
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:NREP-COMMD10 (FusionGDB2 ID:60250) |
Fusion Gene Summary for NREP-COMMD10 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: NREP-COMMD10 | Fusion gene ID: 60250 | Hgene | Tgene | Gene symbol | NREP | COMMD10 | Gene ID | 9315 | 51397 |
| Gene name | neuronal regeneration related protein | COMM domain containing 10 | |
| Synonyms | C5orf13|D4S114|P311|PRO1873|PTZ17|SEZ17 | PTD002 | |
| Cytomap | 5q22.1 | 5q23.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | neuronal regeneration-related proteinneuronal protein 3.1neuronal regeneration related protein homologprotein p311 | COMM domain-containing protein 10 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q16612 | Q9Y6G5 | |
| Ensembl transtripts involved in fusion gene | ENST00000257435, ENST00000379671, ENST00000419114, ENST00000446294, ENST00000447165, ENST00000450761, ENST00000453526, ENST00000455559, ENST00000508870, ENST00000509025, ENST00000509427, ENST00000509979, ENST00000515855, ENST00000395634, ENST00000507742, | ENST00000503424, ENST00000274458, ENST00000515539, | |
| Fusion gene scores | * DoF score | 4 X 4 X 2=32 | 14 X 11 X 6=924 |
| # samples | 4 | 14 | |
| ** MAII score | log2(4/32*10)=0.321928094887362 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(14/924*10)=-2.72246602447109 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: NREP [Title/Abstract] AND COMMD10 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | NREP(111091467)-COMMD10(115469765), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | NREP-COMMD10 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across NREP (5'-gene) Fusion gene breakpoints across NREP (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
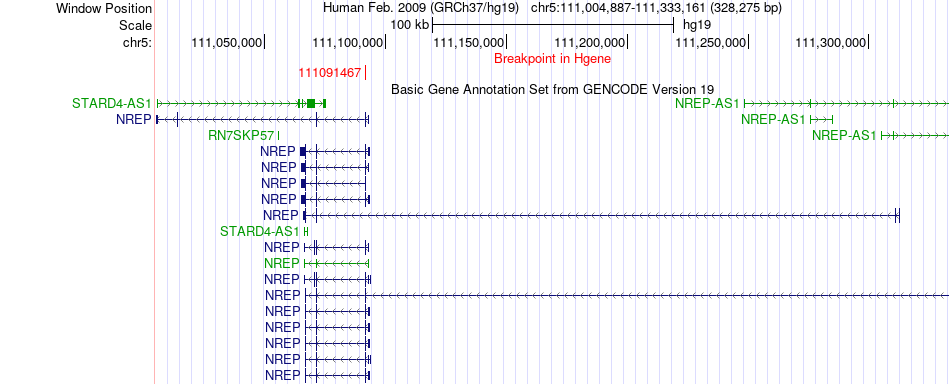 |
 Fusion gene breakpoints across COMMD10 (3'-gene) Fusion gene breakpoints across COMMD10 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
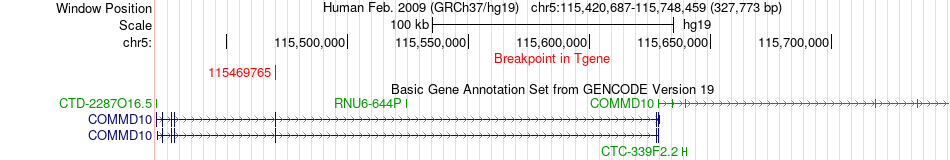 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | KIRC | TCGA-A3-3376-01A | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
Top |
Fusion Gene ORF analysis for NREP-COMMD10 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000257435 | ENST00000503424 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| 5CDS-intron | ENST00000379671 | ENST00000503424 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| 5CDS-intron | ENST00000419114 | ENST00000503424 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| 5CDS-intron | ENST00000446294 | ENST00000503424 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| 5CDS-intron | ENST00000447165 | ENST00000503424 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| 5CDS-intron | ENST00000450761 | ENST00000503424 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| 5CDS-intron | ENST00000453526 | ENST00000503424 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| 5CDS-intron | ENST00000455559 | ENST00000503424 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| 5CDS-intron | ENST00000508870 | ENST00000503424 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| 5CDS-intron | ENST00000509025 | ENST00000503424 