
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:NR6A1-GOLGA1 (FusionGDB2 ID:60189) |
Fusion Gene Summary for NR6A1-GOLGA1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: NR6A1-GOLGA1 | Fusion gene ID: 60189 | Hgene | Tgene | Gene symbol | NR6A1 | GOLGA1 | Gene ID | 2649 | 2800 |
| Gene name | nuclear receptor subfamily 6 group A member 1 | golgin A1 | |
| Synonyms | CT150|GCNF|GCNF1|NR61|RTR|hGCNF|hRTR | golgin-97 | |
| Cytomap | 9q33.3 | 9q33.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | nuclear receptor subfamily 6 group A member 1germ cell nuclear factor variant 1retinoic acid receptor-related testis-associated receptorretinoid receptor-related testis-specific receptor | golgin subfamily A member 1gap junction protein, alpha 4, 37kDgolgi autoantigen, golgin subfamily a, 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q15406 | Q92805 | |
| Ensembl transtripts involved in fusion gene | ENST00000344523, ENST00000373584, ENST00000416460, ENST00000487099, | ENST00000373555, | |
| Fusion gene scores | * DoF score | 4 X 5 X 5=100 | 5 X 6 X 7=210 |
| # samples | 8 | 10 | |
| ** MAII score | log2(8/100*10)=-0.321928094887362 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(10/210*10)=-1.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: NR6A1 [Title/Abstract] AND GOLGA1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | NR6A1(127495586)-GOLGA1(127662826), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | NR6A1-GOLGA1 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. NR6A1-GOLGA1 seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across NR6A1 (5'-gene) Fusion gene breakpoints across NR6A1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
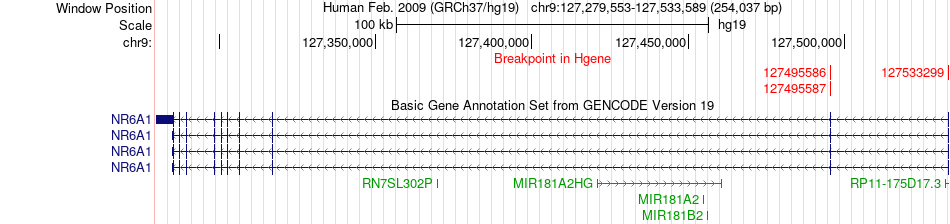 |
 Fusion gene breakpoints across GOLGA1 (3'-gene) Fusion gene breakpoints across GOLGA1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
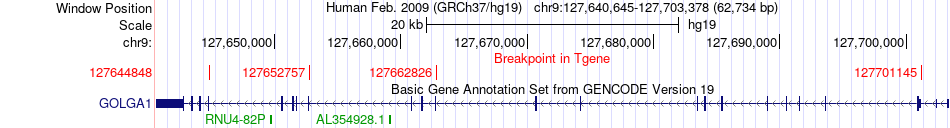 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | CESC | TCGA-VS-A9UT-01A | NR6A1 | chr9 | 127495587 | - | GOLGA1 | chr9 | 127701145 | - |
| ChimerDB4 | OV | TCGA-24-2026 | NR6A1 | chr9 | 127495586 | - | GOLGA1 | chr9 | 127662826 | - |
| ChimerDB4 | OV | TCGA-25-2397-01A | NR6A1 | chr9 | 127495587 | - | GOLGA1 | chr9 | 127652757 | - |
| ChimerDB4 | OV | TCGA-25-2397 | NR6A1 | chr9 | 127495586 | - | GOLGA1 | chr9 | 127652757 | - |
| ChimerDB4 | SKCM | TCGA-ER-A19T-01A | NR6A1 | chr9 | 127533299 | - | GOLGA1 | chr9 | 127644848 | - |
| ChimerDB4 | UCEC | TCGA-B5-A11J | NR6A1 | chr9 | 127495586 | - | GOLGA1 | chr9 | 127662826 | - |
Top |
Fusion Gene ORF analysis for NR6A1-GOLGA1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000344523 | ENST00000373555 | NR6A1 | chr9 | 127495587 | - | GOLGA1 | chr9 | 127701145 | - |
| 5CDS-5UTR | ENST00000373584 | ENST00000373555 | NR6A1 | chr9 | 127495587 | - | GOLGA1 | chr9 | 127701145 | - |
