
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:NQO1-JAG1 (FusionGDB2 ID:60056) |
Fusion Gene Summary for NQO1-JAG1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: NQO1-JAG1 | Fusion gene ID: 60056 | Hgene | Tgene | Gene symbol | NQO1 | JAG1 | Gene ID | 1728 | 182 |
| Gene name | NAD(P)H quinone dehydrogenase 1 | jagged canonical Notch ligand 1 | |
| Synonyms | DHQU|DIA4|DTD|NMOR1|NMORI|QR1 | AGS|AGS1|AHD|AWS|CD339|DCHE|HJ1|JAGL1 | |
| Cytomap | 16q22.1 | 20p12.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | NAD(P)H dehydrogenase [quinone] 1DT-diaphoraseNAD(P)H dehydrogenase, quinone 1NAD(P)H:Quinone acceptor oxidoreductase type 1NAD(P)H:menadione oxidoreductase 1NAD(P)H:quinone oxidoreductase 1NAD(P)H:quinone oxireductaseazoreductasediaphorase (NADH/ | protein jagged-1 | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | P15559 | P78504 | |
| Ensembl transtripts involved in fusion gene | ENST00000320623, ENST00000379046, ENST00000379047, ENST00000439109, ENST00000561500, ENST00000564043, | ENST00000488480, ENST00000254958, ENST00000423891, | |
| Fusion gene scores | * DoF score | 12 X 11 X 5=660 | 8 X 9 X 4=288 |
| # samples | 13 | 8 | |
| ** MAII score | log2(13/660*10)=-2.34395440121736 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(8/288*10)=-1.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: NQO1 [Title/Abstract] AND JAG1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | NQO1(69760336)-JAG1(10621892), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | JAG1 | GO:0001953 | negative regulation of cell-matrix adhesion | 11549580 |
| Tgene | JAG1 | GO:0022408 | negative regulation of cell-cell adhesion | 11549580 |
| Tgene | JAG1 | GO:0030336 | negative regulation of cell migration | 11549580 |
 Fusion gene breakpoints across NQO1 (5'-gene) Fusion gene breakpoints across NQO1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
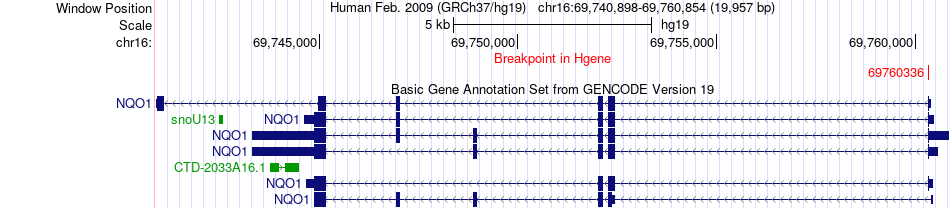 |
 Fusion gene breakpoints across JAG1 (3'-gene) Fusion gene breakpoints across JAG1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
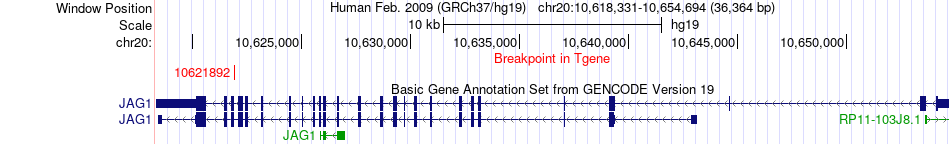 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-55-6987-01A | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - |
Top |
Fusion Gene ORF analysis for NQO1-JAG1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000320623 | ENST00000488480 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - |
| 5CDS-intron | ENST00000379046 | ENST00000488480 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - |
| 5CDS-intron | ENST00000379047 | ENST00000488480 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - |
| 5CDS-intron | ENST00000439109 | ENST00000488480 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - |
| 5CDS-intron | ENST00000561500 | ENST00000488480 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - |
| In-frame | ENST00000320623 | ENST00000254958 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - |
| In-frame | ENST00000320623 | ENST00000423891 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - |
| In-frame | ENST00000379046 | ENST00000254958 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - |
| In-frame | ENST00000379046 | ENST00000423891 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - |
| In-frame | ENST00000379047 | ENST00000254958 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - |
| In-frame | ENST00000379047 | ENST00000423891 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - |
| In-frame | ENST00000439109 | ENST00000254958 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - |
| In-frame | ENST00000439109 | ENST00000423891 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - |
| In-frame | ENST00000561500 | ENST00000254958 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - |
| In-frame | ENST00000561500 | ENST00000423891 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - |
| intron-3CDS | ENST00000564043 | ENST00000254958 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - |
| intron-3CDS | ENST00000564043 | ENST00000423891 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - |
| intron-intron | ENST00000564043 | ENST00000488480 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000561500 | NQO1 | chr16 | 69760336 | - | ENST00000254958 | JAG1 | chr20 | 10621892 | - | 2629 | 74 | 104 | 814 | 236 |
| ENST00000561500 | NQO1 | chr16 | 69760336 | - | ENST00000423891 | JAG1 | chr20 | 10621892 | - | 1050 | 74 | 104 | 814 | 236 |
| ENST00000379047 | NQO1 | chr16 | 69760336 | - | ENST00000254958 | JAG1 | chr20 | 10621892 | - | 2791 | 236 | 92 | 976 | 294 |
| ENST00000379047 | NQO1 | chr16 | 69760336 | - | ENST00000423891 | JAG1 | chr20 | 10621892 | - | 1212 | 236 | 92 | 976 | 294 |
