
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:NPLOC4-SGPL1 (FusionGDB2 ID:59958) |
Fusion Gene Summary for NPLOC4-SGPL1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: NPLOC4-SGPL1 | Fusion gene ID: 59958 | Hgene | Tgene | Gene symbol | NPLOC4 | SGPL1 | Gene ID | 55666 | 8879 |
| Gene name | NPL4 homolog, ubiquitin recognition factor | sphingosine-1-phosphate lyase 1 | |
| Synonyms | NPL4 | NPHS14|S1PL|SPL | |
| Cytomap | 17q25.3 | 10q22.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | nuclear protein localization protein 4 homologNPLOC4 ubiquitin recognition factornuclear protein localization 4 homolog | sphingosine-1-phosphate lyase 1SP-lyase 1SPL 1hSPLsphingosine-1-phosphate aldolase | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q8TAT6 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000331134, ENST00000374747, ENST00000574344, ENST00000539314, ENST00000572760, ENST00000573876, | ENST00000486993, ENST00000373202, | |
| Fusion gene scores | * DoF score | 19 X 17 X 11=3553 | 5 X 5 X 4=100 |
| # samples | 30 | 5 | |
| ** MAII score | log2(30/3553*10)=-3.56600328283534 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/100*10)=-1 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: NPLOC4 [Title/Abstract] AND SGPL1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | NPLOC4(79577236)-SGPL1(72630805), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | SGPL1 | GO:0006631 | fatty acid metabolic process | 24809814 |
| Tgene | SGPL1 | GO:0006672 | ceramide metabolic process | 14570870 |
| Tgene | SGPL1 | GO:0030149 | sphingolipid catabolic process | 24809814 |
| Tgene | SGPL1 | GO:0097190 | apoptotic signaling pathway | 14570870 |
 Fusion gene breakpoints across NPLOC4 (5'-gene) Fusion gene breakpoints across NPLOC4 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
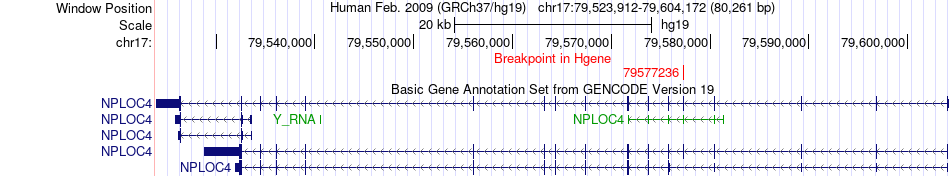 |
 Fusion gene breakpoints across SGPL1 (3'-gene) Fusion gene breakpoints across SGPL1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
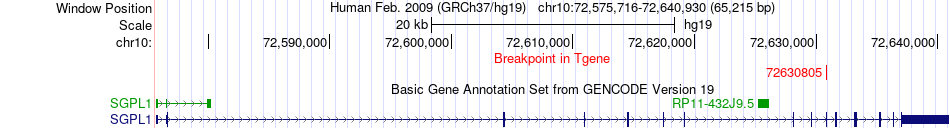 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-D7-A4Z0-01A | NPLOC4 | chr17 | 79577236 | - | SGPL1 | chr10 | 72630805 | + |
Top |
Fusion Gene ORF analysis for NPLOC4-SGPL1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000331134 | ENST00000486993 | NPLOC4 | chr17 | 79577236 | - | SGPL1 | chr10 | 72630805 | + |