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| 5CDS-intron | ENST00000509427 | ENST00000503424 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| 5CDS-intron | ENST00000509979 | ENST00000503424 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| 5CDS-intron | ENST00000515855 | ENST00000503424 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| Frame-shift | ENST00000379671 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| Frame-shift | ENST00000379671 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| Frame-shift | ENST00000446294 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| Frame-shift | ENST00000446294 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| Frame-shift | ENST00000450761 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| Frame-shift | ENST00000450761 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| Frame-shift | ENST00000453526 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| Frame-shift | ENST00000453526 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| Frame-shift | ENST00000508870 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| Frame-shift | ENST00000508870 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| Frame-shift | ENST00000509427 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| Frame-shift | ENST00000509427 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| Frame-shift | ENST00000509979 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| Frame-shift | ENST00000509979 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| In-frame | ENST00000257435 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| In-frame | ENST00000257435 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| In-frame | ENST00000419114 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| In-frame | ENST00000419114 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| In-frame | ENST00000447165 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| In-frame | ENST00000447165 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| In-frame | ENST00000455559 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| In-frame | ENST00000455559 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| In-frame | ENST00000509025 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| In-frame | ENST00000509025 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| In-frame | ENST00000515855 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| In-frame | ENST00000515855 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| intron-3CDS | ENST00000395634 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| intron-3CDS | ENST00000395634 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| intron-3CDS | ENST00000507742 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| intron-3CDS | ENST00000507742 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| intron-intron | ENST00000395634 | ENST00000503424 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
| intron-intron | ENST00000507742 | ENST00000503424 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000509025 | NREP | chr5 | 111091467 | - | ENST00000274458 | COMMD10 | chr5 | 115469765 | + | 1128 | 120 | 866 | 549 | 105 |