| ENST00000320623 | NQO1 | chr16 | 69760336 | - | ENST00000254958 | JAG1 | chr20 | 10621892 | - | 3074 | 519 | 375 | 1259 | 294 |
| ENST00000320623 | NQO1 | chr16 | 69760336 | - | ENST00000423891 | JAG1 | chr20 | 10621892 | - | 1495 | 519 | 375 | 1259 | 294 |
| ENST00000379046 | NQO1 | chr16 | 69760336 | - | ENST00000254958 | JAG1 | chr20 | 10621892 | - | 2707 | 152 | 8 | 892 | 294 |
| ENST00000379046 | NQO1 | chr16 | 69760336 | - | ENST00000423891 | JAG1 | chr20 | 10621892 | - | 1128 | 152 | 8 | 892 | 294 |
| ENST00000439109 | NQO1 | chr16 | 69760336 | - | ENST00000254958 | JAG1 | chr20 | 10621892 | - | 2683 | 128 | 158 | 868 | 236 |
| ENST00000439109 | NQO1 | chr16 | 69760336 | - | ENST00000423891 | JAG1 | chr20 | 10621892 | - | 1104 | 128 | 158 | 868 | 236 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000561500 | ENST00000254958 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - | 0.14948948 | 0.8505105 |
| ENST00000561500 | ENST00000423891 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - | 0.070953906 | 0.9290461 |
| ENST00000379047 | ENST00000254958 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - | 0.000696212 | 0.9993038 |
| ENST00000379047 | ENST00000423891 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - | 0.005296817 | 0.9947031 |
| ENST00000320623 | ENST00000254958 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - | 0.000986805 | 0.9990132 |
| ENST00000320623 | ENST00000423891 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - | 0.010958166 | 0.98904186 |
| ENST00000379046 | ENST00000254958 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - | 0.000700105 | 0.9992999 |
| ENST00000379046 | ENST00000423891 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - | 0.005086717 | 0.9949132 |
| ENST00000439109 | ENST00000254958 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - | 0.06263284 | 0.9373672 |
| ENST00000439109 | ENST00000423891 | NQO1 | chr16 | 69760336 | - | JAG1 | chr20 | 10621892 | - | 0.055590816 | 0.9444092 |
Top |
Fusion Genomic Features for NQO1-JAG1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
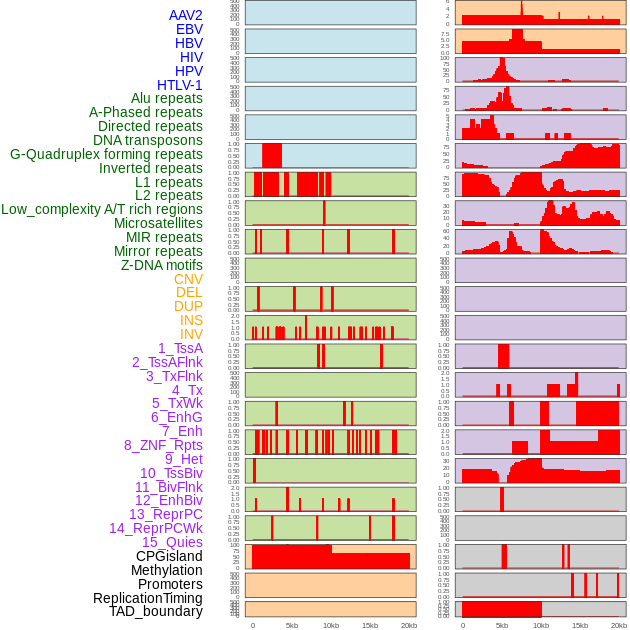 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for NQO1-JAG1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:69760336/chr20:10621892) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NQO1 | JAG1 |
| FUNCTION: The enzyme apparently serves as a quinone reductase in connection with conjugation reactions of hydroquinons involved in detoxification pathways as well as in biosynthetic processes such as the vitamin K-dependent gamma-carboxylation of glutamate residues in prothrombin synthesis. | FUNCTION: Ligand for multiple Notch receptors and involved in the mediation of Notch signaling (PubMed:18660822, PubMed:20437614). May be involved in cell-fate decisions during hematopoiesis (PubMed:9462510). Seems to be involved in early and late stages of mammalian cardiovascular development. Inhibits myoblast differentiation (By similarity). Enhances fibroblast growth factor-induced angiogenesis (in vitro). {ECO:0000250, ECO:0000269|PubMed:18660822, ECO:0000269|PubMed:20437614, ECO:0000269|PubMed:9462510}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 1094_1218 | 972 | 1219.0 | Topological domain | Cytoplasmic | |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 1068_1093 | 972 | 1219.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NQO1 | chr16:69760336 | chr20:10621892 | ENST00000320623 | - | 1 | 6 | 104_107 | 2 | 275.0 | Nucleotide binding | FAD |
| Hgene | NQO1 | chr16:69760336 | chr20:10621892 | ENST00000320623 | - | 1 | 6 | 148_151 | 2 | 275.0 | Nucleotide binding | FAD |
| Hgene | NQO1 | chr16:69760336 | chr20:10621892 | ENST00000320623 | - | 1 | 6 | 18_19 | 2 | 275.0 | Nucleotide binding | FAD |
| Hgene | NQO1 | chr16:69760336 | chr20:10621892 | ENST00000379046 | - | 1 | 5 | 104_107 | 2 | 237.0 | Nucleotide binding | FAD |
| Hgene | NQO1 | chr16:69760336 | chr20:10621892 | ENST00000379046 | - | 1 | 5 | 148_151 | 2 | 237.0 | Nucleotide binding | FAD |
| Hgene | NQO1 | chr16:69760336 | chr20:10621892 | ENST00000379046 | - | 1 | 5 | 18_19 | 2 | 237.0 | Nucleotide binding | FAD |
| Hgene | NQO1 | chr16:69760336 | chr20:10621892 | ENST00000379047 | - | 1 | 5 | 104_107 | 2 | 241.0 | Nucleotide binding | FAD |
| Hgene | NQO1 | chr16:69760336 | chr20:10621892 | ENST00000379047 | - | 1 | 5 | 148_151 | 2 | 241.0 | Nucleotide binding | FAD |
| Hgene | NQO1 | chr16:69760336 | chr20:10621892 | ENST00000379047 | - | 1 | 5 | 18_19 | 2 | 241.0 | Nucleotide binding | FAD |