| 5CDS-intron | ENST00000374747 | ENST00000486993 | NPLOC4 | chr17 | 79577236 | - | SGPL1 | chr10 | 72630805 | + |
| 5UTR-3CDS | ENST00000574344 | ENST00000373202 | NPLOC4 | chr17 | 79577236 | - | SGPL1 | chr10 | 72630805 | + |
| 5UTR-intron | ENST00000574344 | ENST00000486993 | NPLOC4 | chr17 | 79577236 | - | SGPL1 | chr10 | 72630805 | + |
| In-frame | ENST00000331134 | ENST00000373202 | NPLOC4 | chr17 | 79577236 | - | SGPL1 | chr10 | 72630805 | + |
| In-frame | ENST00000374747 | ENST00000373202 | NPLOC4 | chr17 | 79577236 | - | SGPL1 | chr10 | 72630805 | + |
| intron-3CDS | ENST00000539314 | ENST00000373202 | NPLOC4 | chr17 | 79577236 | - | SGPL1 | chr10 | 72630805 | + |
| intron-3CDS | ENST00000572760 | ENST00000373202 | NPLOC4 | chr17 | 79577236 | - | SGPL1 | chr10 | 72630805 | + |
| intron-3CDS | ENST00000573876 | ENST00000373202 | NPLOC4 | chr17 | 79577236 | - | SGPL1 | chr10 | 72630805 | + |
| intron-intron | ENST00000539314 | ENST00000486993 | NPLOC4 | chr17 | 79577236 | - | SGPL1 | chr10 | 72630805 | + |
| intron-intron | ENST00000572760 | ENST00000486993 | NPLOC4 | chr17 | 79577236 | - | SGPL1 | chr10 | 72630805 | + |
| intron-intron | ENST00000573876 | ENST00000486993 | NPLOC4 | chr17 | 79577236 | - | SGPL1 | chr10 | 72630805 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000331134 | NPLOC4 | chr17 | 79577236 | - | ENST00000373202 | SGPL1 | chr10 | 72630805 | + | 5386 | 651 | 3 | 1547 | 514 |
| ENST00000374747 | NPLOC4 | chr17 | 79577236 | - | ENST00000373202 | SGPL1 | chr10 | 72630805 | + | 5300 | 565 | 1 | 1461 | 486 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000331134 | ENST00000373202 | NPLOC4 | chr17 | 79577236 | - | SGPL1 | chr10 | 72630805 | + | 0.001676518 | 0.9983235 |
| ENST00000374747 | ENST00000373202 | NPLOC4 | chr17 | 79577236 | - | SGPL1 | chr10 | 72630805 | + | 0.001620867 | 0.9983791 |
Top |
Fusion Genomic Features for NPLOC4-SGPL1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| NPLOC4 | chr17 | 79577235 | - | SGPL1 | chr10 | 72630804 | + | 0.000768925 | 0.9992311 |
| NPLOC4 | chr17 | 79577235 | - | SGPL1 | chr10 | 72630804 | + | 0.000768925 | 0.9992311 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
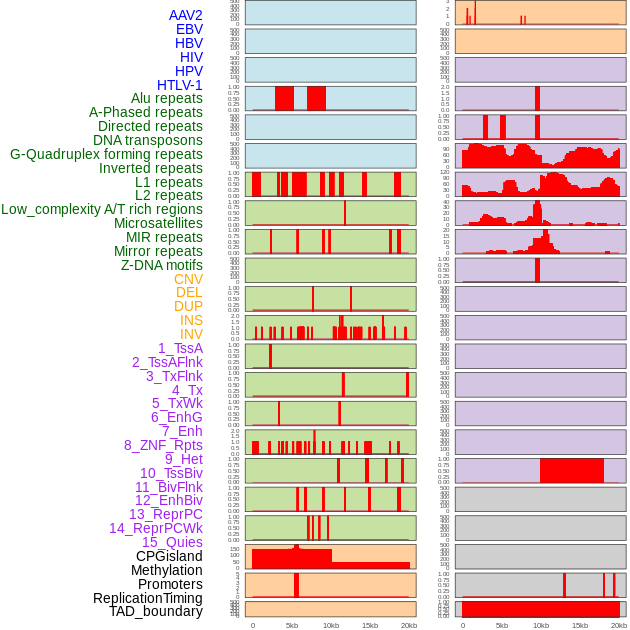 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
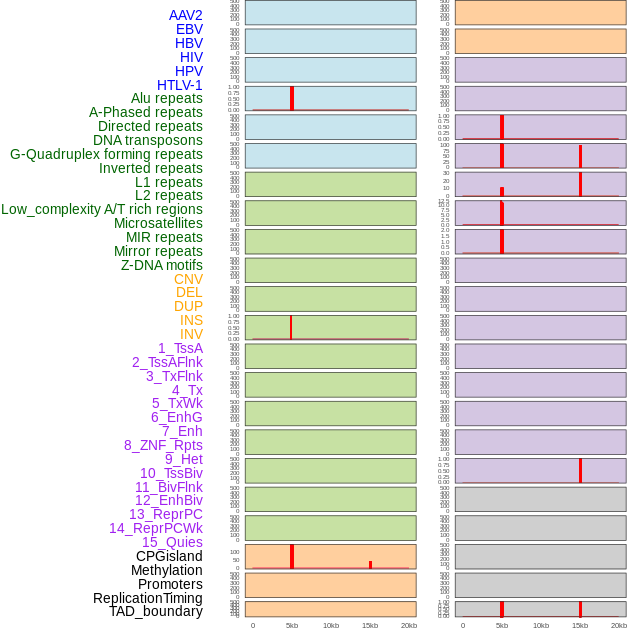 |
Top |