| ENST00000509025 | NREP | chr5 | 111091467 | - | ENST00000515539 | COMMD10 | chr5 | 115469765 | + | 498 | 120 | 117 | 329 | 70 |
| ENST00000257435 | NREP | chr5 | 111091467 | - | ENST00000274458 | COMMD10 | chr5 | 115469765 | + | 1192 | 184 | 930 | 613 | 105 |
| ENST00000257435 | NREP | chr5 | 111091467 | - | ENST00000515539 | COMMD10 | chr5 | 115469765 | + | 562 | 184 | 181 | 393 | 70 |
| ENST00000447165 | NREP | chr5 | 111091467 | - | ENST00000274458 | COMMD10 | chr5 | 115469765 | + | 1490 | 482 | 1228 | 911 | 105 |
| ENST00000447165 | NREP | chr5 | 111091467 | - | ENST00000515539 | COMMD10 | chr5 | 115469765 | + | 860 | 482 | 479 | 691 | 70 |
| ENST00000515855 | NREP | chr5 | 111091467 | - | ENST00000274458 | COMMD10 | chr5 | 115469765 | + | 1128 | 120 | 866 | 549 | 105 |
| ENST00000515855 | NREP | chr5 | 111091467 | - | ENST00000515539 | COMMD10 | chr5 | 115469765 | + | 498 | 120 | 117 | 329 | 70 |
| ENST00000419114 | NREP | chr5 | 111091467 | - | ENST00000274458 | COMMD10 | chr5 | 115469765 | + | 1291 | 283 | 1029 | 712 | 105 |
| ENST00000419114 | NREP | chr5 | 111091467 | - | ENST00000515539 | COMMD10 | chr5 | 115469765 | + | 661 | 283 | 280 | 492 | 70 |
| ENST00000455559 | NREP | chr5 | 111091467 | - | ENST00000274458 | COMMD10 | chr5 | 115469765 | + | 1387 | 379 | 374 | 0 | 125 |
| ENST00000455559 | NREP | chr5 | 111091467 | - | ENST00000515539 | COMMD10 | chr5 | 115469765 | + | 757 | 379 | 374 | 0 | 125 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000509025 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + | 0.27037075 | 0.7296292 |
| ENST00000509025 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + | 0.10034612 | 0.89965385 |
| ENST00000257435 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + | 0.29106155 | 0.7089385 |
| ENST00000257435 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + | 0.061040055 | 0.93895996 |
| ENST00000447165 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + | 0.18336107 | 0.8166389 |
| ENST00000447165 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + | 0.06628071 | 0.9337193 |
| ENST00000515855 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + | 0.27037075 | 0.7296292 |
| ENST00000515855 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + | 0.10034612 | 0.89965385 |
| ENST00000419114 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + | 0.5231782 | 0.4768218 |
| ENST00000419114 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + | 0.19720225 | 0.80279773 |
| ENST00000455559 | ENST00000274458 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + | 0.3317465 | 0.6682535 |
| ENST00000455559 | ENST00000515539 | NREP | chr5 | 111091467 | - | COMMD10 | chr5 | 115469765 | + | 0.1085004 | 0.8914996 |
Top |
Fusion Genomic Features for NREP-COMMD10 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| NREP | chr5 | 111091466 | - | COMMD10 | chr5 | 115469764 | + | 0.6554288 | 0.34457114 |
| NREP | chr5 | 111091466 | - | COMMD10 | chr5 | 115469764 | + | 0.6554288 | 0.34457114 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
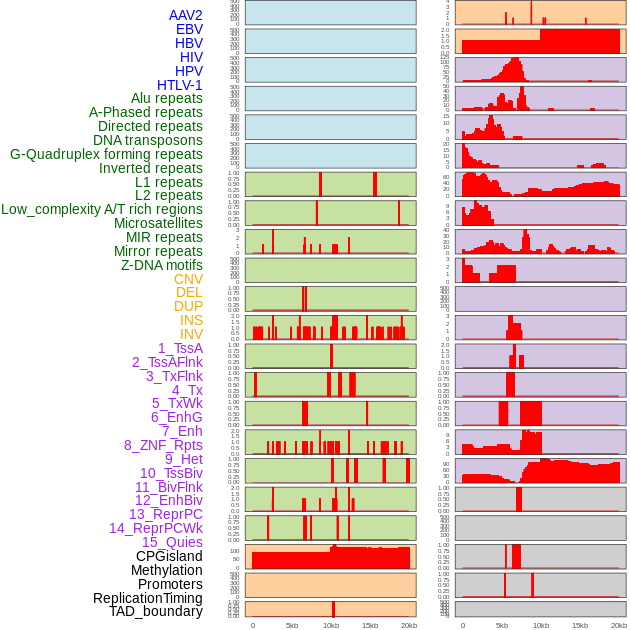 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