| 5CDS-5UTR | ENST00000416460 | ENST00000373555 | NR6A1 | chr9 | 127495587 | - | GOLGA1 | chr9 | 127701145 | - |
| 5CDS-5UTR | ENST00000487099 | ENST00000373555 | NR6A1 | chr9 | 127495587 | - | GOLGA1 | chr9 | 127701145 | - |
| Frame-shift | ENST00000344523 | ENST00000373555 | NR6A1 | chr9 | 127495586 | - | GOLGA1 | chr9 | 127662826 | - |
| Frame-shift | ENST00000344523 | ENST00000373555 | NR6A1 | chr9 | 127495587 | - | GOLGA1 | chr9 | 127652757 | - |
| Frame-shift | ENST00000344523 | ENST00000373555 | NR6A1 | chr9 | 127495586 | - | GOLGA1 | chr9 | 127652757 | - |
| Frame-shift | ENST00000373584 | ENST00000373555 | NR6A1 | chr9 | 127495586 | - | GOLGA1 | chr9 | 127662826 | - |
| Frame-shift | ENST00000373584 | ENST00000373555 | NR6A1 | chr9 | 127495587 | - | GOLGA1 | chr9 | 127652757 | - |
| Frame-shift | ENST00000373584 | ENST00000373555 | NR6A1 | chr9 | 127495586 | - | GOLGA1 | chr9 | 127652757 | - |
| Frame-shift | ENST00000416460 | ENST00000373555 | NR6A1 | chr9 | 127495586 | - | GOLGA1 | chr9 | 127662826 | - |
| Frame-shift | ENST00000416460 | ENST00000373555 | NR6A1 | chr9 | 127495587 | - | GOLGA1 | chr9 | 127652757 | - |
| Frame-shift | ENST00000416460 | ENST00000373555 | NR6A1 | chr9 | 127495586 | - | GOLGA1 | chr9 | 127652757 | - |
| Frame-shift | ENST00000487099 | ENST00000373555 | NR6A1 | chr9 | 127495586 | - | GOLGA1 | chr9 | 127662826 | - |
| Frame-shift | ENST00000487099 | ENST00000373555 | NR6A1 | chr9 | 127495587 | - | GOLGA1 | chr9 | 127652757 | - |
| Frame-shift | ENST00000487099 | ENST00000373555 | NR6A1 | chr9 | 127495586 | - | GOLGA1 | chr9 | 127652757 | - |
| In-frame | ENST00000344523 | ENST00000373555 | NR6A1 | chr9 | 127533299 | - | GOLGA1 | chr9 | 127644848 | - |
| In-frame | ENST00000373584 | ENST00000373555 | NR6A1 | chr9 | 127533299 | - | GOLGA1 | chr9 | 127644848 | - |
| In-frame | ENST00000416460 | ENST00000373555 | NR6A1 | chr9 | 127533299 | - | GOLGA1 | chr9 | 127644848 | - |
| In-frame | ENST00000487099 | ENST00000373555 | NR6A1 | chr9 | 127533299 | - | GOLGA1 | chr9 | 127644848 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000487099 | NR6A1 | chr9 | 127533299 | - | ENST00000373555 | GOLGA1 | chr9 | 127644848 | - | 2820 | 258 | 270 | 656 | 128 |
| ENST00000373584 | NR6A1 | chr9 | 127533299 | - | ENST00000373555 | GOLGA1 | chr9 | 127644848 | - | 2840 | 278 | 290 | 676 | 128 |
| ENST00000416460 | NR6A1 | chr9 | 127533299 | - | ENST00000373555 | GOLGA1 | chr9 | 127644848 | - | 2853 | 291 | 303 | 689 | 128 |
| ENST00000344523 | NR6A1 | chr9 | 127533299 | - | ENST00000373555 | GOLGA1 | chr9 | 127644848 | - | 2830 | 268 | 280 | 666 | 128 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000487099 | ENST00000373555 | NR6A1 | chr9 | 127533299 | - | GOLGA1 | chr9 | 127644848 | - | 0.020481583 | 0.97951835 |