Fusion Protein Features for NPLOC4-SGPL1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:79577236/chr10:72630805) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NPLOC4 | . |
| FUNCTION: The ternary complex containing UFD1, VCP and NPLOC4 binds ubiquitinated proteins and is necessary for the export of misfolded proteins from the ER to the cytoplasm, where they are degraded by the proteasome. The NPLOC4-UFD1-VCP complex regulates spindle disassembly at the end of mitosis and is necessary for the formation of a closed nuclear envelope (By similarity). Acts as a negative regulator of type I interferon production via the complex formed with VCP and UFD1, which binds to DDX58/RIG-I and recruits RNF125 to promote ubiquitination and degradation of DDX58/RIG-I (PubMed:26471729). {ECO:0000250|UniProtKB:Q9ES54, ECO:0000269|PubMed:26471729}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NPLOC4 | chr17:79577236 | chr10:72630805 | ENST00000331134 | - | 5 | 17 | 226_363 | 145 | 609.0 | Domain | MPN |
| Hgene | NPLOC4 | chr17:79577236 | chr10:72630805 | ENST00000374747 | - | 5 | 16 | 226_363 | 145 | 618.0 | Domain | MPN |
| Hgene | NPLOC4 | chr17:79577236 | chr10:72630805 | ENST00000331134 | - | 5 | 17 | 580_608 | 145 | 609.0 | Zinc finger | RanBP2-type |
| Hgene | NPLOC4 | chr17:79577236 | chr10:72630805 | ENST00000374747 | - | 5 | 16 | 580_608 | 145 | 618.0 | Zinc finger | RanBP2-type |
| Tgene | SGPL1 | chr17:79577236 | chr10:72630805 | ENST00000373202 | 8 | 15 | 1_40 | 270 | 569.0 | Topological domain | Lumenal | |
| Tgene | SGPL1 | chr17:79577236 | chr10:72630805 | ENST00000373202 | 8 | 15 | 62_568 | 270 | 569.0 | Topological domain | Cytoplasmic | |
| Tgene | SGPL1 | chr17:79577236 | chr10:72630805 | ENST00000373202 | 8 | 15 | 41_61 | 270 | 569.0 | Transmembrane | Helical%3B Signal-anchor for type III membrane protein |
Top |
Fusion Gene Sequence for NPLOC4-SGPL1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >59958_59958_1_NPLOC4-SGPL1_NPLOC4_chr17_79577236_ENST00000331134_SGPL1_chr10_72630805_ENST00000373202_length(transcript)=5386nt_BP=651nt AACCTGGTTTCCCGGCCTCGCGCCCGAGGGGAGCGTATTGAGCGAGGCACCGGGACGTCGGCGGCTGGCGGAGCCGGGCGGGCGAAGCTG GAGCGGCGGTGGCGGCGGCGTAGGGAGGCCGGGGCCGGGGCTGAGGCCGCGGCCGGGTCTGCGAGGCCCTTGGGGCGGCAGGCGGCGGCG GCCCGGGGCTCGAGCCCGGAGGCAGGAGCAGCCGCCATGGCCGAGAGCATCATAATTCGTGTCCAGTCCCCGGATGGAGTGAAGCGGATC ACAGCAACAAAGAGAGAAACAGCAGCAACATTTTTGAAAAAGGTTGCAAAGGAGTTTGGCTTCCAAAATAATGGCTTCTCGGTTTACATC