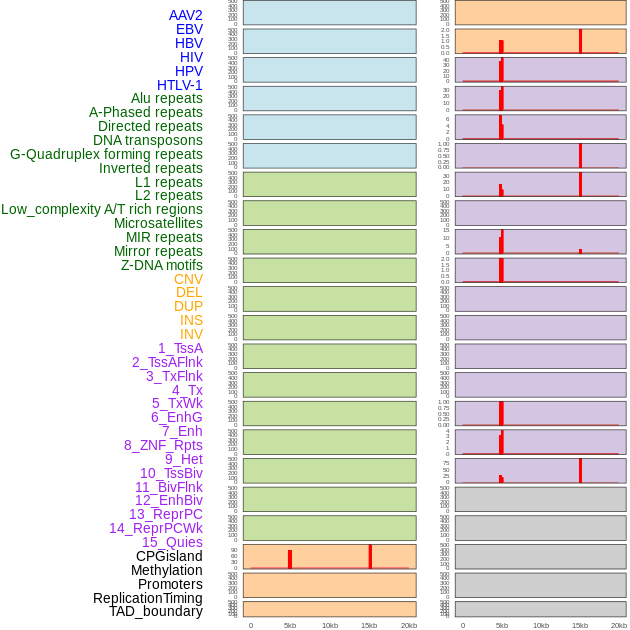 |
Top |
Fusion Protein Features for NREP-COMMD10 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:111091467/chr5:115469765) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NREP | COMMD10 |
| FUNCTION: May have roles in neural function. Ectopic expression augments motility of gliomas. Promotes also axonal regeneration (By similarity). May also have functions in cellular differentiation (By similarity). Induces differentiation of fibroblast into myofibroblast and myofibroblast ameboid migration. Increases retinoic-acid regulation of lipid-droplet biogenesis (By similarity). Down-regulates the expression of TGFB1 and TGFB2 but not of TGFB3 (By similarity). May play a role in the regulation of alveolar generation. {ECO:0000250, ECO:0000269|PubMed:11358844, ECO:0000269|PubMed:16229809}. | FUNCTION: May modulate activity of cullin-RING E3 ubiquitin ligase (CRL) complexes (PubMed:21778237). May down-regulate activation of NF-kappa-B (PubMed:15799966). {ECO:0000269|PubMed:15799966, ECO:0000305|PubMed:21778237}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | COMMD10 | chr5:111091467 | chr5:115469765 | ENST00000274458 | 3 | 7 | 133_202 | 133 | 203.0 | Domain | COMM |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Gene Sequence for NREP-COMMD10 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >60250_60250_1_NREP-COMMD10_NREP_chr5_111091467_ENST00000257435_COMMD10_chr5_115469765_ENST00000274458_length(transcript)=1192nt_BP=184nt CTCCCTCCCCCAATCCCCCCTCCCCCCCCAATCTGTCTTTCTAGCATGTTGCCCTTTTTCAACCACATTTGTGTTTCAGGTGTAGAGAGG AGAGAGAGTGAACAGGGAGCGGGGCTTTTGTCTGTTGGTCTCCCTGGACTGAAGAGAGGGAGAATAGAAGCCCAAGACTAAGATTCTCAA AATGCTAGAGACCGTTGGATGGCAGCTTAACCTTCAGATGGCTCACTCTGCTCAAGCAAAACTAAAATCTCCTCAAGCTGTGTTACAACT CGGAGTGAACAATGAAGATTCAAAGAGCCTGGAGAAAGTTCTTGTGGAATTCAGTCACAAGGAGTTGTTTGATTTCTATAACAAGCTAGA GACTATACAAGCACAGCTGGATTCCCTTACATGATGTTTTCGAAGACTGTTTTTTTCATCACGCTCCTGCCACCTCATTATTTTGCATTG AAGATACATTGCCAGGTTGTGTTTTCTGAAGGATTCAGTGACTTGCTTTCTGTAAATTATATGGCTTATCACTTCTTAGACAAATAACAA CCAATAGAGATCATTGTTAAGAATACTGAGGTTCTAATATACTTTCTTTAGTTCTGTGAGCCAACAGTAATTATTAAGAACACTTTCCCT TTAAAGGAAACAAAAGTGAATACCATATTGTTTTTACTGTCATAGTGTTGCTTTCTTGCCTGTCCTGCTTAGTTTTTACTTGCTGGATGA TACCATAATGTATCAAGGAGCGTCCATGGATACAAGATAAGATGTGTACCTTAGTAGAATACAGAGCTTTGGTAATTACATGAATAAAAT TAAGAAAATAGCCATATACAATCAAATACACTATGGCATTTTTATTTGAATATGATGAGTATATTTTGCTTCGGAAATAATATAGGAAGG AAATGTAAAATAGTGAGTAGTATGGTATCAGTTAATTCCAGTCTGAGCTTCTCTGTCAACTTCAGTTTCTCTCTCAGTTTAATGATTTAA TAATAGTCCAGGTTTTTGTGTGTTTTTCTTTATACTGCAAATTAATAATGATTCACTTTATAGTTTGGGAGACAGAATCAGGTCTTGAAT AAAATAATTGTAATGAGTGCTAAATGGGCACCATTATTCGAATCAGATACCTTTTATATTCTCTTTCCATAAATACGTTGATTTCTGTCA >60250_60250_1_NREP-COMMD10_NREP_chr5_111091467_ENST00000257435_COMMD10_chr5_115469765_ENST00000274458_length(amino acids)=105AA_BP= MIPYYSLFYISFLYYFRSKIYSSYSNKNAIVYLIVYGYFLNFIHVITKALYSTKVHILSCIHGRSLIHYGIIQQVKTKQDRQESNTMTVK -------------------------------------------------------------- >60250_60250_2_NREP-COMMD10_NREP_chr5_111091467_ENST00000257435_COMMD10_chr5_115469765_ENST00000515539_length(transcript)=562nt_BP=184nt