| ENST00000373584 | ENST00000373555 | NR6A1 | chr9 | 127533299 | - | GOLGA1 | chr9 | 127644848 | - | 0.00870422 | 0.9912958 |
| ENST00000416460 | ENST00000373555 | NR6A1 | chr9 | 127533299 | - | GOLGA1 | chr9 | 127644848 | - | 0.009689451 | 0.9903106 |
| ENST00000344523 | ENST00000373555 | NR6A1 | chr9 | 127533299 | - | GOLGA1 | chr9 | 127644848 | - | 0.010252192 | 0.9897479 |
Top |
Fusion Genomic Features for NR6A1-GOLGA1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
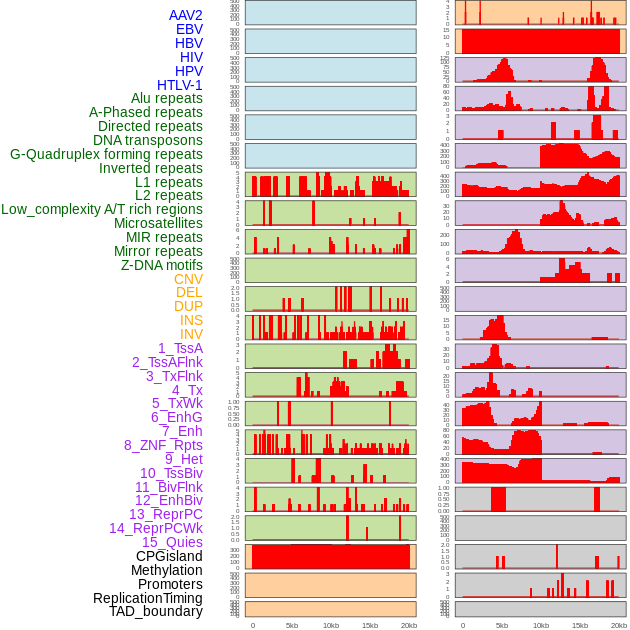 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for NR6A1-GOLGA1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:127495586/chr9:127662826) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NR6A1 | GOLGA1 |
| FUNCTION: Orphan nuclear receptor. Binds to a response element containing the sequence 5'-TCAAGGTCA-3'. May be involved in the regulation of gene expression in germ cell development during gametogenesis (By similarity). {ECO:0000250}. | FUNCTION: Involved in vesicular trafficking at the Golgi apparatus level. Involved in endosome-to-Golgi trafficking. {ECO:0000269|PubMed:29084197}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | GOLGA1 | chr9:127533299 | chr9:127644848 | ENST00000373555 | 18 | 23 | 688_737 | 635 | 768.0 | Domain | GRIP |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NR6A1 | chr9:127533299 | chr9:127644848 | ENST00000344523 | - | 1 | 10 | 57_132 | 33 | 480.0 | DNA binding | Nuclear receptor |
| Hgene | NR6A1 | chr9:127533299 | chr9:127644848 | ENST00000373584 | - | 1 | 10 | 57_132 | 33 | 477.0 | DNA binding | Nuclear receptor |
| Hgene | NR6A1 | chr9:127533299 | chr9:127644848 | ENST00000416460 | - | 1 | 10 | 57_132 | 33 | 476.0 | DNA binding | Nuclear receptor |
| Hgene | NR6A1 | chr9:127533299 | chr9:127644848 | ENST00000487099 | - | 1 | 10 | 57_132 | 33 | 481.0 | DNA binding | Nuclear receptor |
| Hgene | NR6A1 | chr9:127533299 | chr9:127644848 | ENST00000344523 | - | 1 | 10 | 249_480 | 33 | 480.0 | Domain | NR LBD |
| Hgene | NR6A1 | chr9:127533299 | chr9:127644848 | ENST00000373584 | - | 1 | 10 | 249_480 | 33 | 477.0 | Domain | NR LBD |
| Hgene | NR6A1 | chr9:127533299 | chr9:127644848 | ENST00000416460 | - | 1 | 10 | 249_480 | 33 | 476.0 | Domain | NR LBD |
| Hgene | NR6A1 | chr9:127533299 | chr9:127644848 | ENST00000487099 | - | 1 | 10 | 249_480 | 33 | 481.0 | Domain | NR LBD |
| Hgene | NR6A1 | chr9:127533299 | chr9:127644848 | ENST00000344523 | - | 1 | 10 | 60_80 | 33 | 480.0 | Zinc finger | NR C4-type |
| Hgene | NR6A1 | chr9:127533299 | chr9:127644848 | ENST00000344523 | - | 1 | 10 | 96_120 | 33 | 480.0 | Zinc finger | NR C4-type |
| Hgene | NR6A1 | chr9:127533299 | chr9:127644848 | ENST00000373584 | - | 1 | 10 | 60_80 | 33 | 477.0 | Zinc finger | NR C4-type |