AATAGAAACAAGACCGGAGAGATAACAGCCTCCTCCAACAAATCCCTCAACTTGCTAAAAATCAAGCATGGCGATTTGTTGTTCCTGTTT CCCTCGAGCCTTGCTGGGCCCTCATCTGAAATGGAGACGTCAGTTCCACCGGGCTTCAAAGTCTTTGGCGCTCCCAACGTGGTGGAGGAT GAGATTGATCAGTACCTCAGCAAACAGGACGGGAAGATTTACAGAAGCCGAGACCCACAGCTATGCCGCCACGGCCCTTTGGGGAAATGC GTGCACTGCGTCCCTCTAGAGGCAATGAGAAGAGCTATCTCCAGGAACACTGCCATGCTCGTCTGTTCTACCCCACAGTTTCCTCATGGT GTAATAGATCCTGTCCCTGAAGTGGCCAAGCTGGCTGTCAAATACAAAATACCCCTTCATGTCGACGCTTGTCTGGGAGGCTTCCTCATC GTCTTTATGGAGAAAGCAGGATACCCACTGGAGCACCCATTTGATTTCCGGGTGAAAGGTGTAACCAGCATTTCAGCTGACACCCATAAG TATGGCTATGCCCCAAAAGGCTCATCATTGGTGTTGTATAGTGACAAGAAGTACAGGAACTATCAGTTCTTCGTCGATACAGATTGGCAG GGTGGCATCTATGCTTCCCCAACCATCGCAGGCTCACGGCCTGGTGGCATTAGCGCAGCCTGTTGGGCTGCCTTGATGCACTTCGGTGAG AACGGCTATGTTGAAGCTACCAAACAGATCATCAAAACTGCTCGCTTCCTCAAGTCAGAACTGGAAAATATCAAAGGCATCTTTGTTTTT GGGAATCCCCAATTGTCAGTCATTGCTCTGGGATCCCGTGATTTTGACATCTACCGACTATCAAACCTGATGACTGCTAAGGGGTGGAAC TTGAACCAGTTGCAGTTCCCACCCAGTATTCATTTCTGCATCACATTACTACACGCCCGGAAACGAGTAGCTATACAATTCCTAAAGGAC ATTCGAGAATCTGTCACTCAAATCATGAAGAATCCTAAAGCGAAGACCACAGGAATGGGTGCCATCTATGGCATGGCCCAGACAACTGTT GACAGGAATATGGTTGCAGAATTGTCCTCAGTCTTCTTGGACAGCTTGTACAGCACCGACACTGTCACCCAGGGCAGCCAGATGAATGGT TCTCCAAAACCCCACTGAACTTGGACCCTTTCTAGTCTCAAGGGGATTCCAGCCTTCAGAAGGTTCTTGGGATATGGAACAGGCCGTGCA CAACTTTGACATCTGGTCTTGCTCCATAGAGCACAACTCAAGATAGACCATGAGACAGCTTGAGCCTCAGGATTCTTGTTCTTCCTCTTA TCTTCCTTTTGTGGTTTTTAATTTGAAGACCCCAGAGAATTCCATTACATAATGATTTTGCCCTTGTTATAAATGTTACCCTAGGAATTG TTTTAACCATTTCCTTTTCTAAACTCTCTAGCTTTCAACTTTACTTAAACATTGTGTGGTAGCTCTGACCTGTCCTGATTCTTTAGAGAA GCTGGGGTACAGTTTATGAGATAGCTAGAGCTTCTTTGTTATCTCAGGCAGGAGGCGTTTACATAACAGATGTTTCCTCAGCTGGGTGTG AGGTATACTCTAAGCAGGAGGCTTTTTCAGCCTTCTCTCTCTTTTTTTTTTTTTTTTTTTTTTTTGAGATGGAATTTTGCTCTTTTGCCC AGTCTGGAGTGCAGTGGCATGATCTCAGCTCACTGCAACCTCCACCCACTGGGTTCAAGCGATTCTTCTGCCTCAGCCTCCCGAGTAGCT GGGATTACCGGCACCCACCACCACGCCTGGCTAATTTTTCAATTTTCTTTTTCAGTAGAGACGGGTTCACCGTGTTGGCCAGGCTGGTCT TGAACTCCTGACCTCAGGTGATACCCGCCCCCCCGCCTCAGCCTCCCAAAGTGCTGGGATTACAGGCGTGAGCCACCGTGCCTGGCCCTG TCTCTCTTAAGAGTAGGTTCATTGTCTGTCTTAGAGTCACTTCTATTGCAACTCATTTTCTTTTTCCAGGGCACAGATCGACCAAGCTGC CGTTCCCTATTCTGCAGGACAGGACTATTCTAGCATACCTGCTTCGTCCACCCAGGCAGGGTTTGGGGTGGTCTCTTCTGTGCCTGCAGT CCCCATTTGACACTTGGTTGCCACCATCTTTGGAGATTATTGTTTGGAATGATGCTTCCATTGGCTTTTTCTTGTTACCATGGACTAGGA AGAAAACATGGTTTCCAAATAATCTGGGAGCTTTTGGCCATGGTGCCGCCTTCCTGAATTGGCAGTGGTCAGAGCACACCTGAACCCTAT