| Hgene | NQO1 | chr16:69760336 | chr20:10621892 | ENST00000320623 | - | 1 | 6 | 126_128 | 2 | 275.0 | Region | Note=Substrate binding |
| Hgene | NQO1 | chr16:69760336 | chr20:10621892 | ENST00000379046 | - | 1 | 5 | 126_128 | 2 | 237.0 | Region | Note=Substrate binding |
| Hgene | NQO1 | chr16:69760336 | chr20:10621892 | ENST00000379047 | - | 1 | 5 | 126_128 | 2 | 241.0 | Region | Note=Substrate binding |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 185_229 | 972 | 1219.0 | Domain | DSL | |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 230_263 | 972 | 1219.0 | Domain | EGF-like 1 | |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 264_294 | 972 | 1219.0 | Domain | EGF-like 2%3B atypical | |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 296_334 | 972 | 1219.0 | Domain | EGF-like 3 | |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 336_372 | 972 | 1219.0 | Domain | EGF-like 4 | |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 374_410 | 972 | 1219.0 | Domain | EGF-like 5%3B calcium-binding | |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 412_448 | 972 | 1219.0 | Domain | EGF-like 6%3B calcium-binding | |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 450_485 | 972 | 1219.0 | Domain | EGF-like 7%3B calcium-binding | |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 487_523 | 972 | 1219.0 | Domain | EGF-like 8%3B calcium-binding | |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 525_561 | 972 | 1219.0 | Domain | EGF-like 9 | |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 586_627 | 972 | 1219.0 | Domain | EGF-like 10 | |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 629_665 | 972 | 1219.0 | Domain | EGF-like 11%3B calcium-binding | |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 667_703 | 972 | 1219.0 | Domain | EGF-like 12%3B calcium-binding | |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 705_741 | 972 | 1219.0 | Domain | EGF-like 13 | |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 744_780 | 972 | 1219.0 | Domain | EGF-like 14 | |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 782_818 | 972 | 1219.0 | Domain | EGF-like 15%3B calcium-binding | |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 820_856 | 972 | 1219.0 | Domain | EGF-like 16%3B calcium-binding | |
| Tgene | JAG1 | chr16:69760336 | chr20:10621892 | ENST00000254958 | 22 | 26 | 34_1067 | 972 | 1219.0 | Topological domain | Extracellular |
Top |
Fusion Gene Sequence for NQO1-JAG1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >60056_60056_1_NQO1-JAG1_NQO1_chr16_69760336_ENST00000320623_JAG1_chr20_10621892_ENST00000254958_length(transcript)=3074nt_BP=519nt TTTCGTGCCAAGGCTAAAAGGGCATGCCCACTTGATCCCTGGACTCTCTTGGGACGACTTCCACCCTGCATCCTCTTGCACCTCAGGGCA CAGTGCGCAGATGGGCTTGCCTTAGCACCCCCAGCCAGATTTTTGAGGCCTCTGTCACACACACCCCTACAATCCCCTCCCCCAGCCCCG AGAGACTTTTCTTGACTTCCACCAGTTGCTCCGGCGGGTGAGAGTGGAGAGGCCCCTCCTTCATCCCCCAGGCTCCCTCCCTTCCTGGAG CTGCAGCCTCAGCATCCTCCGCCCAGCACCCCAGGATTCAGGCGTTGGGTCCCGCCCTTGTAGGCTGTCCACCTCAAACGGGCCGGACAG GATATATAAGAGAGAATGCACCGTGCACTACACACGCGACTCCCACAAGGTTGCAGCCGGAGCCGCCCAGCTCACCGAGAGCCTAGTTCC GGCCAGGGTCGCCCCGGCAACCACGAGCCCAGCCAATCAGCGCCCCGGACTGCACCAGAGCCATGGTCGGGTCTTACTACGGAGCACATT TGCAGTGAATTGAGGAATTTGAATATTTTGAAGAATGTTTCCGCTGAATATTCAATCTACATCGCTTGCGAGCCTTCCCCTTCAGCGAAC AATGAAATACATGTGGCCATTTCTGCTGAAGATATACGGGATGATGGGAACCCGATCAAGGAAATCACTGACAAAATAATCGATCTTGTT AGTAAACGTGATGGAAACAGCTCGCTGATTGCTGCCGTTGCAGAAGTAAGAGTTCAGAGGCGGCCTCTGAAGAACAGAACAGATTTCCTT GTTCCCTTGCTGAGCTCTGTCTTAACTGTGGCTTGGATCTGTTGCTTGGTGACGGCCTTCTACTGGTGCCTGCGGAAGCGGCGGAAGCCG GGCAGCCACACACACTCAGCCTCTGAGGACAACACCACCAACAACGTGCGGGAGCAGCTGAACCAGATCAAAAACCCCATTGAGAAACAT GGGGCCAACACGGTCCCCATCAAGGATTATGAGAACAAGAACTCCAAAATGTCTAAAATAAGGACACACAATTCTGAAGTAGAAGAGGAC GACATGGACAAACACCAGCAGAAAGCCCGGTTTGCCAAGCAGCCGGCGTACACGCTGGTAGACAGAGAAGAGAAGCCCCCCAACGGCACG CCGACAAAACACCCAAACTGGACAAACAAACAGGACAACAGAGACTTGGAAAGTGCCCAGAGCTTAAACCGAATGGAGTACATCGTATAG CAGACCGCGGGCACTGCCGCCGCTAGGTAGAGTCTGAGGGCTTGTAGTTCTTTAAACTGTCGTGTCATACTCGAGTCTGAGGCCGTTGCT GACTTAGAATCCCTGTGTTAATTTAAGTTTTGACAAGCTGGCTTACACTGGCAATGGTAGTTTCTGTGGTTGGCTGGGAAATCGAGTGCC GCATCTCACAGCTATGCAAAAAGCTAGTCAACAGTACCCTGGTTGTGTGTCCCCTTGCAGCCGACACGGTCTCGGATCAGGCTCCCAGGA GCCTGCCCAGCCCCCTGGTCTTTGAGCTCCCACTTCTGCCAGATGTCCTAATGGTGATGCAGTCTTAGATCATAGTTTTATTTATATTTA TTGACTCTTGAGTTGTTTTTGTATATTGGTTTTATGATGACGTACAAGTAGTTCTGTATTTGAAAGTGCCTTTGCAGCTCAGAACCACAG CAACGATCACAAATGACTTTATTATTTATTTTTTTTAATTGTATTTTTGTTGTTGGGGGAGGGGAGACTTTGATGTCAGCAGTTGCTGGT AAAATGAAGAATTTAAAGAAAAAAATGTCAAAAGTAGAACTTTGTATAGTTATGTAAATAATTCTTTTTTATTAATCACTGTGTATATTT GATTTATTAACTTAATAATCAAGAGCCTTAAAACATCATTCCTTTTTATTTATATGTATGTGTTTAGAATTGAAGGTTTTTGATAGCATT GTAAGCGTATGGCTTTATTTTTTTGAACTCTTCTCATTACTTGTTGCCTATAAGCCAAAATTAAGGTGTTTGAAAATAGTTTATTTTAAA