| Hgene | NR6A1 | chr9:127533299 | chr9:127644848 | ENST00000373584 | - | 1 | 10 | 96_120 | 33 | 477.0 | Zinc finger | NR C4-type |
| Hgene | NR6A1 | chr9:127533299 | chr9:127644848 | ENST00000416460 | - | 1 | 10 | 60_80 | 33 | 476.0 | Zinc finger | NR C4-type |
| Hgene | NR6A1 | chr9:127533299 | chr9:127644848 | ENST00000416460 | - | 1 | 10 | 96_120 | 33 | 476.0 | Zinc finger | NR C4-type |
| Hgene | NR6A1 | chr9:127533299 | chr9:127644848 | ENST00000487099 | - | 1 | 10 | 60_80 | 33 | 481.0 | Zinc finger | NR C4-type |
| Hgene | NR6A1 | chr9:127533299 | chr9:127644848 | ENST00000487099 | - | 1 | 10 | 96_120 | 33 | 481.0 | Zinc finger | NR C4-type |
| Tgene | GOLGA1 | chr9:127533299 | chr9:127644848 | ENST00000373555 | 18 | 23 | 50_657 | 635 | 768.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | GOLGA1 | chr9:127533299 | chr9:127644848 | ENST00000373555 | 18 | 23 | 361_546 | 635 | 768.0 | Compositional bias | Note=Gln-rich |
Top |
Fusion Gene Sequence for NR6A1-GOLGA1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >60189_60189_1_NR6A1-GOLGA1_NR6A1_chr9_127533299_ENST00000344523_GOLGA1_chr9_127644848_ENST00000373555_length(transcript)=2830nt_BP=268nt GGGGCGCGGAGCGGCGCGGAGCCGGGCGGCTCGGGGCCCAGAGAGAGCCGCGGCCGGGAGCTCGCGGGCTCCTGACAACCTCCTCCCCTC GGCGGACGACGACCACGGCGACTAGGGCGCCGGTCATGGCGGAGCAACAAACCCGGCGCGGACCCTAGGCACCACCGCATGGAGCGGGAC GAACCGCCGCCTAGCGGAGGGGGAGGCGGCGGGGGCTCGGCGGGGTTCCTGGAGCCTCCCGCCGCGCTCCCTCCGCCGCCGCGCAACGAC CATAAAGCAGATGCAGCAGCGGATGCTGGAGCTCCGGAAGACTCTGCAGAAGGAGCTGAAAATCAGACCCGATAATGAGCTCTTCGAAGT CCGGGAGAAACCTGGACCTGAGATGGCAAACATGGCGCCTTCCGTCACGAATAACACTGACCTGACAGATGCCCGCGAGATCAACTTTGA GTACCTTAAACATGTGGTTTTAAAATTCATGTCTTGTCGCGAATCCGAGGCTTTTCATCTTATAAAAGCTGTGTCAGTGTTGCTGAACTT TTCCCAAGAGGAGGAGAACATGCTCAAGGAAACTCTGGAATATAAGATGTCATGGTTTGGGTCCAAACCAGCTCCCAAGGGCAGCATCCG GCCGTCTATCTCAAACCCTCGGATACCATGGTCCTAGAGGGGACTACCCAAGGATGGAGCTCCGTGGGTTGACACTTTTTCTGTGAAAAG AACACTGACACACCAGTCTGGGTGGGTTTTTAATCACTGTAACTGCAGTATTTTGTACAAGTGTCTAAACATTGTTTACAAGACTAAGGC CCACTTCCCTGCAGGCTGACCTGAACCTCAGGGGGTAGCTGATCCTGTCATTCTGGTCACCAAACAGGAGGGTCCTGGCACTACCCAGAT TTCCACAGTGCTGCTAATATCCCAGCTCCAGCCAGCACCCCATCTGCACCTGAATCCTCTAACTTCACGGTAGCACTTACAGCTGAAGCC ATCAGCATCTGGCAGGCACACCTGAGTCACCATGTAGCGCTGCTACTGGAGGTAGAGACGGCCCTTTGAGATGGTGCCCAGCAGGCCAAA CCCACCTGCCTCTGCCAGGAACAGCCAACTCCATGGGAACTCTATGGGAGTGGCTTTTAAAAATTCAGATGAGTTAGAAGCTTTTTATCC CTTCCTCTCAAGAAAATATTCTTTCACCCTGTCTCTCAAACCACCTAGAACTTTAGAGGATCCATCTTTAAGGGTCGGTGTGGATGAATG AGAAAATGCACCTTTCTGACAGTATCTCCACTTTACTTAAGAAAACTAGCAAATATATGAAAAGACCCTTAGTACCAAATACACTCAATT GCCTTTTTAATGAATGTACTTGTCTTGGATAGGTTGCTGGTAAACCATTTTAAACTATTTTTTATAGCTGAAGTTCTTCACTACTATAAA CATGTCTTCTGTACTATAAAATCCATTTAACTGGTGTTCTTAAAATCAGAGCGTCCAGAGGAAATTCTTCCTAAAACTAGGATTCCTGTT CCTTTGTCTTCTCACTCGCACTCTGGCACTGCTCCCTCTGAAGTGCAGTGGGATCTCGTGTGCTTTGTCTTGATTCTGTGCTGCGCTGCC GCTGGGCGATGCAGACCACCTGTCTTCTACTGAAGGACAGTCCGCTGTCTCCAGTGGGGGCAGCAGCTGTCCCCCAGCCTCGATGGAGAC