CCTGGGCTGGTGATGAGCAGAAATCAGACCTTTTTCTATGCTTTTTTGAATATCAGAGTAGGATGAACACCCAGATTCAAATATGTCACC AAAGTTGGTGGTGGTCCTTCCCTGCACCCTTGCGTTAAGCCATTATGTAATGAAAATGTGTTTGCTTGAAGGAACAGCTCAAAGCACCTT CACAAGTTGCCTTGACTTACCCTAGGTGGGTGTGAAAGAGCACCCGTAGCAAGGAAAATTTTCTCTATTAGTGTGTTCTTCTGCCTCTTC CCCCTTGATTCAGCTTTCAGAGGTACTATGGCAGTTTTGCCTCAGGTGCTGAACATTTCTCAGCCCTGGCTAAAAGGGAGCAGCACAGGG AGAGAAACAGGATAGGAAAGCAGAATGGCGAGCAGCCTATGGCCCAGGGCCTGTAATCCCTTCCCAAGACTAGCTGCTCAGGGTGGTGCA GGGACAGGACCAGACCCTGCGCCTATTTCCTGCCTTCTTTCCCCTATAGGGAACTCTGTAGGCTGAGCCACTGTCCTGCTCTTATGACAT TATATCTTGTGCCTTTCTCCTCAGCAGTGAGCAGTGAGCTACTCCTGGCCCAGGCCCTAGGGGAAATGGATCAGTCTTTGAGGTTTCTAT TTGGGGAGGGGAGTACTTAAGATGAGTCAAAAGACACTTTCCTCTGTTCCATTCCCCATCTCAGGGACTCCTGAATATTCAGCCTCTCCA GGCTGGTGTCTTCTAGTTTCCCCCACTGGGAATGCTGGCTGGGAGAGCCATGACTACCAGACTTTTCCTCAGGCTCCTTGGCATGTTAGT CTGAATTGTTCTTGAGCACTGTACTACTGACCCAACAACTGTGACTAGCTGGCCACGCCATTCAGGGCTGGTGTGGCATTTATGTGTGTG TGTGTGTGTGTGTGTGTTTTTCCTGTTTGCCCAGCAGTGCATTGTGGGTTCCAAGAGTGGGTAGTGTGTGTATGTGTGTGTGTCAGAGGG AGACCTGGCAGGCACCTCTTTGAGAGTAGCTGTGGTCAGAGCTGTTTGGTCAGTGCATTATGTTGAATGAGGTCCAGGAACCCAGAGCCA CCCAGCAGACACCACTGTGGCTTGCCAGCTGCCAAGATGGAGAAGCATGTGCCCCTGTAGAGCGTCTCCCCAGAACCAGACCCCGAGCCA CTCGCTTCCTCTGTGCTGTGACAACATTGGTGCCAGGGGAGATGGTGTTTTTCAAAGGGACCTACTGTAGCCACTTTAATTTACAATTAA GAGCCTTAGTTTGACTTAACACTTTTGTAGGCTTTTCATTGTGTATTTTTGTGTATGTGTGCATATAGCAGCTACTCTGTAGCAGAGGTG GGTAGAGACACTTAATAGTATCATGTCGCATGCAGATGTCACATCGGCCTCTGCAAAAACTGTACTGTCTTGTTTCTGCATTAGACTTAA GTAGTCATGTGAATATACTGCTATGTCACTTTTAATATTACGAGTTTTATACTTGGAAAATGGTACTTGCTTCTTTTAAATCTCTGTCTT CTCTAACCTCCCCCTTCCCATTTCAATGCTCCCTTCCTAATTTCAGCAATAATCTCAAAAAGCAATTAAATAGTTAAATGACCCTAATTG TAATTACTGTGGATGGTTGCATTCATTTGATTACTTGGGCACACACGAGATGACAAATGGGGCAGTGGCCATGCTTGAATGGGCTCCTGG TGAGAGATTGCCCCCTGGTGGTGAAACAATCGTGTGTGCCCACTGATACCAAGACCAATGAAAGAGACACAGTTAAGCAGCAATCCATCT CATTTCCAGGCACTTCAATAGGTCGCTGATTGGTCCTTGCACCAGCAGTGGTAGTCGTACCTATTTCAGAGAGGTCTGAAATTCAGGTTC TTAGTTTGCCAGGGACAGGCCCTATCTTATATTTTTTTCCATCTTCATCATCCACTTCTGCTTACAGTTTGCTGCTTACAATAACTTAAT GATGGATTGAGTTATCTGGGTGGTCTCTAGCCATCTGGGCAGTGTGGTTCTGTCTAACCAAAGGGCATTGGCCTCAAACCCTGCATTTGG TTTAGGGGCTAACAGAGCTCCTCAGATAATCTTCACACACATGTAACTGCTGGAGATCTTATTCTATTATGAATAAGAAACGAGAAGTTT TTCCAAAGTGTTAGTCAGGATCTGAAGGCTGTCATTCAGATAACCCAGCTTTTCCTTTTGGCTTTTAGCCCATTCAGACTTTGCCAGAGT CAAGCCAAGGATTGCTTTTTTGCTACAGTTTTCTGCCAAATGGCCTAGTTCCTGAGTACCTGGAAACCAGAGAGAAAGAGGATCCAGGAT