CTCCCTCCCCCAATCCCCCCTCCCCCCCCAATCTGTCTTTCTAGCATGTTGCCCTTTTTCAACCACATTTGTGTTTCAGGTGTAGAGAGG AGAGAGAGTGAACAGGGAGCGGGGCTTTTGTCTGTTGGTCTCCCTGGACTGAAGAGAGGGAGAATAGAAGCCCAAGACTAAGATTCTCAA AATGCTAGAGACCGTTGGATGGCAGCTTAACCTTCAGATGGCTCACTCTGCTCAAGCAAAACTAAAATCTCCTCAAGCTGTGTTACAACT CGGAGTGAACAATGAAGATTCAAAGAGCCTGGAGAAAGTTCTTGTGGAATTCAGTCACAAGGAGTTGTTTGATTTCTATAACAAGCTAGA GACTATACAAGCACAGCTGGATTCCCTTACATGATGTTTTCGAAGACTGTTTTTTTCATCACGCTCCTGCCACCTCATTATTTTGCATTG AAGATACATTGCCAGGTTGTGTTTTCTGAAGGATTCAGTGACTTGCTTTCTGTAAATTATATGGCTTATCACTTCTTAGACAAATAACAA >60250_60250_2_NREP-COMMD10_NREP_chr5_111091467_ENST00000257435_COMMD10_chr5_115469765_ENST00000515539_length(amino acids)=70AA_BP=1 -------------------------------------------------------------- >60250_60250_3_NREP-COMMD10_NREP_chr5_111091467_ENST00000419114_COMMD10_chr5_115469765_ENST00000274458_length(transcript)=1291nt_BP=283nt AGTCTGGCTCCTAGGCGGCTGCACAGTGGCAGCAAGGCTGGGGCGCTGGCTCCGCGCCAGCTCCATCAGAATTAGCTCATTGTTCACTAG CACCCTAGCGGGAGAGAGAGGGAAAGGGAGAGAGAGAGACTCTTAGAGAAAGAGAGAGACACACACACTCTTGGAGAGAGAGCGCGAGGT GTAGAGAGGAGAGAGAGTGAACAGGGAGCGGGGCTTTTGTCTGTTGGTCTCCCTGGACTGAAGAGAGGGAGAATAGAAGCCCAAGACTAA GATTCTCAAAATGCTAGAGACCGTTGGATGGCAGCTTAACCTTCAGATGGCTCACTCTGCTCAAGCAAAACTAAAATCTCCTCAAGCTGT GTTACAACTCGGAGTGAACAATGAAGATTCAAAGAGCCTGGAGAAAGTTCTTGTGGAATTCAGTCACAAGGAGTTGTTTGATTTCTATAA CAAGCTAGAGACTATACAAGCACAGCTGGATTCCCTTACATGATGTTTTCGAAGACTGTTTTTTTCATCACGCTCCTGCCACCTCATTAT TTTGCATTGAAGATACATTGCCAGGTTGTGTTTTCTGAAGGATTCAGTGACTTGCTTTCTGTAAATTATATGGCTTATCACTTCTTAGAC AAATAACAACCAATAGAGATCATTGTTAAGAATACTGAGGTTCTAATATACTTTCTTTAGTTCTGTGAGCCAACAGTAATTATTAAGAAC ACTTTCCCTTTAAAGGAAACAAAAGTGAATACCATATTGTTTTTACTGTCATAGTGTTGCTTTCTTGCCTGTCCTGCTTAGTTTTTACTT GCTGGATGATACCATAATGTATCAAGGAGCGTCCATGGATACAAGATAAGATGTGTACCTTAGTAGAATACAGAGCTTTGGTAATTACAT GAATAAAATTAAGAAAATAGCCATATACAATCAAATACACTATGGCATTTTTATTTGAATATGATGAGTATATTTTGCTTCGGAAATAAT ATAGGAAGGAAATGTAAAATAGTGAGTAGTATGGTATCAGTTAATTCCAGTCTGAGCTTCTCTGTCAACTTCAGTTTCTCTCTCAGTTTA ATGATTTAATAATAGTCCAGGTTTTTGTGTGTTTTTCTTTATACTGCAAATTAATAATGATTCACTTTATAGTTTGGGAGACAGAATCAG GTCTTGAATAAAATAATTGTAATGAGTGCTAAATGGGCACCATTATTCGAATCAGATACCTTTTATATTCTCTTTCCATAAATACGTTGA >60250_60250_3_NREP-COMMD10_NREP_chr5_111091467_ENST00000419114_COMMD10_chr5_115469765_ENST00000274458_length(amino acids)=105AA_BP= MIPYYSLFYISFLYYFRSKIYSSYSNKNAIVYLIVYGYFLNFIHVITKALYSTKVHILSCIHGRSLIHYGIIQQVKTKQDRQESNTMTVK -------------------------------------------------------------- >60250_60250_4_NREP-COMMD10_NREP_chr5_111091467_ENST00000419114_COMMD10_chr5_115469765_ENST00000515539_length(transcript)=661nt_BP=283nt AGTCTGGCTCCTAGGCGGCTGCACAGTGGCAGCAAGGCTGGGGCGCTGGCTCCGCGCCAGCTCCATCAGAATTAGCTCATTGTTCACTAG CACCCTAGCGGGAGAGAGAGGGAAAGGGAGAGAGAGAGACTCTTAGAGAAAGAGAGAGACACACACACTCTTGGAGAGAGAGCGCGAGGT GTAGAGAGGAGAGAGAGTGAACAGGGAGCGGGGCTTTTGTCTGTTGGTCTCCCTGGACTGAAGAGAGGGAGAATAGAAGCCCAAGACTAA GATTCTCAAAATGCTAGAGACCGTTGGATGGCAGCTTAACCTTCAGATGGCTCACTCTGCTCAAGCAAAACTAAAATCTCCTCAAGCTGT GTTACAACTCGGAGTGAACAATGAAGATTCAAAGAGCCTGGAGAAAGTTCTTGTGGAATTCAGTCACAAGGAGTTGTTTGATTTCTATAA CAAGCTAGAGACTATACAAGCACAGCTGGATTCCCTTACATGATGTTTTCGAAGACTGTTTTTTTCATCACGCTCCTGCCACCTCATTAT TTTGCATTGAAGATACATTGCCAGGTTGTGTTTTCTGAAGGATTCAGTGACTTGCTTTCTGTAAATTATATGGCTTATCACTTCTTAGAC >60250_60250_4_NREP-COMMD10_NREP_chr5_111091467_ENST00000419114_COMMD10_chr5_115469765_ENST00000515539_length(amino acids)=70AA_BP=1 -------------------------------------------------------------- >60250_60250_5_NREP-COMMD10_NREP_chr5_111091467_ENST00000447165_COMMD10_chr5_115469765_ENST00000274458_length(transcript)=1490nt_BP=482nt ACTCTGAGGCATTACACTACACGGTGGAAGGCTGATTCCTATTCTGTCGTCTTGATTGCAAATATACTGCCAGTAGTATGTCTGGGAACA AAGAGCAGCCAGAAAACAATCTATTTTTATCACTTGGTTCTACTTTTAAGTTGCTAAACCTTTTGAACTCATGCAATTTAAAGATGAACC AAACACTAAGAATAGCCCAGGACTCTGCAAAGCCAAGTACCAGGGATTCCTTTGTCCCTTAGTTAAGCCTATAGGCCACTTGGAAAGCTC TTTCAGAAATCCCAGTAGTTAACAATAATTTGCATGTAACTTATCTTAACCACAGAAAATGGTTATTCGTCTGTAACCTCTAAAGAGGCA TTTTTCGTTTGCTGACCTGTGGTGAGGCGATGTGTCTACTGTATTTAAACATATTTTGCAGGTTGGTCTCCCTGGACTGAAGAGAGGGAG AATAGAAGCCCAAGACTAAGATTCTCAAAATGCTAGAGACCGTTGGATGGCAGCTTAACCTTCAGATGGCTCACTCTGCTCAAGCAAAAC TAAAATCTCCTCAAGCTGTGTTACAACTCGGAGTGAACAATGAAGATTCAAAGAGCCTGGAGAAAGTTCTTGTGGAATTCAGTCACAAGG AGTTGTTTGATTTCTATAACAAGCTAGAGACTATACAAGCACAGCTGGATTCCCTTACATGATGTTTTCGAAGACTGTTTTTTTCATCAC GCTCCTGCCACCTCATTATTTTGCATTGAAGATACATTGCCAGGTTGTGTTTTCTGAAGGATTCAGTGACTTGCTTTCTGTAAATTATAT GGCTTATCACTTCTTAGACAAATAACAACCAATAGAGATCATTGTTAAGAATACTGAGGTTCTAATATACTTTCTTTAGTTCTGTGAGCC AACAGTAATTATTAAGAACACTTTCCCTTTAAAGGAAACAAAAGTGAATACCATATTGTTTTTACTGTCATAGTGTTGCTTTCTTGCCTG TCCTGCTTAGTTTTTACTTGCTGGATGATACCATAATGTATCAAGGAGCGTCCATGGATACAAGATAAGATGTGTACCTTAGTAGAATAC AGAGCTTTGGTAATTACATGAATAAAATTAAGAAAATAGCCATATACAATCAAATACACTATGGCATTTTTATTTGAATATGATGAGTAT ATTTTGCTTCGGAAATAATATAGGAAGGAAATGTAAAATAGTGAGTAGTATGGTATCAGTTAATTCCAGTCTGAGCTTCTCTGTCAACTT CAGTTTCTCTCTCAGTTTAATGATTTAATAATAGTCCAGGTTTTTGTGTGTTTTTCTTTATACTGCAAATTAATAATGATTCACTTTATA GTTTGGGAGACAGAATCAGGTCTTGAATAAAATAATTGTAATGAGTGCTAAATGGGCACCATTATTCGAATCAGATACCTTTTATATTCT >60250_60250_5_NREP-COMMD10_NREP_chr5_111091467_ENST00000447165_COMMD10_chr5_115469765_ENST00000274458_length(amino acids)=105AA_BP= MIPYYSLFYISFLYYFRSKIYSSYSNKNAIVYLIVYGYFLNFIHVITKALYSTKVHILSCIHGRSLIHYGIIQQVKTKQDRQESNTMTVK -------------------------------------------------------------- >60250_60250_6_NREP-COMMD10_NREP_chr5_111091467_ENST00000447165_COMMD10_chr5_115469765_ENST00000515539_length(transcript)=860nt_BP=482nt ACTCTGAGGCATTACACTACACGGTGGAAGGCTGATTCCTATTCTGTCGTCTTGATTGCAAATATACTGCCAGTAGTATGTCTGGGAACA AAGAGCAGCCAGAAAACAATCTATTTTTATCACTTGGTTCTACTTTTAAGTTGCTAAACCTTTTGAACTCATGCAATTTAAAGATGAACC AAACACTAAGAATAGCCCAGGACTCTGCAAAGCCAAGTACCAGGGATTCCTTTGTCCCTTAGTTAAGCCTATAGGCCACTTGGAAAGCTC TTTCAGAAATCCCAGTAGTTAACAATAATTTGCATGTAACTTATCTTAACCACAGAAAATGGTTATTCGTCTGTAACCTCTAAAGAGGCA TTTTTCGTTTGCTGACCTGTGGTGAGGCGATGTGTCTACTGTATTTAAACATATTTTGCAGGTTGGTCTCCCTGGACTGAAGAGAGGGAG AATAGAAGCCCAAGACTAAGATTCTCAAAATGCTAGAGACCGTTGGATGGCAGCTTAACCTTCAGATGGCTCACTCTGCTCAAGCAAAAC TAAAATCTCCTCAAGCTGTGTTACAACTCGGAGTGAACAATGAAGATTCAAAGAGCCTGGAGAAAGTTCTTGTGGAATTCAGTCACAAGG AGTTGTTTGATTTCTATAACAAGCTAGAGACTATACAAGCACAGCTGGATTCCCTTACATGATGTTTTCGAAGACTGTTTTTTTCATCAC GCTCCTGCCACCTCATTATTTTGCATTGAAGATACATTGCCAGGTTGTGTTTTCTGAAGGATTCAGTGACTTGCTTTCTGTAAATTATAT >60250_60250_6_NREP-COMMD10_NREP_chr5_111091467_ENST00000447165_COMMD10_chr5_115469765_ENST00000515539_length(amino