ACAATAGGATGGGCTTCTGTGCCCAGAATACTGATGGAATTTTTTTGTACGACGTCAGATGTTTAAAACACCTTCTATAGCATCACTTAA AACACGTTTTAAGGACTGACTGAGGCAGTTTGAGGATTAGTTTAGAACAGGTTTTTTTGTTTGTTTGTTTTTTGTTTTTCTGCTTTAGAC TTGAAAAGAGACAGGCAGGTGATCTGCTGCAGAGCAGTAAGGGAACAAGTTGAGCTATGACTTAACATAGCCAAAATGTGAGTGGTTGAA TATGATTAAAAATATCAAATTAATTGTGTGAACTTGGAAGCACACCAATCTTACTTTGTAAATTCTGATTTCTTTTCACCATTCGTACAT AATACTGAACCACTTGTAGATTTGATTTTTTTTTTTAATCTACTGCATTTAGGGAGTATTCTAATAAGCTAGTTGAATACTTGAACCATA AAATGTCCAGTAAGATCACTGTTTAGATTTGCCATAGAGTACACTGCCTGCCTTAAGTGAGGAAATCAAAGTGCTATTACGAAGTTCAAG ATCAAAAAGGCTTATAAAACAGAGTAATCTTGTTGGTTCACCATTGAGACCGTGAAGATACTTTGTATTGTCCTATTAGTGTTATATGAA CATACAAATGCATCTTTGATGTGTTGTTCTTGGCAATAAATTTTGAAAAGTAATATTTATTAAATTTTTTTGTATGAAAACATGGAACAG TGTGGCCTCTTCTGAGCTTACGTAGTTCTACCGGCTTTGCCATGTGCTTCTGCCACCCTGCTGAGTCTGTTCTGGTAATCGGGGTATAAT AGGCTCTGCCTGACAGAGGGATGGAGGAAGAACTGAAAGGCTTTTCAACCACAAAACTCATCTGGAGTTCTCAAAGACCTGGGGCTGCTG TGAAGCTGGAACTGCGGGAGCCCCATCTAGGGGAGCCTTGATTCCCTTGTTATTCAACAGCAAGTGTGAATACTGCTTGAATAAACACCA >60056_60056_1_NQO1-JAG1_NQO1_chr16_69760336_ENST00000320623_JAG1_chr20_10621892_ENST00000254958_length(amino acids)=294AA_BP=48 MHRALHTRLPQGCSRSRPAHREPSSGQGRPGNHEPSQSAPRTAPEPWSGLTTEHICSELRNLNILKNVSAEYSIYIACEPSPSANNEIHV AISAEDIRDDGNPIKEITDKIIDLVSKRDGNSSLIAAVAEVRVQRRPLKNRTDFLVPLLSSVLTVAWICCLVTAFYWCLRKRRKPGSHTH SASEDNTTNNVREQLNQIKNPIEKHGANTVPIKDYENKNSKMSKIRTHNSEVEEDDMDKHQQKARFAKQPAYTLVDREEKPPNGTPTKHP -------------------------------------------------------------- >60056_60056_2_NQO1-JAG1_NQO1_chr16_69760336_ENST00000320623_JAG1_chr20_10621892_ENST00000423891_length(transcript)=1495nt_BP=519nt TTTCGTGCCAAGGCTAAAAGGGCATGCCCACTTGATCCCTGGACTCTCTTGGGACGACTTCCACCCTGCATCCTCTTGCACCTCAGGGCA CAGTGCGCAGATGGGCTTGCCTTAGCACCCCCAGCCAGATTTTTGAGGCCTCTGTCACACACACCCCTACAATCCCCTCCCCCAGCCCCG AGAGACTTTTCTTGACTTCCACCAGTTGCTCCGGCGGGTGAGAGTGGAGAGGCCCCTCCTTCATCCCCCAGGCTCCCTCCCTTCCTGGAG CTGCAGCCTCAGCATCCTCCGCCCAGCACCCCAGGATTCAGGCGTTGGGTCCCGCCCTTGTAGGCTGTCCACCTCAAACGGGCCGGACAG GATATATAAGAGAGAATGCACCGTGCACTACACACGCGACTCCCACAAGGTTGCAGCCGGAGCCGCCCAGCTCACCGAGAGCCTAGTTCC GGCCAGGGTCGCCCCGGCAACCACGAGCCCAGCCAATCAGCGCCCCGGACTGCACCAGAGCCATGGTCGGGTCTTACTACGGAGCACATT TGCAGTGAATTGAGGAATTTGAATATTTTGAAGAATGTTTCCGCTGAATATTCAATCTACATCGCTTGCGAGCCTTCCCCTTCAGCGAAC AATGAAATACATGTGGCCATTTCTGCTGAAGATATACGGGATGATGGGAACCCGATCAAGGAAATCACTGACAAAATAATCGATCTTGTT AGTAAACGTGATGGAAACAGCTCGCTGATTGCTGCCGTTGCAGAAGTAAGAGTTCAGAGGCGGCCTCTGAAGAACAGAACAGATTTCCTT GTTCCCTTGCTGAGCTCTGTCTTAACTGTGGCTTGGATCTGTTGCTTGGTGACGGCCTTCTACTGGTGCCTGCGGAAGCGGCGGAAGCCG GGCAGCCACACACACTCAGCCTCTGAGGACAACACCACCAACAACGTGCGGGAGCAGCTGAACCAGATCAAAAACCCCATTGAGAAACAT GGGGCCAACACGGTCCCCATCAAGGATTATGAGAACAAGAACTCCAAAATGTCTAAAATAAGGACACACAATTCTGAAGTAGAAGAGGAC GACATGGACAAACACCAGCAGAAAGCCCGGTTTGCCAAGCAGCCGGCGTACACGCTGGTAGACAGAGAAGAGAAGCCCCCCAACGGCACG CCGACAAAACACCCAAACTGGACAAACAAACAGGACAACAGAGACTTGGAAAGTGCCCAGAGCTTAAACCGAATGGAGTACATCGTATAG CAGACCGCGGGCACTGCCGCCGCTAGTGTGGCCTCTTCTGAGCTTACGTAGTTCTACCGGCTTTGCCATGTGCTTCTGCCACCCTGCTGA GTCTGTTCTGGTAATCGGGGTATAATAGGCTCTGCCTGACAGAGGGATGGAGGAAGAACTGAAAGGCTTTTCAACCACAAAACTCATCTG >60056_60056_2_NQO1-JAG1_NQO1_chr16_69760336_ENST00000320623_JAG1_chr20_10621892_ENST00000423891_length(amino acids)=294AA_BP=48 MHRALHTRLPQGCSRSRPAHREPSSGQGRPGNHEPSQSAPRTAPEPWSGLTTEHICSELRNLNILKNVSAEYSIYIACEPSPSANNEIHV AISAEDIRDDGNPIKEITDKIIDLVSKRDGNSSLIAAVAEVRVQRRPLKNRTDFLVPLLSSVLTVAWICCLVTAFYWCLRKRRKPGSHTH SASEDNTTNNVREQLNQIKNPIEKHGANTVPIKDYENKNSKMSKIRTHNSEVEEDDMDKHQQKARFAKQPAYTLVDREEKPPNGTPTKHP -------------------------------------------------------------- >60056_60056_3_NQO1-JAG1_NQO1_chr16_69760336_ENST00000379046_JAG1_chr20_10621892_ENST00000254958_length(transcript)=2707nt_BP=152nt AAGAGAGAATGCACCGTGCACTACACACGCGACTCCCACAAGGTTGCAGCCGGAGCCGCCCAGCTCACCGAGAGCCTAGTTCCGGCCAGG GTCGCCCCGGCAACCACGAGCCCAGCCAATCAGCGCCCCGGACTGCACCAGAGCCATGGTCGGGTCTTACTACGGAGCACATTTGCAGTG AATTGAGGAATTTGAATATTTTGAAGAATGTTTCCGCTGAATATTCAATCTACATCGCTTGCGAGCCTTCCCCTTCAGCGAACAATGAAA TACATGTGGCCATTTCTGCTGAAGATATACGGGATGATGGGAACCCGATCAAGGAAATCACTGACAAAATAATCGATCTTGTTAGTAAAC GTGATGGAAACAGCTCGCTGATTGCTGCCGTTGCAGAAGTAAGAGTTCAGAGGCGGCCTCTGAAGAACAGAACAGATTTCCTTGTTCCCT TGCTGAGCTCTGTCTTAACTGTGGCTTGGATCTGTTGCTTGGTGACGGCCTTCTACTGGTGCCTGCGGAAGCGGCGGAAGCCGGGCAGCC ACACACACTCAGCCTCTGAGGACAACACCACCAACAACGTGCGGGAGCAGCTGAACCAGATCAAAAACCCCATTGAGAAACATGGGGCCA ACACGGTCCCCATCAAGGATTATGAGAACAAGAACTCCAAAATGTCTAAAATAAGGACACACAATTCTGAAGTAGAAGAGGACGACATGG ACAAACACCAGCAGAAAGCCCGGTTTGCCAAGCAGCCGGCGTACACGCTGGTAGACAGAGAAGAGAAGCCCCCCAACGGCACGCCGACAA AACACCCAAACTGGACAAACAAACAGGACAACAGAGACTTGGAAAGTGCCCAGAGCTTAAACCGAATGGAGTACATCGTATAGCAGACCG CGGGCACTGCCGCCGCTAGGTAGAGTCTGAGGGCTTGTAGTTCTTTAAACTGTCGTGTCATACTCGAGTCTGAGGCCGTTGCTGACTTAG