GTACTTGGATGAGGAGGCCTGGCTTATCTAGGAAGTCGTGTCTGGGGTGCTTATTGCTGCTCCATACAGCTGTACGTCAGCCCCTTGGCC TTCTCTGTAGGTTCTTGGCAGCAATGAGCAGCTTTCACTCAGTGACACAAGTAATTACTGAGTCCTAATTTGATAGCCACCAACTGTACC TGGGTAGGCAAAGTCAGATTTTTGAGAACCTTTTTCCTGATTTGAAGTTTTAATTACCTTATTTTCTTTTATGCTTTCCTCTGTCTTGTA >59958_59958_1_NPLOC4-SGPL1_NPLOC4_chr17_79577236_ENST00000331134_SGPL1_chr10_72630805_ENST00000373202_length(amino acids)=514AA_BP=216 MVSRPRARGERIERGTGTSAAGGAGRAKLERRWRRRREAGAGAEAAAGSARPLGRQAAAARGSSPEAGAAAMAESIIIRVQSPDGVKRIT ATKRETAATFLKKVAKEFGFQNNGFSVYINRNKTGEITASSNKSLNLLKIKHGDLLFLFPSSLAGPSSEMETSVPPGFKVFGAPNVVEDE IDQYLSKQDGKIYRSRDPQLCRHGPLGKCVHCVPLEAMRRAISRNTAMLVCSTPQFPHGVIDPVPEVAKLAVKYKIPLHVDACLGGFLIV FMEKAGYPLEHPFDFRVKGVTSISADTHKYGYAPKGSSLVLYSDKKYRNYQFFVDTDWQGGIYASPTIAGSRPGGISAACWAALMHFGEN GYVEATKQIIKTARFLKSELENIKGIFVFGNPQLSVIALGSRDFDIYRLSNLMTAKGWNLNQLQFPPSIHFCITLLHARKRVAIQFLKDI -------------------------------------------------------------- >59958_59958_2_NPLOC4-SGPL1_NPLOC4_chr17_79577236_ENST00000374747_SGPL1_chr10_72630805_ENST00000373202_length(transcript)=5300nt_BP=565nt GCTGGAGCGGCGGTGGCGGCGGCGTAGGGAGGCCGGGGCCGGGGCTGAGGCCGCGGCCGGGTCTGCGAGGCCCTTGGGGCGGCAGGCGGC GGCGGCCCGGGGCTCGAGCCCGGAGGCAGGAGCAGCCGCCATGGCCGAGAGCATCATAATTCGTGTCCAGTCCCCGGATGGAGTGAAGCG GATCACAGCAACAAAGAGAGAAACAGCAGCAACATTTTTGAAAAAGGTTGCAAAGGAGTTTGGCTTCCAAAATAATGGCTTCTCGGTTTA CATCAATAGAAACAAGACCGGAGAGATAACAGCCTCCTCCAACAAATCCCTCAACTTGCTAAAAATCAAGCATGGCGATTTGTTGTTCCT GTTTCCCTCGAGCCTTGCTGGGCCCTCATCTGAAATGGAGACGTCAGTTCCACCGGGCTTCAAAGTCTTTGGCGCTCCCAACGTGGTGGA GGATGAGATTGATCAGTACCTCAGCAAACAGGACGGGAAGATTTACAGAAGCCGAGACCCACAGCTATGCCGCCACGGCCCTTTGGGGAA ATGCGTGCACTGCGTCCCTCTAGAGGCAATGAGAAGAGCTATCTCCAGGAACACTGCCATGCTCGTCTGTTCTACCCCACAGTTTCCTCA TGGTGTAATAGATCCTGTCCCTGAAGTGGCCAAGCTGGCTGTCAAATACAAAATACCCCTTCATGTCGACGCTTGTCTGGGAGGCTTCCT CATCGTCTTTATGGAGAAAGCAGGATACCCACTGGAGCACCCATTTGATTTCCGGGTGAAAGGTGTAACCAGCATTTCAGCTGACACCCA TAAGTATGGCTATGCCCCAAAAGGCTCATCATTGGTGTTGTATAGTGACAAGAAGTACAGGAACTATCAGTTCTTCGTCGATACAGATTG GCAGGGTGGCATCTATGCTTCCCCAACCATCGCAGGCTCACGGCCTGGTGGCATTAGCGCAGCCTGTTGGGCTGCCTTGATGCACTTCGG TGAGAACGGCTATGTTGAAGCTACCAAACAGATCATCAAAACTGCTCGCTTCCTCAAGTCAGAACTGGAAAATATCAAAGGCATCTTTGT TTTTGGGAATCCCCAATTGTCAGTCATTGCTCTGGGATCCCGTGATTTTGACATCTACCGACTATCAAACCTGATGACTGCTAAGGGGTG GAACTTGAACCAGTTGCAGTTCCCACCCAGTATTCATTTCTGCATCACATTACTACACGCCCGGAAACGAGTAGCTATACAATTCCTAAA GGACATTCGAGAATCTGTCACTCAAATCATGAAGAATCCTAAAGCGAAGACCACAGGAATGGGTGCCATCTATGGCATGGCCCAGACAAC