGGCCGGGTCTGCGAGGCCCTTGGGGCGGCAGGCGGCGGCG GCCCGGGGCTCGAGCCCGGAGGCAGGAGCAGCCGCCATGGCCGAGAGCATCATAATTCGTGTCCAGTCCCCGGATGGAGTGAAGCGGATC ACAGCAACAAAGAGAGAAACAGCAGCAACATTTTTGAAAAAGGTTGCAAAGGAGTTTGGCTTCCAAAATAATGGCTTCTCGGTTTACATC AATAGAAACAAGACCGGAGAGATAACAGCCTCCTCCAACAAATCCCTCAACTTGCTAAAAATCAAGCATGGCGATTTGTTGTTCCTGTTT CCCTCGAGCCTTGCTGGGCCCTCATCTGAAATGGAGACGTCAGTTCCACCGGGCTTCAAAGTCTTTGGCGCTCCCAACGTGGTGGAGGAT GAGATTGATCAGTACCTCAGCAAACAGGACGGGAAGATTTACAGAAGCCGAGACCCACAGCTATGCCGCCACGGCCCTTTGGGGAAATGC GTGCACTGCGTCCCTCTAGAGCCATTCGATGAGGACTATCTAAACCATCTCGAGCCTCCCGTGAAGCACATGTCCTTCCACGCCTACATC CGGAAGCTGACTGGAGGGGCTGACAAGGGGAAGTTTGTTGCCCTGGAGAACATCAGCTGCAAGATTAAGTCAGGGTGCGAGGGGCACCTC CCGTGGCCGAATGGCATCTGTACTAAGTGCCAGCCGAGCGCCATCACGCTGAACAGACAGAAGTACAGGCATGTGGACAATATCATGTTT GAGAATCACACCGTCGCTGACCGCTTTCTTGACTTCTGGAGAAAGACAGGGAACCAGCATTTTGGGTACTTATACGGACGGTACACGGAG CACAAAGACATTCCCCTTGGCATCAGGGCTGAAGTGGCTGCGATTTATGAGCCACCTCAGATTGGTACACAGAACAGCTTGGAGCTTCTT GAGGATCCAAAAGCTGAAGTGGTCGATGAAATTGCTGCCAAACTTGGCCTGCGGAAGCCAAAACAACTTCCCGACAAGTGGCAGCACGAT CTTTTCGACAGTGGCTTCGGCGGTGGTGCCGGCGTGGAGACAGGTGGGAAACTGCTGGTGTCCAATCTGGATTTTGGAGTCTCAGACGCC GATATTCAGGAACTCTTTGCTGAATTTGGAACGCTGAAGAAGGCGGCTGTGCACTATGATCGCTCTGGTCGCAGCTTAGGAACAGCAGAC GTGCACTTTGAGCGGAAGGCAGATGCCCTGAAGGCCATGAAGCAGTACAACGGCGTCCCTCTGGATGGCCGCCCCATGAACATTCAGCTT GTCACGTCACAGATTGACGCACAGCGGAGGCCTGCACAGAGCGTAAACAGAGGTGGCATGACTAGAAACCGTGGCGCTGGAGGTTTTGGT GGTGGTGGAGGCACCCGGAGAGGCACCCGCGGAGGCGCCCGTGGAAGAGGCAGAGGTGCCGGCAGGAATTCAAAGCAGCAGCTTTCGGCA GAGGAGCTGGATGCCCAGCTGGACGCCTATAATGCGAGAATGGACACCAGTTAAACAGACCAGCAAATCCGCGTGCGGAACAGGACCCAG GCGTCTCCTCTTGCTCCCTGGTTGGGGGGCGGTGGCTGGGGCTGTGCGGCCAATGATGGATTTGTTTCTTTTATGTTTTAAAATAGGATT TAAAAACTCATGTAAAGGTTTTTTTTTTTTCTTTTTTTTTTTTTTTAATTCTGAAACAGACCTGTTTTGTACCGAGTTATTTTTGGGATA >59941_59941_2_NPLOC4-ALYREF_NPLOC4_chr17_79567367_ENST00000331134_ALYREF_chr17_79848696_ENST00000505490_length(amino acids)=556AA_BP=20 MVSRPRARGERIERGTGTSAAGGAGRAKLERRWRRRREAGAGAEAAAGSARPLGRQAAAARGSSPEAGAAAMAESIIIRVQSPDGVKRIT