ACTGGGGCAGTCTGCCTTGTCTGTGGAGCTGCTCTCTCTCCCTCATCCCACCCCAAATACTTAAAATGACACTACACCCAGACGGCGCCC AGCTGGCTGCAGCACTTGTAGCATGCACATGACTCTGGTAGTAACCAACAAAAACTTGTTTTATGGATTCCTCGTTTACTGAGGAAAGGG AACATGCTGGTTCTGGAAAAGCCACAATATTGAATCTAAAAGGAAACCGTTTATTGTTTGATGAAAGTTTCACTGGTTAAATAAAAAACT AAATTAATAACTGGAGCCTCTAAATTTATTATCCATTATCCAGTGATGGAAAGTTGTATTTCTCAATCATGCTTAGGGCCAAAATAGGTA TATAAAATGTGTCACAGAAAAACACGCATTTGCAACGTTAACCTAACGAAATTTCCATGAAGAACCAAGTCAGGGCAGCATCTCCTTAGT CCCAGCTCAGGCTCTCTGCCTTCCAGAGGCCGCTTCTCCAGTGACTAACCTCCTCCTCTGGCTCCTCCTTGCAGACAGTTATCCCTTGTT TAGAACACGAATTTCCATTTACCTGGTGGGAACACGAAACAGGAGTCTCTTCTGTTCTGCAAGTTTGATGGGTAAGAGGTAGCCTTTTTT CAAAGTAGGATTTCCTTTTTCAACTGTTCCAGGAAAGAATCTCTAAGACTGGGTAGCTCACAGCCAGCCAAAGGCAGCTACATTTCCACA GAAGCCCATCCGCTGCCTCCGTGGCTTCTCCAGCCATTGAACTGGTCCCACACGCACCCCAGGCCCCACTCCTCGGCAGTTTCAGGTGTA GCTGTGGGGCCCGTTCCTAGGTCTGTACTCACTTTAGGGAGGCTTCACTGACTAGGCTTTCCTCCTGCATGTTGAATTTCCTTCAGCTTT AAGAGGAAGAGTGGAATAAATATTCTAAGTGATTTAATGCACTTTGACTTGTATAAAACTTTCTGTGTTAGCGACGGTATCTATAGCCCT TTATACGAGCGATGGATCTTGAGCTCTCCTTCCATGTTGTAAATAGGGATTGTATTCTTGAAAACTGCTGTAGCAAATTCATCTGTGGTG >60189_60189_1_NR6A1-GOLGA1_NR6A1_chr9_127533299_ENST00000344523_GOLGA1_chr9_127644848_ENST00000373555_length(amino acids)=128AA_BP= MQQRMLELRKTLQKELKIRPDNELFEVREKPGPEMANMAPSVTNNTDLTDAREINFEYLKHVVLKFMSCRESEAFHLIKAVSVLLNFSQE -------------------------------------------------------------- >60189_60189_2_NR6A1-GOLGA1_NR6A1_chr9_127533299_ENST00000373584_GOLGA1_chr9_127644848_ENST00000373555_length(transcript)=2840nt_BP=278nt GCGGCGCGGAGGGGCGCGGAGCGGCGCGGAGCCGGGCGGCTCGGGGCCCAGAGAGAGCCGCGGCCGGGAGCTCGCGGGCTCCTGACAACC TCCTCCCCTCGGCGGACGACGACCACGGCGACTAGGGCGCCGGTCATGGCGGAGCAACAAACCCGGCGCGGACCCTAGGCACCACCGCAT GGAGCGGGACGAACCGCCGCCTAGCGGAGGGGGAGGCGGCGGGGGCTCGGCGGGGTTCCTGGAGCCTCCCGCCGCGCTCCCTCCGCCGCC GCGCAACGACCATAAAGCAGATGCAGCAGCGGATGCTGGAGCTCCGGAAGACTCTGCAGAAGGAGCTGAAAATCAGACCCGATAATGAGC TCTTCGAAGTCCGGGAGAAACCTGGACCTGAGATGGCAAACATGGCGCCTTCCGTCACGAATAACACTGACCTGACAGATGCCCGCGAGA TCAACTTTGAGTACCTTAAACATGTGGTTTTAAAATTCATGTCTTGTCGCGAATCCGAGGCTTTTCATCTTATAAAAGCTGTGTCAGTGT TGCTGAACTTTTCCCAAGAGGAGGAGAACATGCTCAAGGAAACTCTGGAATATAAGATGTCATGGTTTGGGTCCAAACCAGCTCCCAAGG GCAGCATCCGGCCGTCTATCTCAAACCCTCGGATACCATGGTCCTAGAGGGGACTACCCAAGGATGGAGCTCCGTGGGTTGACACTTTTT CTGTGAAAAGAACACTGACACACCAGTCTGGGTGGGTTTTTAATCACTGTAACTGCAGTATTTTGTACAAGTGTCTAAACATTGTTTACA AGACTAAGGCCCACTTCCCTGCAGGCTGACCTGAACCTCAGGGGGTAGCTGATCCTGTCATTCTGGTCACCAAACAGGAGGGTCCTGGCA CTACCCAGATTTCCACAGTGCTGCTAATATCCCAGCTCCAGCCAGCACCCCATCTGCACCTGAATCCTCTAACTTCACGGTAGCACTTAC AGCTGAAGCCATCAGCATCTGGCAGGCACACCTGAGTCACCATGTAGCGCTGCTACTGGAGGTAGAGACGGCCCTTTGAGATGGTGCCCA GCAGGCCAAACCCACCTGCCTCTGCCAGGAACAGCCAACTCCATGGGAACTCTATGGGAGTGGCTTTTAAAAATTCAGATGAGTTAGAAG CTTTTTATCCCTTCCTCTCAAGAAAATATTCTTTCACCCTGTCTCTCAAACCACCTAGAACTTTAGAGGATCCATCTTTAAGGGTCGGTG