TGTTGACAGGAATATGGTTGCAGAATTGTCCTCAGTCTTCTTGGACAGCTTGTACAGCACCGACACTGTCACCCAGGGCAGCCAGATGAA TGGTTCTCCAAAACCCCACTGAACTTGGACCCTTTCTAGTCTCAAGGGGATTCCAGCCTTCAGAAGGTTCTTGGGATATGGAACAGGCCG TGCACAACTTTGACATCTGGTCTTGCTCCATAGAGCACAACTCAAGATAGACCATGAGACAGCTTGAGCCTCAGGATTCTTGTTCTTCCT CTTATCTTCCTTTTGTGGTTTTTAATTTGAAGACCCCAGAGAATTCCATTACATAATGATTTTGCCCTTGTTATAAATGTTACCCTAGGA ATTGTTTTAACCATTTCCTTTTCTAAACTCTCTAGCTTTCAACTTTACTTAAACATTGTGTGGTAGCTCTGACCTGTCCTGATTCTTTAG AGAAGCTGGGGTACAGTTTATGAGATAGCTAGAGCTTCTTTGTTATCTCAGGCAGGAGGCGTTTACATAACAGATGTTTCCTCAGCTGGG TGTGAGGTATACTCTAAGCAGGAGGCTTTTTCAGCCTTCTCTCTCTTTTTTTTTTTTTTTTTTTTTTTTGAGATGGAATTTTGCTCTTTT GCCCAGTCTGGAGTGCAGTGGCATGATCTCAGCTCACTGCAACCTCCACCCACTGGGTTCAAGCGATTCTTCTGCCTCAGCCTCCCGAGT AGCTGGGATTACCGGCACCCACCACCACGCCTGGCTAATTTTTCAATTTTCTTTTTCAGTAGAGACGGGTTCACCGTGTTGGCCAGGCTG GTCTTGAACTCCTGACCTCAGGTGATACCCGCCCCCCCGCCTCAGCCTCCCAAAGTGCTGGGATTACAGGCGTGAGCCACCGTGCCTGGC CCTGTCTCTCTTAAGAGTAGGTTCATTGTCTGTCTTAGAGTCACTTCTATTGCAACTCATTTTCTTTTTCCAGGGCACAGATCGACCAAG CTGCCGTTCCCTATTCTGCAGGACAGGACTATTCTAGCATACCTGCTTCGTCCACCCAGGCAGGGTTTGGGGTGGTCTCTTCTGTGCCTG CAGTCCCCATTTGACACTTGGTTGCCACCATCTTTGGAGATTATTGTTTGGAATGATGCTTCCATTGGCTTTTTCTTGTTACCATGGACT AGGAAGAAAACATGGTTTCCAAATAATCTGGGAGCTTTTGGCCATGGTGCCGCCTTCCTGAATTGGCAGTGGTCAGAGCACACCTGAACC CTATCCTGGGCTGGTGATGAGCAGAAATCAGACCTTTTTCTATGCTTTTTTGAATATCAGAGTAGGATGAACACCCAGATTCAAATATGT CACCAAAGTTGGTGGTGGTCCTTCCCTGCACCCTTGCGTTAAGCCATTATGTAATGAAAATGTGTTTGCTTGAAGGAACAGCTCAAAGCA CCTTCACAAGTTGCCTTGACTTACCCTAGGTGGGTGTGAAAGAGCACCCGTAGCAAGGAAAATTTTCTCTATTAGTGTGTTCTTCTGCCT CTTCCCCCTTGATTCAGCTTTCAGAGGTACTATGGCAGTTTTGCCTCAGGTGCTGAACATTTCTCAGCCCTGGCTAAAAGGGAGCAGCAC AGGGAGAGAAACAGGATAGGAAAGCAGAATGGCGAGCAGCCTATGGCCCAGGGCCTGTAATCCCTTCCCAAGACTAGCTGCTCAGGGTGG TGCAGGGACAGGACCAGACCCTGCGCCTATTTCCTGCCTTCTTTCCCCTATAGGGAACTCTGTAGGCTGAGCCACTGTCCTGCTCTTATG ACATTATATCTTGTGCCTTTCTCCTCAGCAGTGAGCAGTGAGCTACTCCTGGCCCAGGCCCTAGGGGAAATGGATCAGTCTTTGAGGTTT CTATTTGGGGAGGGGAGTACTTAAGATGAGTCAAAAGACACTTTCCTCTGTTCCATTCCCCATCTCAGGGACTCCTGAATATTCAGCCTC TCCAGGCTGGTGTCTTCTAGTTTCCCCCACTGGGAATGCTGGCTGGGAGAGCCATGACTACCAGACTTTTCCTCAGGCTCCTTGGCATGT TAGTCTGAATTGTTCTTGAGCACTGTACTACTGACCCAACAACTGTGACTAGCTGGCCACGCCATTCAGGGCTGGTGTGGCATTTATGTG TGTGTGTGTGTGTGTGTGTGTTTTTCCTGTTTGCCCAGCAGTGCATTGTGGGTTCCAAGAGTGGGTAGTGTGTGTATGTGTGTGTGTCAG AGGGAGACCTGGCAGGCACCTCTTTGAGAGTAGCTGTGGTCAGAGCTGTTTGGTCAGTGCATTATGTTGAATGAGGTCCAGGAACCCAGA