acids)=70AA_BP=1 -------------------------------------------------------------- >60250_60250_7_NREP-COMMD10_NREP_chr5_111091467_ENST00000455559_COMMD10_chr5_115469765_ENST00000274458_length(transcript)=1387nt_BP=379nt ACGCGCAGCCACAGGACCTCTGGGCCCGGCCTTCAGGGGCGCTCCCCGCCCCGGGCTTCCCCGCGGAAGGGAGTGGGGCGCGCGTACCCA GCTGAGGGGCAGGGTCCGACCCGAGGCCGTGTGCAAATGCATCCTTCGCGGACTCTTTGTGTGTCTGAGCGCGGCTCCGCGCCGCCGCAG GCACCATTTTCTGCTTCGCTCAGGACAGGCACATAAAAGGAAGGCGGCTGCCGCCCGTCGCCGTCCTCTTTTCCTCAGATGCCCTCTGCT GCAGGTGTAGAGAGGAGAGAGAGTGAACAGGGAGCGGGGCTTTTGTCTGTTGGTCTCCCTGGACTGAAGAGAGGGAGAATAGAAGCCCAA GACTAAGATTCTCAAAATGCTAGAGACCGTTGGATGGCAGCTTAACCTTCAGATGGCTCACTCTGCTCAAGCAAAACTAAAATCTCCTCA AGCTGTGTTACAACTCGGAGTGAACAATGAAGATTCAAAGAGCCTGGAGAAAGTTCTTGTGGAATTCAGTCACAAGGAGTTGTTTGATTT CTATAACAAGCTAGAGACTATACAAGCACAGCTGGATTCCCTTACATGATGTTTTCGAAGACTGTTTTTTTCATCACGCTCCTGCCACCT CATTATTTTGCATTGAAGATACATTGCCAGGTTGTGTTTTCTGAAGGATTCAGTGACTTGCTTTCTGTAAATTATATGGCTTATCACTTC TTAGACAAATAACAACCAATAGAGATCATTGTTAAGAATACTGAGGTTCTAATATACTTTCTTTAGTTCTGTGAGCCAACAGTAATTATT AAGAACACTTTCCCTTTAAAGGAAACAAAAGTGAATACCATATTGTTTTTACTGTCATAGTGTTGCTTTCTTGCCTGTCCTGCTTAGTTT TTACTTGCTGGATGATACCATAATGTATCAAGGAGCGTCCATGGATACAAGATAAGATGTGTACCTTAGTAGAATACAGAGCTTTGGTAA TTACATGAATAAAATTAAGAAAATAGCCATATACAATCAAATACACTATGGCATTTTTATTTGAATATGATGAGTATATTTTGCTTCGGA AATAATATAGGAAGGAAATGTAAAATAGTGAGTAGTATGGTATCAGTTAATTCCAGTCTGAGCTTCTCTGTCAACTTCAGTTTCTCTCTC AGTTTAATGATTTAATAATAGTCCAGGTTTTTGTGTGTTTTTCTTTATACTGCAAATTAATAATGATTCACTTTATAGTTTGGGAGACAG AATCAGGTCTTGAATAAAATAATTGTAATGAGTGCTAAATGGGCACCATTATTCGAATCAGATACCTTTTATATTCTCTTTCCATAAATA >60250_60250_7_NREP-COMMD10_NREP_chr5_111091467_ENST00000455559_COMMD10_chr5_115469765_ENST00000274458_length(amino acids)=125AA_BP= MRILVLGFYSPSLQSRETNRQKPRSLFTLSPLYTCSRGHLRKRGRRRAAAAFLLCACPERSRKWCLRRRGAALRHTKSPRRMHLHTASGR -------------------------------------------------------------- >60250_60250_8_NREP-COMMD10_NREP_chr5_111091467_ENST00000455559_COMMD10_chr5_115469765_ENST00000515539_length(transcript)=757nt_BP=379nt ACGCGCAGCCACAGGACCTCTGGGCCCGGCCTTCAGGGGCGCTCCCCGCCCCGGGCTTCCCCGCGGAAGGGAGTGGGGCGCGCGTACCCA GCTGAGGGGCAGGGTCCGACCCGAGGCCGTGTGCAAATGCATCCTTCGCGGACTCTTTGTGTGTCTGAGCGCGGCTCCGCGCCGCCGCAG GCACCATTTTCTGCTTCGCTCAGGACAGGCACATAAAAGGAAGGCGGCTGCCGCCCGTCGCCGTCCTCTTTTCCTCAGATGCCCTCTGCT GCAGGTGTAGAGAGGAGAGAGAGTGAACAGGGAGCGGGGCTTTTGTCTGTTGGTCTCCCTGGACTGAAGAGAGGGAGAATAGAAGCCCAA GACTAAGATTCTCAAAATGCTAGAGACCGTTGGATGGCAGCTTAACCTTCAGATGGCTCACTCTGCTCAAGCAAAACTAAAATCTCCTCA AGCTGTGTTACAACTCGGAGTGAACAATGAAGATTCAAAGAGCCTGGAGAAAGTTCTTGTGGAATTCAGTCACAAGGAGTTGTTTGATTT CTATAACAAGCTAGAGACTATACAAGCACAGCTGGATTCCCTTACATGATGTTTTCGAAGACTGTTTTTTTCATCACGCTCCTGCCACCT CATTATTTTGCATTGAAGATACATTGCCAGGTTGTGTTTTCTGAAGGATTCAGTGACTTGCTTTCTGTAAATTATATGGCTTATCACTTC >60250_60250_8_NREP-COMMD10_NREP_chr5_111091467_ENST00000455559_COMMD10_chr5_115469765_ENST00000515539_length(amino acids)=125AA_BP= MRILVLGFYSPSLQSRETNRQKPRSLFTLSPLYTCSRGHLRKRGRRRAAAAFLLCACPERSRKWCLRRRGAALRHTKSPRRMHLHTASGR -------------------------------------------------------------- >60250_60250_9_NREP-COMMD10_NREP_chr5_111091467_ENST00000509025_COMMD10_chr5_115469765_ENST00000274458_length(transcript)=1128nt_BP=120nt