ATKRETAATFLKKVAKEFGFQNNGFSVYINRNKTGEITASSNKSLNLLKIKHGDLLFLFPSSLAGPSSEMETSVPPGFKVFGAPNVVEDE IDQYLSKQDGKIYRSRDPQLCRHGPLGKCVHCVPLEPFDEDYLNHLEPPVKHMSFHAYIRKLTGGADKGKFVALENISCKIKSGCEGHLP WPNGICTKCQPSAITLNRQKYRHVDNIMFENHTVADRFLDFWRKTGNQHFGYLYGRYTEHKDIPLGIRAEVAAIYEPPQIGTQNSLELLE DPKAEVVDEIAAKLGLRKPKQLPDKWQHDLFDSGFGGGAGVETGGKLLVSNLDFGVSDADIQELFAEFGTLKKAAVHYDRSGRSLGTADV HFERKADALKAMKQYNGVPLDGRPMNIQLVTSQIDAQRRPAQSVNRGGMTRNRGAGGFGGGGGTRRGTRGGARGRGRGAGRNSKQQLSAE -------------------------------------------------------------- >59941_59941_3_NPLOC4-ALYREF_NPLOC4_chr17_79567367_ENST00000374747_ALYREF_chr17_79848696_ENST00000331204_length(transcript)=1882nt_BP=1051nt GCTGGAGCGGCGGTGGCGGCGGCGTAGGGAGGCCGGGGCCGGGGCTGAGGCCGCGGCCGGGTCTGCGAGGCCCTTGGGGCGGCAGGCGGC GGCGGCCCGGGGCTCGAGCCCGGAGGCAGGAGCAGCCGCCATGGCCGAGAGCATCATAATTCGTGTCCAGTCCCCGGATGGAGTGAAGCG GATCACAGCAACAAAGAGAGAAACAGCAGCAACATTTTTGAAAAAGGTTGCAAAGGAGTTTGGCTTCCAAAATAATGGCTTCTCGGTTTA CATCAATAGAAACAAGACCGGAGAGATAACAGCCTCCTCCAACAAATCCCTCAACTTGCTAAAAATCAAGCATGGCGATTTGTTGTTCCT GTTTCCCTCGAGCCTTGCTGGGCCCTCATCTGAAATGGAGACGTCAGTTCCACCGGGCTTCAAAGTCTTTGGCGCTCCCAACGTGGTGGA GGATGAGATTGATCAGTACCTCAGCAAACAGGACGGGAAGATTTACAGAAGCCGAGACCCACAGCTATGCCGCCACGGCCCTTTGGGGAA ATGCGTGCACTGCGTCCCTCTAGAGCCATTCGATGAGGACTATCTAAACCATCTCGAGCCTCCCGTGAAGCACATGTCCTTCCACGCCTA CATCCGGAAGCTGACTGGAGGGGCTGACAAGGGGAAGTTTGTTGCCCTGGAGAACATCAGCTGCAAGATTAAGTCAGGGTGCGAGGGGCA CCTCCCGTGGCCGAATGGCATCTGTACTAAGTGCCAGCCGAGCGCCATCACGCTGAACAGACAGAAGTACAGGCATGTGGACAATATCAT GTTTGAGAATCACACCGTCGCTGACCGCTTTCTTGACTTCTGGAGAAAGACAGGGAACCAGCATTTTGGGTACTTATACGGACGGTACAC GGAGCACAAAGACATTCCCCTTGGCATCAGGGCTGAAGTGGCTGCGATTTATGAGCCACCTCAGATTGGTACACAGAACAGCTTGGAGCT TCTTGAGGATCCAAAAGCTGAAGTGGTCGATGAAATTGCTGCCAAACTTGGCCTGCGGAAGCCAAAACAACTTCCCGACAAGTGGCAGCA CGATCTTTTCGACAGTGGCTTCGGCGGTGGTGCCGGCGTGGAGACAGGTGGGAAACTGCTGGTGTCCAATCTGGATTTTGGAGTCTCAGA CGCCGATATTCAGGAACTCTTTGCTGAATTTGGAACGCTGAAGAAGGCGGCTGTGCACTATGATCGCTCTGGTCGCAGCTTAGGAACAGC AGACGTGCACTTTGAGCGGAAGGCAGATGCCCTGAAGGCCATGAAGCAGTACAACGGCGTCCCTCTGGATGGCCGCCCCATGAACATTCA GCTTGTCACGTCACAGATTGACGCACAGCGGAGGCCTGCACAGAGCGTAAACAGAGGTGGCATGACTAGAAACCGTGGCGCTGGAGGTTT