TGGATGAATGAGAAAATGCACCTTTCTGACAGTATCTCCACTTTACTTAAGAAAACTAGCAAATATATGAAAAGACCCTTAGTACCAAAT ACACTCAATTGCCTTTTTAATGAATGTACTTGTCTTGGATAGGTTGCTGGTAAACCATTTTAAACTATTTTTTATAGCTGAAGTTCTTCA CTACTATAAACATGTCTTCTGTACTATAAAATCCATTTAACTGGTGTTCTTAAAATCAGAGCGTCCAGAGGAAATTCTTCCTAAAACTAG GATTCCTGTTCCTTTGTCTTCTCACTCGCACTCTGGCACTGCTCCCTCTGAAGTGCAGTGGGATCTCGTGTGCTTTGTCTTGATTCTGTG CTGCGCTGCCGCTGGGCGATGCAGACCACCTGTCTTCTACTGAAGGACAGTCCGCTGTCTCCAGTGGGGGCAGCAGCTGTCCCCCAGCCT CGATGGAGACACTGGGGCAGTCTGCCTTGTCTGTGGAGCTGCTCTCTCTCCCTCATCCCACCCCAAATACTTAAAATGACACTACACCCA GACGGCGCCCAGCTGGCTGCAGCACTTGTAGCATGCACATGACTCTGGTAGTAACCAACAAAAACTTGTTTTATGGATTCCTCGTTTACT GAGGAAAGGGAACATGCTGGTTCTGGAAAAGCCACAATATTGAATCTAAAAGGAAACCGTTTATTGTTTGATGAAAGTTTCACTGGTTAA ATAAAAAACTAAATTAATAACTGGAGCCTCTAAATTTATTATCCATTATCCAGTGATGGAAAGTTGTATTTCTCAATCATGCTTAGGGCC AAAATAGGTATATAAAATGTGTCACAGAAAAACACGCATTTGCAACGTTAACCTAACGAAATTTCCATGAAGAACCAAGTCAGGGCAGCA TCTCCTTAGTCCCAGCTCAGGCTCTCTGCCTTCCAGAGGCCGCTTCTCCAGTGACTAACCTCCTCCTCTGGCTCCTCCTTGCAGACAGTT ATCCCTTGTTTAGAACACGAATTTCCATTTACCTGGTGGGAACACGAAACAGGAGTCTCTTCTGTTCTGCAAGTTTGATGGGTAAGAGGT AGCCTTTTTTCAAAGTAGGATTTCCTTTTTCAACTGTTCCAGGAAAGAATCTCTAAGACTGGGTAGCTCACAGCCAGCCAAAGGCAGCTA CATTTCCACAGAAGCCCATCCGCTGCCTCCGTGGCTTCTCCAGCCATTGAACTGGTCCCACACGCACCCCAGGCCCCACTCCTCGGCAGT TTCAGGTGTAGCTGTGGGGCCCGTTCCTAGGTCTGTACTCACTTTAGGGAGGCTTCACTGACTAGGCTTTCCTCCTGCATGTTGAATTTC CTTCAGCTTTAAGAGGAAGAGTGGAATAAATATTCTAAGTGATTTAATGCACTTTGACTTGTATAAAACTTTCTGTGTTAGCGACGGTAT CTATAGCCCTTTATACGAGCGATGGATCTTGAGCTCTCCTTCCATGTTGTAAATAGGGATTGTATTCTTGAAAACTGCTGTAGCAAATTC >60189_60189_2_NR6A1-GOLGA1_NR6A1_chr9_127533299_ENST00000373584_GOLGA1_chr9_127644848_ENST00000373555_length(amino acids)=128AA_BP= MQQRMLELRKTLQKELKIRPDNELFEVREKPGPEMANMAPSVTNNTDLTDAREINFEYLKHVVLKFMSCRESEAFHLIKAVSVLLNFSQE -------------------------------------------------------------- >60189_60189_3_NR6A1-GOLGA1_NR6A1_chr9_127533299_ENST00000416460_GOLGA1_chr9_127644848_ENST00000373555_length(transcript)=2853nt_BP=291nt GTGCTGAGGGGGCGCGGCGCGGAGGGGCGCGGAGCGGCGCGGAGCCGGGCGGCTCGGGGCCCAGAGAGAGCCGCGGCCGGGAGCTCGCGG GCTCCTGACAACCTCCTCCCCTCGGCGGACGACGACCACGGCGACTAGGGCGCCGGTCATGGCGGAGCAACAAACCCGGCGCGGACCCTA GGCACCACCGCATGGAGCGGGACGAACCGCCGCCTAGCGGAGGGGGAGGCGGCGGGGGCTCGGCGGGGTTCCTGGAGCCTCCCGCCGCGC TCCCTCCGCCGCCGCGCAACGACCATAAAGCAGATGCAGCAGCGGATGCTGGAGCTCCGGAAGACTCTGCAGAAGGAGCTGAAAATCAGA CCCGATAATGAGCTCTTCGAAGTCCGGGAGAAACCTGGACCTGAGATGGCAAACATGGCGCCTTCCGTCACGAATAACACTGACCTGACA GATGCCCGCGAGATCAACTTTGAGTACCTTAAACATGTGGTTTTAAAATTCATGTCTTGTCGCGAATCCGAGGCTTTTCATCTTATAAAA GCTGTGTCAGTGTTGCTGAACTTTTCCCAAGAGGAGGAGAACATGCTCAAGGAAACTCTGGAATATAAGATGTCATGGTTTGGGTCCAAA CCAGCTCCCAAGGGCAGCATCCGGCCGTCTATCTCAAACCCTCGGATACCATGGTCCTAGAGGGGACTACCCAAGGATGGAGCTCCGTGG GTTGACACTTTTTCTGTGAAAAGAACACTGACACACCAGTCTGGGTGGGTTTTTAATCACTGTAACTGCAGTATTTTGTACAAGTGTCTA AACATTGTTTACAAGACTAAGGCCCACTTCCCTGCAGGCTGACCTGAACCTCAGGGGGTAGCTGATCCTGTCATTCTGGTCACCAAACAG