AATCCCTGTGTTAATTTAAGTTTTGACAAGCTGGCTTACACTGGCAATGGTAGTTTCTGTGGTTGGCTGGGAAATCGAGTGCCGCATCTC ACAGCTATGCAAAAAGCTAGTCAACAGTACCCTGGTTGTGTGTCCCCTTGCAGCCGACACGGTCTCGGATCAGGCTCCCAGGAGCCTGCC CAGCCCCCTGGTCTTTGAGCTCCCACTTCTGCCAGATGTCCTAATGGTGATGCAGTCTTAGATCATAGTTTTATTTATATTTATTGACTC TTGAGTTGTTTTTGTATATTGGTTTTATGATGACGTACAAGTAGTTCTGTATTTGAAAGTGCCTTTGCAGCTCAGAACCACAGCAACGAT CACAAATGACTTTATTATTTATTTTTTTTAATTGTATTTTTGTTGTTGGGGGAGGGGAGACTTTGATGTCAGCAGTTGCTGGTAAAATGA AGAATTTAAAGAAAAAAATGTCAAAAGTAGAACTTTGTATAGTTATGTAAATAATTCTTTTTTATTAATCACTGTGTATATTTGATTTAT TAACTTAATAATCAAGAGCCTTAAAACATCATTCCTTTTTATTTATATGTATGTGTTTAGAATTGAAGGTTTTTGATAGCATTGTAAGCG TATGGCTTTATTTTTTTGAACTCTTCTCATTACTTGTTGCCTATAAGCCAAAATTAAGGTGTTTGAAAATAGTTTATTTTAAAACAATAG GATGGGCTTCTGTGCCCAGAATACTGATGGAATTTTTTTGTACGACGTCAGATGTTTAAAACACCTTCTATAGCATCACTTAAAACACGT TTTAAGGACTGACTGAGGCAGTTTGAGGATTAGTTTAGAACAGGTTTTTTTGTTTGTTTGTTTTTTGTTTTTCTGCTTTAGACTTGAAAA GAGACAGGCAGGTGATCTGCTGCAGAGCAGTAAGGGAACAAGTTGAGCTATGACTTAACATAGCCAAAATGTGAGTGGTTGAATATGATT AAAAATATCAAATTAATTGTGTGAACTTGGAAGCACACCAATCTTACTTTGTAAATTCTGATTTCTTTTCACCATTCGTACATAATACTG AACCACTTGTAGATTTGATTTTTTTTTTTAATCTACTGCATTTAGGGAGTATTCTAATAAGCTAGTTGAATACTTGAACCATAAAATGTC CAGTAAGATCACTGTTTAGATTTGCCATAGAGTACACTGCCTGCCTTAAGTGAGGAAATCAAAGTGCTATTACGAAGTTCAAGATCAAAA AGGCTTATAAAACAGAGTAATCTTGTTGGTTCACCATTGAGACCGTGAAGATACTTTGTATTGTCCTATTAGTGTTATATGAACATACAA ATGCATCTTTGATGTGTTGTTCTTGGCAATAAATTTTGAAAAGTAATATTTATTAAATTTTTTTGTATGAAAACATGGAACAGTGTGGCC TCTTCTGAGCTTACGTAGTTCTACCGGCTTTGCCATGTGCTTCTGCCACCCTGCTGAGTCTGTTCTGGTAATCGGGGTATAATAGGCTCT GCCTGACAGAGGGATGGAGGAAGAACTGAAAGGCTTTTCAACCACAAAACTCATCTGGAGTTCTCAAAGACCTGGGGCTGCTGTGAAGCT GGAACTGCGGGAGCCCCATCTAGGGGAGCCTTGATTCCCTTGTTATTCAACAGCAAGTGTGAATACTGCTTGAATAAACACCACTGGATT >60056_60056_3_NQO1-JAG1_NQO1_chr16_69760336_ENST00000379046_JAG1_chr20_10621892_ENST00000254958_length(amino acids)=294AA_BP=48 MHRALHTRLPQGCSRSRPAHREPSSGQGRPGNHEPSQSAPRTAPEPWSGLTTEHICSELRNLNILKNVSAEYSIYIACEPSPSANNEIHV AISAEDIRDDGNPIKEITDKIIDLVSKRDGNSSLIAAVAEVRVQRRPLKNRTDFLVPLLSSVLTVAWICCLVTAFYWCLRKRRKPGSHTH SASEDNTTNNVREQLNQIKNPIEKHGANTVPIKDYENKNSKMSKIRTHNSEVEEDDMDKHQQKARFAKQPAYTLVDREEKPPNGTPTKHP -------------------------------------------------------------- >60056_60056_4_NQO1-JAG1_NQO1_chr16_69760336_ENST00000379046_JAG1_chr20_10621892_ENST00000423891_length(transcript)=1128nt_BP=152nt AAGAGAGAATGCACCGTGCACTACACACGCGACTCCCACAAGGTTGCAGCCGGAGCCGCCCAGCTCACCGAGAGCCTAGTTCCGGCCAGG GTCGCCCCGGCAACCACGAGCCCAGCCAATCAGCGCCCCGGACTGCACCAGAGCCATGGTCGGGTCTTACTACGGAGCACATTTGCAGTG AATTGAGGAATTTGAATATTTTGAAGAATGTTTCCGCTGAATATTCAATCTACATCGCTTGCGAGCCTTCCCCTTCAGCGAACAATGAAA TACATGTGGCCATTTCTGCTGAAGATATACGGGATGATGGGAACCCGATCAAGGAAATCACTGACAAAATAATCGATCTTGTTAGTAAAC GTGATGGAAACAGCTCGCTGATTGCTGCCGTTGCAGAAGTAAGAGTTCAGAGGCGGCCTCTGAAGAACAGAACAGATTTCCTTGTTCCCT TGCTGAGCTCTGTCTTAACTGTGGCTTGGATCTGTTGCTTGGTGACGGCCTTCTACTGGTGCCTGCGGAAGCGGCGGAAGCCGGGCAGCC ACACACACTCAGCCTCTGAGGACAACACCACCAACAACGTGCGGGAGCAGCTGAACCAGATCAAAAACCCCATTGAGAAACATGGGGCCA ACACGGTCCCCATCAAGGATTATGAGAACAAGAACTCCAAAATGTCTAAAATAAGGACACACAATTCTGAAGTAGAAGAGGACGACATGG ACAAACACCAGCAGAAAGCCCGGTTTGCCAAGCAGCCGGCGTACACGCTGGTAGACAGAGAAGAGAAGCCCCCCAACGGCACGCCGACAA AACACCCAAACTGGACAAACAAACAGGACAACAGAGACTTGGAAAGTGCCCAGAGCTTAAACCGAATGGAGTACATCGTATAGCAGACCG CGGGCACTGCCGCCGCTAGTGTGGCCTCTTCTGAGCTTACGTAGTTCTACCGGCTTTGCCATGTGCTTCTGCCACCCTGCTGAGTCTGTT CTGGTAATCGGGGTATAATAGGCTCTGCCTGACAGAGGGATGGAGGAAGAACTGAAAGGCTTTTCAACCACAAAACTCATCTGGAGTTCT >60056_60056_4_NQO1-JAG1_NQO1_chr16_69760336_ENST00000379046_JAG1_chr20_10621892_ENST00000423891_length(amino acids)=294AA_BP=48 MHRALHTRLPQGCSRSRPAHREPSSGQGRPGNHEPSQSAPRTAPEPWSGLTTEHICSELRNLNILKNVSAEYSIYIACEPSPSANNEIHV AISAEDIRDDGNPIKEITDKIIDLVSKRDGNSSLIAAVAEVRVQRRPLKNRTDFLVPLLSSVLTVAWICCLVTAFYWCLRKRRKPGSHTH SASEDNTTNNVREQLNQIKNPIEKHGANTVPIKDYENKNSKMSKIRTHNSEVEEDDMDKHQQKARFAKQPAYTLVDREEKPPNGTPTKHP -------------------------------------------------------------- >60056_60056_5_NQO1-JAG1_NQO1_chr16_69760336_ENST00000379047_JAG1_chr20_10621892_ENST00000254958_length(transcript)=2791nt_BP=236nt ATCCTCCGCCCAGCACCCCAGGATTCAGGCGTTGGGTCCCGCCCTTGTAGGCTGTCCACCTCAAACGGGCCGGACAGGATATATAAGAGA GAATGCACCGTGCACTACACACGCGACTCCCACAAGGTTGCAGCCGGAGCCGCCCAGCTCACCGAGAGCCTAGTTCCGGCCAGGGTCGCC CCGGCAACCACGAGCCCAGCCAATCAGCGCCCCGGACTGCACCAGAGCCATGGTCGGGTCTTACTACGGAGCACATTTGCAGTGAATTGA GGAATTTGAATATTTTGAAGAATGTTTCCGCTGAATATTCAATCTACATCGCTTGCGAGCCTTCCCCTTCAGCGAACAATGAAATACATG TGGCCATTTCTGCTGAAGATATACGGGATGATGGGAACCCGATCAAGGAAATCACTGACAAAATAATCGATCTTGTTAGTAAACGTGATG GAAACAGCTCGCTGATTGCTGCCGTTGCAGAAGTAAGAGTTCAGAGGCGGCCTCTGAAGAACAGAACAGATTTCCTTGTTCCCTTGCTGA GCTCTGTCTTAACTGTGGCTTGGATCTGTTGCTTGGTGACGGCCTTCTACTGGTGCCTGCGGAAGCGGCGGAAGCCGGGCAGCCACACAC