TGGTGGTGGTGGAGGCACCCGGAGAGGCACCCGCGGAGGCGCCCGTGGAAGAGGCAGAGGTGCCGGCAGGAATTCAAAGCAGCAGCTTTC GGCAGAGGAGCTGGATGCCCAGCTGGACGCCTATAATGCGAGAATGGACACCAGTTAAACAGACCAGCAAATCCGCGTGCGGAACAGGAC CCAGGCGTCTCCTCTTGCTCCCTGGTTGGGGGGCGGTGGCTGGGGCTGTGCGGCCAATGATGGATTTGTTTCTTTTATGTTTTAAAATAG GATTTAAAAACTCATGTAAAGGTTTTTTTTTTTTCTTTTTTTTTTTTTTTAATTCTGAAACAGACCTGTTTTGTACCGAGTTATTTTTGG >59941_59941_3_NPLOC4-ALYREF_NPLOC4_chr17_79567367_ENST00000374747_ALYREF_chr17_79848696_ENST00000331204_length(amino acids)=528AA_BP=2 LERRWRRRREAGAGAEAAAGSARPLGRQAAAARGSSPEAGAAAMAESIIIRVQSPDGVKRITATKRETAATFLKKVAKEFGFQNNGFSVY INRNKTGEITASSNKSLNLLKIKHGDLLFLFPSSLAGPSSEMETSVPPGFKVFGAPNVVEDEIDQYLSKQDGKIYRSRDPQLCRHGPLGK CVHCVPLEPFDEDYLNHLEPPVKHMSFHAYIRKLTGGADKGKFVALENISCKIKSGCEGHLPWPNGICTKCQPSAITLNRQKYRHVDNIM FENHTVADRFLDFWRKTGNQHFGYLYGRYTEHKDIPLGIRAEVAAIYEPPQIGTQNSLELLEDPKAEVVDEIAAKLGLRKPKQLPDKWQH DLFDSGFGGGAGVETGGKLLVSNLDFGVSDADIQELFAEFGTLKKAAVHYDRSGRSLGTADVHFERKADALKAMKQYNGVPLDGRPMNIQ -------------------------------------------------------------- >59941_59941_4_NPLOC4-ALYREF_NPLOC4_chr17_79567367_ENST00000374747_ALYREF_chr17_79848696_ENST00000505490_length(transcript)=1875nt_BP=1051nt GCTGGAGCGGCGGTGGCGGCGGCGTAGGGAGGCCGGGGCCGGGGCTGAGGCCGCGGCCGGGTCTGCGAGGCCCTTGGGGCGGCAGGCGGC GGCGGCCCGGGGCTCGAGCCCGGAGGCAGGAGCAGCCGCCATGGCCGAGAGCATCATAATTCGTGTCCAGTCCCCGGATGGAGTGAAGCG GATCACAGCAACAAAGAGAGAAACAGCAGCAACATTTTTGAAAAAGGTTGCAAAGGAGTTTGGCTTCCAAAATAATGGCTTCTCGGTTTA CATCAATAGAAACAAGACCGGAGAGATAACAGCCTCCTCCAACAAATCCCTCAACTTGCTAAAAATCAAGCATGGCGATTTGTTGTTCCT GTTTCCCTCGAGCCTTGCTGGGCCCTCATCTGAAATGGAGACGTCAGTTCCACCGGGCTTCAAAGTCTTTGGCGCTCCCAACGTGGTGGA GGATGAGATTGATCAGTACCTCAGCAAACAGGACGGGAAGATTTACAGAAGCCGAGACCCACAGCTATGCCGCCACGGCCCTTTGGGGAA ATGCGTGCACTGCGTCCCTCTAGAGCCATTCGATGAGGACTATCTAAACCATCTCGAGCCTCCCGTGAAGCACATGTCCTTCCACGCCTA CATCCGGAAGCTGACTGGAGGGGCTGACAAGGGGAAGTTTGTTGCCCTGGAGAACATCAGCTGCAAGATTAAGTCAGGGTGCGAGGGGCA CCTCCCGTGGCCGAATGGCATCTGTACTAAGTGCCAGCCGAGCGCCATCACGCTGAACAGACAGAAGTACAGGCATGTGGACAATATCAT