GAGGGTCCTGGCACTACCCAGATTTCCACAGTGCTGCTAATATCCCAGCTCCAGCCAGCACCCCATCTGCACCTGAATCCTCTAACTTCA CGGTAGCACTTACAGCTGAAGCCATCAGCATCTGGCAGGCACACCTGAGTCACCATGTAGCGCTGCTACTGGAGGTAGAGACGGCCCTTT GAGATGGTGCCCAGCAGGCCAAACCCACCTGCCTCTGCCAGGAACAGCCAACTCCATGGGAACTCTATGGGAGTGGCTTTTAAAAATTCA GATGAGTTAGAAGCTTTTTATCCCTTCCTCTCAAGAAAATATTCTTTCACCCTGTCTCTCAAACCACCTAGAACTTTAGAGGATCCATCT TTAAGGGTCGGTGTGGATGAATGAGAAAATGCACCTTTCTGACAGTATCTCCACTTTACTTAAGAAAACTAGCAAATATATGAAAAGACC CTTAGTACCAAATACACTCAATTGCCTTTTTAATGAATGTACTTGTCTTGGATAGGTTGCTGGTAAACCATTTTAAACTATTTTTTATAG CTGAAGTTCTTCACTACTATAAACATGTCTTCTGTACTATAAAATCCATTTAACTGGTGTTCTTAAAATCAGAGCGTCCAGAGGAAATTC TTCCTAAAACTAGGATTCCTGTTCCTTTGTCTTCTCACTCGCACTCTGGCACTGCTCCCTCTGAAGTGCAGTGGGATCTCGTGTGCTTTG TCTTGATTCTGTGCTGCGCTGCCGCTGGGCGATGCAGACCACCTGTCTTCTACTGAAGGACAGTCCGCTGTCTCCAGTGGGGGCAGCAGC TGTCCCCCAGCCTCGATGGAGACACTGGGGCAGTCTGCCTTGTCTGTGGAGCTGCTCTCTCTCCCTCATCCCACCCCAAATACTTAAAAT GACACTACACCCAGACGGCGCCCAGCTGGCTGCAGCACTTGTAGCATGCACATGACTCTGGTAGTAACCAACAAAAACTTGTTTTATGGA TTCCTCGTTTACTGAGGAAAGGGAACATGCTGGTTCTGGAAAAGCCACAATATTGAATCTAAAAGGAAACCGTTTATTGTTTGATGAAAG TTTCACTGGTTAAATAAAAAACTAAATTAATAACTGGAGCCTCTAAATTTATTATCCATTATCCAGTGATGGAAAGTTGTATTTCTCAAT CATGCTTAGGGCCAAAATAGGTATATAAAATGTGTCACAGAAAAACACGCATTTGCAACGTTAACCTAACGAAATTTCCATGAAGAACCA AGTCAGGGCAGCATCTCCTTAGTCCCAGCTCAGGCTCTCTGCCTTCCAGAGGCCGCTTCTCCAGTGACTAACCTCCTCCTCTGGCTCCTC CTTGCAGACAGTTATCCCTTGTTTAGAACACGAATTTCCATTTACCTGGTGGGAACACGAAACAGGAGTCTCTTCTGTTCTGCAAGTTTG ATGGGTAAGAGGTAGCCTTTTTTCAAAGTAGGATTTCCTTTTTCAACTGTTCCAGGAAAGAATCTCTAAGACTGGGTAGCTCACAGCCAG CCAAAGGCAGCTACATTTCCACAGAAGCCCATCCGCTGCCTCCGTGGCTTCTCCAGCCATTGAACTGGTCCCACACGCACCCCAGGCCCC ACTCCTCGGCAGTTTCAGGTGTAGCTGTGGGGCCCGTTCCTAGGTCTGTACTCACTTTAGGGAGGCTTCACTGACTAGGCTTTCCTCCTG CATGTTGAATTTCCTTCAGCTTTAAGAGGAAGAGTGGAATAAATATTCTAAGTGATTTAATGCACTTTGACTTGTATAAAACTTTCTGTG TTAGCGACGGTATCTATAGCCCTTTATACGAGCGATGGATCTTGAGCTCTCCTTCCATGTTGTAAATAGGGATTGTATTCTTGAAAACTG >60189_60189_3_NR6A1-GOLGA1_NR6A1_chr9_127533299_ENST00000416460_GOLGA1_chr9_127644848_ENST00000373555_length(amino acids)=128AA_BP= MQQRMLELRKTLQKELKIRPDNELFEVREKPGPEMANMAPSVTNNTDLTDAREINFEYLKHVVLKFMSCRESEAFHLIKAVSVLLNFSQE -------------------------------------------------------------- >60189_60189_4_NR6A1-GOLGA1_NR6A1_chr9_127533299_ENST00000487099_GOLGA1_chr9_127644848_ENST00000373555_length(transcript)=2820nt_BP=258nt GCGGCGCGGAGCCGGGCGGCTCGGGGCCCAGAGAGAGCCGCGGCCGGGAGCTCGCGGGCTCCTGACAACCTCCTCCCCTCGGCGGACGAC GACCACGGCGACTAGGGCGCCGGTCATGGCGGAGCAACAAACCCGGCGCGGACCCTAGGCACCACCGCATGGAGCGGGACGAACCGCCGC CTAGCGGAGGGGGAGGCGGCGGGGGCTCGGCGGGGTTCCTGGAGCCTCCCGCCGCGCTCCCTCCGCCGCCGCGCAACGACCATAAAGCAG ATGCAGCAGCGGATGCTGGAGCTCCGGAAGACTCTGCAGAAGGAGCTGAAAATCAGACCCGATAATGAGCTCTTCGAAGTCCGGGAGAAA CCTGGACCTGAGATGGCAAACATGGCGCCTTCCGTCACGAATAACACTGACCTGACAGATGCCCGCGAGATCAACTTTGAGTACCTTAAA