ACTCAGCCTCTGAGGACAACACCACCAACAACGTGCGGGAGCAGCTGAACCAGATCAAAAACCCCATTGAGAAACATGGGGCCAACACGG TCCCCATCAAGGATTATGAGAACAAGAACTCCAAAATGTCTAAAATAAGGACACACAATTCTGAAGTAGAAGAGGACGACATGGACAAAC ACCAGCAGAAAGCCCGGTTTGCCAAGCAGCCGGCGTACACGCTGGTAGACAGAGAAGAGAAGCCCCCCAACGGCACGCCGACAAAACACC CAAACTGGACAAACAAACAGGACAACAGAGACTTGGAAAGTGCCCAGAGCTTAAACCGAATGGAGTACATCGTATAGCAGACCGCGGGCA CTGCCGCCGCTAGGTAGAGTCTGAGGGCTTGTAGTTCTTTAAACTGTCGTGTCATACTCGAGTCTGAGGCCGTTGCTGACTTAGAATCCC TGTGTTAATTTAAGTTTTGACAAGCTGGCTTACACTGGCAATGGTAGTTTCTGTGGTTGGCTGGGAAATCGAGTGCCGCATCTCACAGCT ATGCAAAAAGCTAGTCAACAGTACCCTGGTTGTGTGTCCCCTTGCAGCCGACACGGTCTCGGATCAGGCTCCCAGGAGCCTGCCCAGCCC CCTGGTCTTTGAGCTCCCACTTCTGCCAGATGTCCTAATGGTGATGCAGTCTTAGATCATAGTTTTATTTATATTTATTGACTCTTGAGT TGTTTTTGTATATTGGTTTTATGATGACGTACAAGTAGTTCTGTATTTGAAAGTGCCTTTGCAGCTCAGAACCACAGCAACGATCACAAA TGACTTTATTATTTATTTTTTTTAATTGTATTTTTGTTGTTGGGGGAGGGGAGACTTTGATGTCAGCAGTTGCTGGTAAAATGAAGAATT TAAAGAAAAAAATGTCAAAAGTAGAACTTTGTATAGTTATGTAAATAATTCTTTTTTATTAATCACTGTGTATATTTGATTTATTAACTT AATAATCAAGAGCCTTAAAACATCATTCCTTTTTATTTATATGTATGTGTTTAGAATTGAAGGTTTTTGATAGCATTGTAAGCGTATGGC TTTATTTTTTTGAACTCTTCTCATTACTTGTTGCCTATAAGCCAAAATTAAGGTGTTTGAAAATAGTTTATTTTAAAACAATAGGATGGG CTTCTGTGCCCAGAATACTGATGGAATTTTTTTGTACGACGTCAGATGTTTAAAACACCTTCTATAGCATCACTTAAAACACGTTTTAAG GACTGACTGAGGCAGTTTGAGGATTAGTTTAGAACAGGTTTTTTTGTTTGTTTGTTTTTTGTTTTTCTGCTTTAGACTTGAAAAGAGACA GGCAGGTGATCTGCTGCAGAGCAGTAAGGGAACAAGTTGAGCTATGACTTAACATAGCCAAAATGTGAGTGGTTGAATATGATTAAAAAT ATCAAATTAATTGTGTGAACTTGGAAGCACACCAATCTTACTTTGTAAATTCTGATTTCTTTTCACCATTCGTACATAATACTGAACCAC TTGTAGATTTGATTTTTTTTTTTAATCTACTGCATTTAGGGAGTATTCTAATAAGCTAGTTGAATACTTGAACCATAAAATGTCCAGTAA GATCACTGTTTAGATTTGCCATAGAGTACACTGCCTGCCTTAAGTGAGGAAATCAAAGTGCTATTACGAAGTTCAAGATCAAAAAGGCTT ATAAAACAGAGTAATCTTGTTGGTTCACCATTGAGACCGTGAAGATACTTTGTATTGTCCTATTAGTGTTATATGAACATACAAATGCAT CTTTGATGTGTTGTTCTTGGCAATAAATTTTGAAAAGTAATATTTATTAAATTTTTTTGTATGAAAACATGGAACAGTGTGGCCTCTTCT GAGCTTACGTAGTTCTACCGGCTTTGCCATGTGCTTCTGCCACCCTGCTGAGTCTGTTCTGGTAATCGGGGTATAATAGGCTCTGCCTGA CAGAGGGATGGAGGAAGAACTGAAAGGCTTTTCAACCACAAAACTCATCTGGAGTTCTCAAAGACCTGGGGCTGCTGTGAAGCTGGAACT GCGGGAGCCCCATCTAGGGGAGCCTTGATTCCCTTGTTATTCAACAGCAAGTGTGAATACTGCTTGAATAAACACCACTGGATTAATGGC >60056_60056_5_NQO1-JAG1_NQO1_chr16_69760336_ENST00000379047_JAG1_chr20_10621892_ENST00000254958_length(amino acids)=294AA_BP=48 MHRALHTRLPQGCSRSRPAHREPSSGQGRPGNHEPSQSAPRTAPEPWSGLTTEHICSELRNLNILKNVSAEYSIYIACEPSPSANNEIHV AISAEDIRDDGNPIKEITDKIIDLVSKRDGNSSLIAAVAEVRVQRRPLKNRTDFLVPLLSSVLTVAWICCLVTAFYWCLRKRRKPGSHTH SASEDNTTNNVREQLNQIKNPIEKHGANTVPIKDYENKNSKMSKIRTHNSEVEEDDMDKHQQKARFAKQPAYTLVDREEKPPNGTPTKHP -------------------------------------------------------------- >60056_60056_6_NQO1-JAG1_NQO1_chr16_69760336_ENST00000379047_JAG1_chr20_10621892_ENST00000423891_length(transcript)=1212nt_BP=236nt ATCCTCCGCCCAGCACCCCAGGATTCAGGCGTTGGGTCCCGCCCTTGTAGGCTGTCCACCTCAAACGGGCCGGACAGGATATATAAGAGA GAATGCACCGTGCACTACACACGCGACTCCCACAAGGTTGCAGCCGGAGCCGCCCAGCTCACCGAGAGCCTAGTTCCGGCCAGGGTCGCC CCGGCAACCACGAGCCCAGCCAATCAGCGCCCCGGACTGCACCAGAGCCATGGTCGGGTCTTACTACGGAGCACATTTGCAGTGAATTGA GGAATTTGAATATTTTGAAGAATGTTTCCGCTGAATATTCAATCTACATCGCTTGCGAGCCTTCCCCTTCAGCGAACAATGAAATACATG TGGCCATTTCTGCTGAAGATATACGGGATGATGGGAACCCGATCAAGGAAATCACTGACAAAATAATCGATCTTGTTAGTAAACGTGATG GAAACAGCTCGCTGATTGCTGCCGTTGCAGAAGTAAGAGTTCAGAGGCGGCCTCTGAAGAACAGAACAGATTTCCTTGTTCCCTTGCTGA GCTCTGTCTTAACTGTGGCTTGGATCTGTTGCTTGGTGACGGCCTTCTACTGGTGCCTGCGGAAGCGGCGGAAGCCGGGCAGCCACACAC ACTCAGCCTCTGAGGACAACACCACCAACAACGTGCGGGAGCAGCTGAACCAGATCAAAAACCCCATTGAGAAACATGGGGCCAACACGG TCCCCATCAAGGATTATGAGAACAAGAACTCCAAAATGTCTAAAATAAGGACACACAATTCTGAAGTAGAAGAGGACGACATGGACAAAC ACCAGCAGAAAGCCCGGTTTGCCAAGCAGCCGGCGTACACGCTGGTAGACAGAGAAGAGAAGCCCCCCAACGGCACGCCGACAAAACACC CAAACTGGACAAACAAACAGGACAACAGAGACTTGGAAAGTGCCCAGAGCTTAAACCGAATGGAGTACATCGTATAGCAGACCGCGGGCA CTGCCGCCGCTAGTGTGGCCTCTTCTGAGCTTACGTAGTTCTACCGGCTTTGCCATGTGCTTCTGCCACCCTGCTGAGTCTGTTCTGGTA ATCGGGGTATAATAGGCTCTGCCTGACAGAGGGATGGAGGAAGAACTGAAAGGCTTTTCAACCACAAAACTCATCTGGAGTTCTCAAAGA >60056_60056_6_NQO1-JAG1_NQO1_chr16_69760336_ENST00000379047_JAG1_chr20_10621892_ENST00000423891_length(amino acids)=294AA_BP=48 MHRALHTRLPQGCSRSRPAHREPSSGQGRPGNHEPSQSAPRTAPEPWSGLTTEHICSELRNLNILKNVSAEYSIYIACEPSPSANNEIHV AISAEDIRDDGNPIKEITDKIIDLVSKRDGNSSLIAAVAEVRVQRRPLKNRTDFLVPLLSSVLTVAWICCLVTAFYWCLRKRRKPGSHTH SASEDNTTNNVREQLNQIKNPIEKHGANTVPIKDYENKNSKMSKIRTHNSEVEEDDMDKHQQKARFAKQPAYTLVDREEKPPNGTPTKHP -------------------------------------------------------------- >60056_60056_7_NQO1-JAG1_NQO1_chr16_69760336_ENST00000439109_JAG1_chr20_10621892_ENST00000254958_length(transcript)=2683nt_BP=128nt ACACGCGACTCCCACAAGGTTGCAGCCGGAGCCGCCCAGCTCACCGAGAGCCTAGTTCCGGCCAGGGTCGCCCCGGCAACCACGAGCCCA