GTTTGAGAATCACACCGTCGCTGACCGCTTTCTTGACTTCTGGAGAAAGACAGGGAACCAGCATTTTGGGTACTTATACGGACGGTACAC GGAGCACAAAGACATTCCCCTTGGCATCAGGGCTGAAGTGGCTGCGATTTATGAGCCACCTCAGATTGGTACACAGAACAGCTTGGAGCT TCTTGAGGATCCAAAAGCTGAAGTGGTCGATGAAATTGCTGCCAAACTTGGCCTGCGGAAGCCAAAACAACTTCCCGACAAGTGGCAGCA CGATCTTTTCGACAGTGGCTTCGGCGGTGGTGCCGGCGTGGAGACAGGTGGGAAACTGCTGGTGTCCAATCTGGATTTTGGAGTCTCAGA CGCCGATATTCAGGAACTCTTTGCTGAATTTGGAACGCTGAAGAAGGCGGCTGTGCACTATGATCGCTCTGGTCGCAGCTTAGGAACAGC AGACGTGCACTTTGAGCGGAAGGCAGATGCCCTGAAGGCCATGAAGCAGTACAACGGCGTCCCTCTGGATGGCCGCCCCATGAACATTCA GCTTGTCACGTCACAGATTGACGCACAGCGGAGGCCTGCACAGAGCGTAAACAGAGGTGGCATGACTAGAAACCGTGGCGCTGGAGGTTT TGGTGGTGGTGGAGGCACCCGGAGAGGCACCCGCGGAGGCGCCCGTGGAAGAGGCAGAGGTGCCGGCAGGAATTCAAAGCAGCAGCTTTC GGCAGAGGAGCTGGATGCCCAGCTGGACGCCTATAATGCGAGAATGGACACCAGTTAAACAGACCAGCAAATCCGCGTGCGGAACAGGAC CCAGGCGTCTCCTCTTGCTCCCTGGTTGGGGGGCGGTGGCTGGGGCTGTGCGGCCAATGATGGATTTGTTTCTTTTATGTTTTAAAATAG GATTTAAAAACTCATGTAAAGGTTTTTTTTTTTTCTTTTTTTTTTTTTTTAATTCTGAAACAGACCTGTTTTGTACCGAGTTATTTTTGG >59941_59941_4_NPLOC4-ALYREF_NPLOC4_chr17_79567367_ENST00000374747_ALYREF_chr17_79848696_ENST00000505490_length(amino acids)=528AA_BP=2 LERRWRRRREAGAGAEAAAGSARPLGRQAAAARGSSPEAGAAAMAESIIIRVQSPDGVKRITATKRETAATFLKKVAKEFGFQNNGFSVY INRNKTGEITASSNKSLNLLKIKHGDLLFLFPSSLAGPSSEMETSVPPGFKVFGAPNVVEDEIDQYLSKQDGKIYRSRDPQLCRHGPLGK CVHCVPLEPFDEDYLNHLEPPVKHMSFHAYIRKLTGGADKGKFVALENISCKIKSGCEGHLPWPNGICTKCQPSAITLNRQKYRHVDNIM FENHTVADRFLDFWRKTGNQHFGYLYGRYTEHKDIPLGIRAEVAAIYEPPQIGTQNSLELLEDPKAEVVDEIAAKLGLRKPKQLPDKWQH DLFDSGFGGGAGVETGGKLLVSNLDFGVSDADIQELFAEFGTLKKAAVHYDRSGRSLGTADVHFERKADALKAMKQYNGVPLDGRPMNIQ -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for NPLOC4-ALYREF |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | ALYREF | chr17:79567367 | chr17:79848696 | ENST00000331204 | 0 | 6 | 85_186 | 79.0 | 258.0 | HHV-8 ORF57 protein and with ICP27 from HHV-1 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for NPLOC4-ALYREF |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for NPLOC4-ALYREF |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