GCCACCCAGCAGACACCACTGTGGCTTGCCAGCTGCCAAGATGGAGAAGCATGTGCCCCTGTAGAGCGTCTCCCCAGAACCAGACCCCGA GCCACTCGCTTCCTCTGTGCTGTGACAACATTGGTGCCAGGGGAGATGGTGTTTTTCAAAGGGACCTACTGTAGCCACTTTAATTTACAA TTAAGAGCCTTAGTTTGACTTAACACTTTTGTAGGCTTTTCATTGTGTATTTTTGTGTATGTGTGCATATAGCAGCTACTCTGTAGCAGA GGTGGGTAGAGACACTTAATAGTATCATGTCGCATGCAGATGTCACATCGGCCTCTGCAAAAACTGTACTGTCTTGTTTCTGCATTAGAC TTAAGTAGTCATGTGAATATACTGCTATGTCACTTTTAATATTACGAGTTTTATACTTGGAAAATGGTACTTGCTTCTTTTAAATCTCTG TCTTCTCTAACCTCCCCCTTCCCATTTCAATGCTCCCTTCCTAATTTCAGCAATAATCTCAAAAAGCAATTAAATAGTTAAATGACCCTA ATTGTAATTACTGTGGATGGTTGCATTCATTTGATTACTTGGGCACACACGAGATGACAAATGGGGCAGTGGCCATGCTTGAATGGGCTC CTGGTGAGAGATTGCCCCCTGGTGGTGAAACAATCGTGTGTGCCCACTGATACCAAGACCAATGAAAGAGACACAGTTAAGCAGCAATCC ATCTCATTTCCAGGCACTTCAATAGGTCGCTGATTGGTCCTTGCACCAGCAGTGGTAGTCGTACCTATTTCAGAGAGGTCTGAAATTCAG GTTCTTAGTTTGCCAGGGACAGGCCCTATCTTATATTTTTTTCCATCTTCATCATCCACTTCTGCTTACAGTTTGCTGCTTACAATAACT TAATGATGGATTGAGTTATCTGGGTGGTCTCTAGCCATCTGGGCAGTGTGGTTCTGTCTAACCAAAGGGCATTGGCCTCAAACCCTGCAT TTGGTTTAGGGGCTAACAGAGCTCCTCAGATAATCTTCACACACATGTAACTGCTGGAGATCTTATTCTATTATGAATAAGAAACGAGAA GTTTTTCCAAAGTGTTAGTCAGGATCTGAAGGCTGTCATTCAGATAACCCAGCTTTTCCTTTTGGCTTTTAGCCCATTCAGACTTTGCCA GAGTCAAGCCAAGGATTGCTTTTTTGCTACAGTTTTCTGCCAAATGGCCTAGTTCCTGAGTACCTGGAAACCAGAGAGAAAGAGGATCCA GGATGTACTTGGATGAGGAGGCCTGGCTTATCTAGGAAGTCGTGTCTGGGGTGCTTATTGCTGCTCCATACAGCTGTACGTCAGCCCCTT GGCCTTCTCTGTAGGTTCTTGGCAGCAATGAGCAGCTTTCACTCAGTGACACAAGTAATTACTGAGTCCTAATTTGATAGCCACCAACTG TACCTGGGTAGGCAAAGTCAGATTTTTGAGAACCTTTTTCCTGATTTGAAGTTTTAATTACCTTATTTTCTTTTATGCTTTCCTCTGTCT >59958_59958_2_NPLOC4-SGPL1_NPLOC4_chr17_79577236_ENST00000374747_SGPL1_chr10_72630805_ENST00000373202_length(amino acids)=486AA_BP=188 LERRWRRRREAGAGAEAAAGSARPLGRQAAAARGSSPEAGAAAMAESIIIRVQSPDGVKRITATKRETAATFLKKVAKEFGFQNNGFSVY INRNKTGEITASSNKSLNLLKIKHGDLLFLFPSSLAGPSSEMETSVPPGFKVFGAPNVVEDEIDQYLSKQDGKIYRSRDPQLCRHGPLGK CVHCVPLEAMRRAISRNTAMLVCSTPQFPHGVIDPVPEVAKLAVKYKIPLHVDACLGGFLIVFMEKAGYPLEHPFDFRVKGVTSISADTH KYGYAPKGSSLVLYSDKKYRNYQFFVDTDWQGGIYASPTIAGSRPGGISAACWAALMHFGENGYVEATKQIIKTARFLKSELENIKGIFV FGNPQLSVIALGSRDFDIYRLSNLMTAKGWNLNQLQFPPSIHFCITLLHARKRVAIQFLKDIRESVTQIMKNPKAKTTGMGAIYGMAQTT -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for NPLOC4-SGPL1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for NPLOC4-SGPL1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for NPLOC4-SGPL1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