GCCAATCAGCGCCCCGGACTGCACCAGAGCCATGGTCGGGTCTTACTACGGAGCACATTTGCAGTGAATTGAGGAATTTGAATATTTTGA AGAATGTTTCCGCTGAATATTCAATCTACATCGCTTGCGAGCCTTCCCCTTCAGCGAACAATGAAATACATGTGGCCATTTCTGCTGAAG ATATACGGGATGATGGGAACCCGATCAAGGAAATCACTGACAAAATAATCGATCTTGTTAGTAAACGTGATGGAAACAGCTCGCTGATTG CTGCCGTTGCAGAAGTAAGAGTTCAGAGGCGGCCTCTGAAGAACAGAACAGATTTCCTTGTTCCCTTGCTGAGCTCTGTCTTAACTGTGG CTTGGATCTGTTGCTTGGTGACGGCCTTCTACTGGTGCCTGCGGAAGCGGCGGAAGCCGGGCAGCCACACACACTCAGCCTCTGAGGACA ACACCACCAACAACGTGCGGGAGCAGCTGAACCAGATCAAAAACCCCATTGAGAAACATGGGGCCAACACGGTCCCCATCAAGGATTATG AGAACAAGAACTCCAAAATGTCTAAAATAAGGACACACAATTCTGAAGTAGAAGAGGACGACATGGACAAACACCAGCAGAAAGCCCGGT TTGCCAAGCAGCCGGCGTACACGCTGGTAGACAGAGAAGAGAAGCCCCCCAACGGCACGCCGACAAAACACCCAAACTGGACAAACAAAC AGGACAACAGAGACTTGGAAAGTGCCCAGAGCTTAAACCGAATGGAGTACATCGTATAGCAGACCGCGGGCACTGCCGCCGCTAGGTAGA GTCTGAGGGCTTGTAGTTCTTTAAACTGTCGTGTCATACTCGAGTCTGAGGCCGTTGCTGACTTAGAATCCCTGTGTTAATTTAAGTTTT GACAAGCTGGCTTACACTGGCAATGGTAGTTTCTGTGGTTGGCTGGGAAATCGAGTGCCGCATCTCACAGCTATGCAAAAAGCTAGTCAA CAGTACCCTGGTTGTGTGTCCCCTTGCAGCCGACACGGTCTCGGATCAGGCTCCCAGGAGCCTGCCCAGCCCCCTGGTCTTTGAGCTCCC ACTTCTGCCAGATGTCCTAATGGTGATGCAGTCTTAGATCATAGTTTTATTTATATTTATTGACTCTTGAGTTGTTTTTGTATATTGGTT TTATGATGACGTACAAGTAGTTCTGTATTTGAAAGTGCCTTTGCAGCTCAGAACCACAGCAACGATCACAAATGACTTTATTATTTATTT TTTTTAATTGTATTTTTGTTGTTGGGGGAGGGGAGACTTTGATGTCAGCAGTTGCTGGTAAAATGAAGAATTTAAAGAAAAAAATGTCAA AAGTAGAACTTTGTATAGTTATGTAAATAATTCTTTTTTATTAATCACTGTGTATATTTGATTTATTAACTTAATAATCAAGAGCCTTAA AACATCATTCCTTTTTATTTATATGTATGTGTTTAGAATTGAAGGTTTTTGATAGCATTGTAAGCGTATGGCTTTATTTTTTTGAACTCT TCTCATTACTTGTTGCCTATAAGCCAAAATTAAGGTGTTTGAAAATAGTTTATTTTAAAACAATAGGATGGGCTTCTGTGCCCAGAATAC TGATGGAATTTTTTTGTACGACGTCAGATGTTTAAAACACCTTCTATAGCATCACTTAAAACACGTTTTAAGGACTGACTGAGGCAGTTT GAGGATTAGTTTAGAACAGGTTTTTTTGTTTGTTTGTTTTTTGTTTTTCTGCTTTAGACTTGAAAAGAGACAGGCAGGTGATCTGCTGCA GAGCAGTAAGGGAACAAGTTGAGCTATGACTTAACATAGCCAAAATGTGAGTGGTTGAATATGATTAAAAATATCAAATTAATTGTGTGA ACTTGGAAGCACACCAATCTTACTTTGTAAATTCTGATTTCTTTTCACCATTCGTACATAATACTGAACCACTTGTAGATTTGATTTTTT TTTTTAATCTACTGCATTTAGGGAGTATTCTAATAAGCTAGTTGAATACTTGAACCATAAAATGTCCAGTAAGATCACTGTTTAGATTTG CCATAGAGTACACTGCCTGCCTTAAGTGAGGAAATCAAAGTGCTATTACGAAGTTCAAGATCAAAAAGGCTTATAAAACAGAGTAATCTT GTTGGTTCACCATTGAGACCGTGAAGATACTTTGTATTGTCCTATTAGTGTTATATGAACATACAAATGCATCTTTGATGTGTTGTTCTT GGCAATAAATTTTGAAAAGTAATATTTATTAAATTTTTTTGTATGAAAACATGGAACAGTGTGGCCTCTTCTGAGCTTACGTAGTTCTAC CGGCTTTGCCATGTGCTTCTGCCACCCTGCTGAGTCTGTTCTGGTAATCGGGGTATAATAGGCTCTGCCTGACAGAGGGATGGAGGAAGA ACTGAAAGGCTTTTCAACCACAAAACTCATCTGGAGTTCTCAAAGACCTGGGGCTGCTGTGAAGCTGGAACTGCGGGAGCCCCATCTAGG >60056_60056_7_NQO1-JAG1_NQO1_chr16_69760336_ENST00000439109_JAG1_chr20_10621892_ENST00000254958_length(amino acids)=236AA_BP= MRNLNILKNVSAEYSIYIACEPSPSANNEIHVAISAEDIRDDGNPIKEITDKIIDLVSKRDGNSSLIAAVAEVRVQRRPLKNRTDFLVPL LSSVLTVAWICCLVTAFYWCLRKRRKPGSHTHSASEDNTTNNVREQLNQIKNPIEKHGANTVPIKDYENKNSKMSKIRTHNSEVEEDDMD -------------------------------------------------------------- >60056_60056_8_NQO1-JAG1_NQO1_chr16_69760336_ENST00000439109_JAG1_chr20_10621892_ENST00000423891_length(transcript)=1104nt_BP=128nt ACACGCGACTCCCACAAGGTTGCAGCCGGAGCCGCCCAGCTCACCGAGAGCCTAGTTCCGGCCAGGGTCGCCCCGGCAACCACGAGCCCA GCCAATCAGCGCCCCGGACTGCACCAGAGCCATGGTCGGGTCTTACTACGGAGCACATTTGCAGTGAATTGAGGAATTTGAATATTTTGA AGAATGTTTCCGCTGAATATTCAATCTACATCGCTTGCGAGCCTTCCCCTTCAGCGAACAATGAAATACATGTGGCCATTTCTGCTGAAG ATATACGGGATGATGGGAACCCGATCAAGGAAATCACTGACAAAATAATCGATCTTGTTAGTAAACGTGATGGAAACAGCTCGCTGATTG CTGCCGTTGCAGAAGTAAGAGTTCAGAGGCGGCCTCTGAAGAACAGAACAGATTTCCTTGTTCCCTTGCTGAGCTCTGTCTTAACTGTGG CTTGGATCTGTTGCTTGGTGACGGCCTTCTACTGGTGCCTGCGGAAGCGGCGGAAGCCGGGCAGCCACACACACTCAGCCTCTGAGGACA ACACCACCAACAACGTGCGGGAGCAGCTGAACCAGATCAAAAACCCCATTGAGAAACATGGGGCCAACACGGTCCCCATCAAGGATTATG AGAACAAGAACTCCAAAATGTCTAAAATAAGGACACACAATTCTGAAGTAGAAGAGGACGACATGGACAAACACCAGCAGAAAGCCCGGT TTGCCAAGCAGCCGGCGTACACGCTGGTAGACAGAGAAGAGAAGCCCCCCAACGGCACGCCGACAAAACACCCAAACTGGACAAACAAAC AGGACAACAGAGACTTGGAAAGTGCCCAGAGCTTAAACCGAATGGAGTACATCGTATAGCAGACCGCGGGCACTGCCGCCGCTAGTGTGG CCTCTTCTGAGCTTACGTAGTTCTACCGGCTTTGCCATGTGCTTCTGCCACCCTGCTGAGTCTGTTCTGGTAATCGGGGTATAATAGGCT CTGCCTGACAGAGGGATGGAGGAAGAACTGAAAGGCTTTTCAACCACAAAACTCATCTGGAGTTCTCAAAGACCTGGGGCTGCTGTGAAG >60056_60056_8_NQO1-JAG1_NQO1_chr16_69760336_ENST00000439109_JAG1_chr20_10621892_ENST00000423891_length(amino acids)=236AA_BP= MRNLNILKNVSAEYSIYIACEPSPSANNEIHVAISAEDIRDDGNPIKEITDKIIDLVSKRDGNSSLIAAVAEVRVQRRPLKNRTDFLVPL LSSVLTVAWICCLVTAFYWCLRKRRKPGSHTHSASEDNTTNNVREQLNQIKNPIEKHGANTVPIKDYENKNSKMSKIRTHNSEVEEDDMD -------------------------------------------------------------- >60056_60056_9_NQO1-JAG1_NQO1_chr16_69760336_ENST00000561500_JAG1_chr20_10621892_ENST00000254958_length(transcript)=2629nt_BP=74nt