CATGTGGTTTTAAAATTCATGTCTTGTCGCGAATCCGAGGCTTTTCATCTTATAAAAGCTGTGTCAGTGTTGCTGAACTTTTCCCAAGAG GAGGAGAACATGCTCAAGGAAACTCTGGAATATAAGATGTCATGGTTTGGGTCCAAACCAGCTCCCAAGGGCAGCATCCGGCCGTCTATC TCAAACCCTCGGATACCATGGTCCTAGAGGGGACTACCCAAGGATGGAGCTCCGTGGGTTGACACTTTTTCTGTGAAAAGAACACTGACA CACCAGTCTGGGTGGGTTTTTAATCACTGTAACTGCAGTATTTTGTACAAGTGTCTAAACATTGTTTACAAGACTAAGGCCCACTTCCCT GCAGGCTGACCTGAACCTCAGGGGGTAGCTGATCCTGTCATTCTGGTCACCAAACAGGAGGGTCCTGGCACTACCCAGATTTCCACAGTG CTGCTAATATCCCAGCTCCAGCCAGCACCCCATCTGCACCTGAATCCTCTAACTTCACGGTAGCACTTACAGCTGAAGCCATCAGCATCT GGCAGGCACACCTGAGTCACCATGTAGCGCTGCTACTGGAGGTAGAGACGGCCCTTTGAGATGGTGCCCAGCAGGCCAAACCCACCTGCC TCTGCCAGGAACAGCCAACTCCATGGGAACTCTATGGGAGTGGCTTTTAAAAATTCAGATGAGTTAGAAGCTTTTTATCCCTTCCTCTCA AGAAAATATTCTTTCACCCTGTCTCTCAAACCACCTAGAACTTTAGAGGATCCATCTTTAAGGGTCGGTGTGGATGAATGAGAAAATGCA CCTTTCTGACAGTATCTCCACTTTACTTAAGAAAACTAGCAAATATATGAAAAGACCCTTAGTACCAAATACACTCAATTGCCTTTTTAA TGAATGTACTTGTCTTGGATAGGTTGCTGGTAAACCATTTTAAACTATTTTTTATAGCTGAAGTTCTTCACTACTATAAACATGTCTTCT GTACTATAAAATCCATTTAACTGGTGTTCTTAAAATCAGAGCGTCCAGAGGAAATTCTTCCTAAAACTAGGATTCCTGTTCCTTTGTCTT CTCACTCGCACTCTGGCACTGCTCCCTCTGAAGTGCAGTGGGATCTCGTGTGCTTTGTCTTGATTCTGTGCTGCGCTGCCGCTGGGCGAT GCAGACCACCTGTCTTCTACTGAAGGACAGTCCGCTGTCTCCAGTGGGGGCAGCAGCTGTCCCCCAGCCTCGATGGAGACACTGGGGCAG TCTGCCTTGTCTGTGGAGCTGCTCTCTCTCCCTCATCCCACCCCAAATACTTAAAATGACACTACACCCAGACGGCGCCCAGCTGGCTGC AGCACTTGTAGCATGCACATGACTCTGGTAGTAACCAACAAAAACTTGTTTTATGGATTCCTCGTTTACTGAGGAAAGGGAACATGCTGG TTCTGGAAAAGCCACAATATTGAATCTAAAAGGAAACCGTTTATTGTTTGATGAAAGTTTCACTGGTTAAATAAAAAACTAAATTAATAA CTGGAGCCTCTAAATTTATTATCCATTATCCAGTGATGGAAAGTTGTATTTCTCAATCATGCTTAGGGCCAAAATAGGTATATAAAATGT GTCACAGAAAAACACGCATTTGCAACGTTAACCTAACGAAATTTCCATGAAGAACCAAGTCAGGGCAGCATCTCCTTAGTCCCAGCTCAG GCTCTCTGCCTTCCAGAGGCCGCTTCTCCAGTGACTAACCTCCTCCTCTGGCTCCTCCTTGCAGACAGTTATCCCTTGTTTAGAACACGA ATTTCCATTTACCTGGTGGGAACACGAAACAGGAGTCTCTTCTGTTCTGCAAGTTTGATGGGTAAGAGGTAGCCTTTTTTCAAAGTAGGA TTTCCTTTTTCAACTGTTCCAGGAAAGAATCTCTAAGACTGGGTAGCTCACAGCCAGCCAAAGGCAGCTACATTTCCACAGAAGCCCATC CGCTGCCTCCGTGGCTTCTCCAGCCATTGAACTGGTCCCACACGCACCCCAGGCCCCACTCCTCGGCAGTTTCAGGTGTAGCTGTGGGGC CCGTTCCTAGGTCTGTACTCACTTTAGGGAGGCTTCACTGACTAGGCTTTCCTCCTGCATGTTGAATTTCCTTCAGCTTTAAGAGGAAGA GTGGAATAAATATTCTAAGTGATTTAATGCACTTTGACTTGTATAAAACTTTCTGTGTTAGCGACGGTATCTATAGCCCTTTATACGAGC GATGGATCTTGAGCTCTCCTTCCATGTTGTAAATAGGGATTGTATTCTTGAAAACTGCTGTAGCAAATTCATCTGTGGTGCAATACACTT >60189_60189_4_NR6A1-GOLGA1_NR6A1_chr9_127533299_ENST00000487099_GOLGA1_chr9_127644848_ENST00000373555_length(amino acids)=128AA_BP= MQQRMLELRKTLQKELKIRPDNELFEVREKPGPEMANMAPSVTNNTDLTDAREINFEYLKHVVLKFMSCRESEAFHLIKAVSVLLNFSQE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for NR6A1-GOLGA1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for NR6A1-GOLGA1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for NR6A1-GOLGA1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