GTTCCGGCCAGGGTCGCCCCGGCAACCACGAGCCCAGCCAATCAGCGCCCCGGACTGCACCAGAGCCATGGTCGGGTCTTACTACGGAGC ACATTTGCAGTGAATTGAGGAATTTGAATATTTTGAAGAATGTTTCCGCTGAATATTCAATCTACATCGCTTGCGAGCCTTCCCCTTCAG CGAACAATGAAATACATGTGGCCATTTCTGCTGAAGATATACGGGATGATGGGAACCCGATCAAGGAAATCACTGACAAAATAATCGATC TTGTTAGTAAACGTGATGGAAACAGCTCGCTGATTGCTGCCGTTGCAGAAGTAAGAGTTCAGAGGCGGCCTCTGAAGAACAGAACAGATT TCCTTGTTCCCTTGCTGAGCTCTGTCTTAACTGTGGCTTGGATCTGTTGCTTGGTGACGGCCTTCTACTGGTGCCTGCGGAAGCGGCGGA AGCCGGGCAGCCACACACACTCAGCCTCTGAGGACAACACCACCAACAACGTGCGGGAGCAGCTGAACCAGATCAAAAACCCCATTGAGA AACATGGGGCCAACACGGTCCCCATCAAGGATTATGAGAACAAGAACTCCAAAATGTCTAAAATAAGGACACACAATTCTGAAGTAGAAG AGGACGACATGGACAAACACCAGCAGAAAGCCCGGTTTGCCAAGCAGCCGGCGTACACGCTGGTAGACAGAGAAGAGAAGCCCCCCAACG GCACGCCGACAAAACACCCAAACTGGACAAACAAACAGGACAACAGAGACTTGGAAAGTGCCCAGAGCTTAAACCGAATGGAGTACATCG TATAGCAGACCGCGGGCACTGCCGCCGCTAGGTAGAGTCTGAGGGCTTGTAGTTCTTTAAACTGTCGTGTCATACTCGAGTCTGAGGCCG TTGCTGACTTAGAATCCCTGTGTTAATTTAAGTTTTGACAAGCTGGCTTACACTGGCAATGGTAGTTTCTGTGGTTGGCTGGGAAATCGA GTGCCGCATCTCACAGCTATGCAAAAAGCTAGTCAACAGTACCCTGGTTGTGTGTCCCCTTGCAGCCGACACGGTCTCGGATCAGGCTCC CAGGAGCCTGCCCAGCCCCCTGGTCTTTGAGCTCCCACTTCTGCCAGATGTCCTAATGGTGATGCAGTCTTAGATCATAGTTTTATTTAT ATTTATTGACTCTTGAGTTGTTTTTGTATATTGGTTTTATGATGACGTACAAGTAGTTCTGTATTTGAAAGTGCCTTTGCAGCTCAGAAC CACAGCAACGATCACAAATGACTTTATTATTTATTTTTTTTAATTGTATTTTTGTTGTTGGGGGAGGGGAGACTTTGATGTCAGCAGTTG CTGGTAAAATGAAGAATTTAAAGAAAAAAATGTCAAAAGTAGAACTTTGTATAGTTATGTAAATAATTCTTTTTTATTAATCACTGTGTA TATTTGATTTATTAACTTAATAATCAAGAGCCTTAAAACATCATTCCTTTTTATTTATATGTATGTGTTTAGAATTGAAGGTTTTTGATA GCATTGTAAGCGTATGGCTTTATTTTTTTGAACTCTTCTCATTACTTGTTGCCTATAAGCCAAAATTAAGGTGTTTGAAAATAGTTTATT TTAAAACAATAGGATGGGCTTCTGTGCCCAGAATACTGATGGAATTTTTTTGTACGACGTCAGATGTTTAAAACACCTTCTATAGCATCA CTTAAAACACGTTTTAAGGACTGACTGAGGCAGTTTGAGGATTAGTTTAGAACAGGTTTTTTTGTTTGTTTGTTTTTTGTTTTTCTGCTT TAGACTTGAAAAGAGACAGGCAGGTGATCTGCTGCAGAGCAGTAAGGGAACAAGTTGAGCTATGACTTAACATAGCCAAAATGTGAGTGG TTGAATATGATTAAAAATATCAAATTAATTGTGTGAACTTGGAAGCACACCAATCTTACTTTGTAAATTCTGATTTCTTTTCACCATTCG TACATAATACTGAACCACTTGTAGATTTGATTTTTTTTTTTAATCTACTGCATTTAGGGAGTATTCTAATAAGCTAGTTGAATACTTGAA CCATAAAATGTCCAGTAAGATCACTGTTTAGATTTGCCATAGAGTACACTGCCTGCCTTAAGTGAGGAAATCAAAGTGCTATTACGAAGT TCAAGATCAAAAAGGCTTATAAAACAGAGTAATCTTGTTGGTTCACCATTGAGACCGTGAAGATACTTTGTATTGTCCTATTAGTGTTAT ATGAACATACAAATGCATCTTTGATGTGTTGTTCTTGGCAATAAATTTTGAAAAGTAATATTTATTAAATTTTTTTGTATGAAAACATGG AACAGTGTGGCCTCTTCTGAGCTTACGTAGTTCTACCGGCTTTGCCATGTGCTTCTGCCACCCTGCTGAGTCTGTTCTGGTAATCGGGGT ATAATAGGCTCTGCCTGACAGAGGGATGGAGGAAGAACTGAAAGGCTTTTCAACCACAAAACTCATCTGGAGTTCTCAAAGACCTGGGGC TGCTGTGAAGCTGGAACTGCGGGAGCCCCATCTAGGGGAGCCTTGATTCCCTTGTTATTCAACAGCAAGTGTGAATACTGCTTGAATAAA >60056_60056_9_NQO1-JAG1_NQO1_chr16_69760336_ENST00000561500_JAG1_chr20_10621892_ENST00000254958_length(amino acids)=236AA_BP= MRNLNILKNVSAEYSIYIACEPSPSANNEIHVAISAEDIRDDGNPIKEITDKIIDLVSKRDGNSSLIAAVAEVRVQRRPLKNRTDFLVPL LSSVLTVAWICCLVTAFYWCLRKRRKPGSHTHSASEDNTTNNVREQLNQIKNPIEKHGANTVPIKDYENKNSKMSKIRTHNSEVEEDDMD -------------------------------------------------------------- >60056_60056_10_NQO1-JAG1_NQO1_chr16_69760336_ENST00000561500_JAG1_chr20_10621892_ENST00000423891_length(transcript)=1050nt_BP=74nt GTTCCGGCCAGGGTCGCCCCGGCAACCACGAGCCCAGCCAATCAGCGCCCCGGACTGCACCAGAGCCATGGTCGGGTCTTACTACGGAGC ACATTTGCAGTGAATTGAGGAATTTGAATATTTTGAAGAATGTTTCCGCTGAATATTCAATCTACATCGCTTGCGAGCCTTCCCCTTCAG CGAACAATGAAATACATGTGGCCATTTCTGCTGAAGATATACGGGATGATGGGAACCCGATCAAGGAAATCACTGACAAAATAATCGATC TTGTTAGTAAACGTGATGGAAACAGCTCGCTGATTGCTGCCGTTGCAGAAGTAAGAGTTCAGAGGCGGCCTCTGAAGAACAGAACAGATT TCCTTGTTCCCTTGCTGAGCTCTGTCTTAACTGTGGCTTGGATCTGTTGCTTGGTGACGGCCTTCTACTGGTGCCTGCGGAAGCGGCGGA AGCCGGGCAGCCACACACACTCAGCCTCTGAGGACAACACCACCAACAACGTGCGGGAGCAGCTGAACCAGATCAAAAACCCCATTGAGA AACATGGGGCCAACACGGTCCCCATCAAGGATTATGAGAACAAGAACTCCAAAATGTCTAAAATAAGGACACACAATTCTGAAGTAGAAG AGGACGACATGGACAAACACCAGCAGAAAGCCCGGTTTGCCAAGCAGCCGGCGTACACGCTGGTAGACAGAGAAGAGAAGCCCCCCAACG GCACGCCGACAAAACACCCAAACTGGACAAACAAACAGGACAACAGAGACTTGGAAAGTGCCCAGAGCTTAAACCGAATGGAGTACATCG TATAGCAGACCGCGGGCACTGCCGCCGCTAGTGTGGCCTCTTCTGAGCTTACGTAGTTCTACCGGCTTTGCCATGTGCTTCTGCCACCCT GCTGAGTCTGTTCTGGTAATCGGGGTATAATAGGCTCTGCCTGACAGAGGGATGGAGGAAGAACTGAAAGGCTTTTCAACCACAAAACTC >60056_60056_10_NQO1-JAG1_NQO1_chr16_69760336_ENST00000561500_JAG1_chr20_10621892_ENST00000423891_length(amino acids)=236AA_BP= MRNLNILKNVSAEYSIYIACEPSPSANNEIHVAISAEDIRDDGNPIKEITDKIIDLVSKRDGNSSLIAAVAEVRVQRRPLKNRTDFLVPL LSSVLTVAWICCLVTAFYWCLRKRRKPGSHTHSASEDNTTNNVREQLNQIKNPIEKHGANTVPIKDYENKNSKMSKIRTHNSEVEEDDMD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for NQO1-JAG1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for NQO1-JAG1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for NQO1-JAG1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