ACATTTGTGTTTCAGGTGTAGAGAGGAGAGAGAGTGAACAGGGAGCGGGGCTTTTGTCTGTTGGTCTCCCTGGACTGAAGAGAGGGAGAA TAGAAGCCCAAGACTAAGATTCTCAAAATGCTAGAGACCGTTGGATGGCAGCTTAACCTTCAGATGGCTCACTCTGCTCAAGCAAAACTA AAATCTCCTCAAGCTGTGTTACAACTCGGAGTGAACAATGAAGATTCAAAGAGCCTGGAGAAAGTTCTTGTGGAATTCAGTCACAAGGAG TTGTTTGATTTCTATAACAAGCTAGAGACTATACAAGCACAGCTGGATTCCCTTACATGATGTTTTCGAAGACTGTTTTTTTCATCACGC TCCTGCCACCTCATTATTTTGCATTGAAGATACATTGCCAGGTTGTGTTTTCTGAAGGATTCAGTGACTTGCTTTCTGTAAATTATATGG CTTATCACTTCTTAGACAAATAACAACCAATAGAGATCATTGTTAAGAATACTGAGGTTCTAATATACTTTCTTTAGTTCTGTGAGCCAA CAGTAATTATTAAGAACACTTTCCCTTTAAAGGAAACAAAAGTGAATACCATATTGTTTTTACTGTCATAGTGTTGCTTTCTTGCCTGTC CTGCTTAGTTTTTACTTGCTGGATGATACCATAATGTATCAAGGAGCGTCCATGGATACAAGATAAGATGTGTACCTTAGTAGAATACAG AGCTTTGGTAATTACATGAATAAAATTAAGAAAATAGCCATATACAATCAAATACACTATGGCATTTTTATTTGAATATGATGAGTATAT TTTGCTTCGGAAATAATATAGGAAGGAAATGTAAAATAGTGAGTAGTATGGTATCAGTTAATTCCAGTCTGAGCTTCTCTGTCAACTTCA GTTTCTCTCTCAGTTTAATGATTTAATAATAGTCCAGGTTTTTGTGTGTTTTTCTTTATACTGCAAATTAATAATGATTCACTTTATAGT TTGGGAGACAGAATCAGGTCTTGAATAAAATAATTGTAATGAGTGCTAAATGGGCACCATTATTCGAATCAGATACCTTTTATATTCTCT >60250_60250_9_NREP-COMMD10_NREP_chr5_111091467_ENST00000509025_COMMD10_chr5_115469765_ENST00000274458_length(amino acids)=105AA_BP= MIPYYSLFYISFLYYFRSKIYSSYSNKNAIVYLIVYGYFLNFIHVITKALYSTKVHILSCIHGRSLIHYGIIQQVKTKQDRQESNTMTVK -------------------------------------------------------------- >60250_60250_10_NREP-COMMD10_NREP_chr5_111091467_ENST00000509025_COMMD10_chr5_115469765_ENST00000515539_length(transcript)=498nt_BP=120nt ACATTTGTGTTTCAGGTGTAGAGAGGAGAGAGAGTGAACAGGGAGCGGGGCTTTTGTCTGTTGGTCTCCCTGGACTGAAGAGAGGGAGAA TAGAAGCCCAAGACTAAGATTCTCAAAATGCTAGAGACCGTTGGATGGCAGCTTAACCTTCAGATGGCTCACTCTGCTCAAGCAAAACTA AAATCTCCTCAAGCTGTGTTACAACTCGGAGTGAACAATGAAGATTCAAAGAGCCTGGAGAAAGTTCTTGTGGAATTCAGTCACAAGGAG TTGTTTGATTTCTATAACAAGCTAGAGACTATACAAGCACAGCTGGATTCCCTTACATGATGTTTTCGAAGACTGTTTTTTTCATCACGC TCCTGCCACCTCATTATTTTGCATTGAAGATACATTGCCAGGTTGTGTTTTCTGAAGGATTCAGTGACTTGCTTTCTGTAAATTATATGG >60250_60250_10_NREP-COMMD10_NREP_chr5_111091467_ENST00000509025_COMMD10_chr5_115469765_ENST00000515539_length(amino acids)=70AA_BP=1 -------------------------------------------------------------- >60250_60250_11_NREP-COMMD10_NREP_chr5_111091467_ENST00000515855_COMMD10_chr5_115469765_ENST00000274458_length(transcript)=1128nt_BP=120nt ACATTTGTGTTTCAGGTGTAGAGAGGAGAGAGAGTGAACAGGGAGCGGGGCTTTTGTCTGTTGGTCTCCCTGGACTGAAGAGAGGGAGAA TAGAAGCCCAAGACTAAGATTCTCAAAATGCTAGAGACCGTTGGATGGCAGCTTAACCTTCAGATGGCTCACTCTGCTCAAGCAAAACTA AAATCTCCTCAAGCTGTGTTACAACTCGGAGTGAACAATGAAGATTCAAAGAGCCTGGAGAAAGTTCTTGTGGAATTCAGTCACAAGGAG TTGTTTGATTTCTATAACAAGCTAGAGACTATACAAGCACAGCTGGATTCCCTTACATGATGTTTTCGAAGACTGTTTTTTTCATCACGC TCCTGCCACCTCATTATTTTGCATTGAAGATACATTGCCAGGTTGTGTTTTCTGAAGGATTCAGTGACTTGCTTTCTGTAAATTATATGG CTTATCACTTCTTAGACAAATAACAACCAATAGAGATCATTGTTAAGAATACTGAGGTTCTAATATACTTTCTTTAGTTCTGTGAGCCAA CAGTAATTATTAAGAACACTTTCCCTTTAAAGGAAACAAAAGTGAATACCATATTGTTTTTACTGTCATAGTGTTGCTTTCTTGCCTGTC CTGCTTAGTTTTTACTTGCTGGATGATACCATAATGTATCAAGGAGCGTCCATGGATACAAGATAAGATGTGTACCTTAGTAGAATACAG AGCTTTGGTAATTACATGAATAAAATTAAGAAAATAGCCATATACAATCAAATACACTATGGCATTTTTATTTGAATATGATGAGTATAT TTTGCTTCGGAAATAATATAGGAAGGAAATGTAAAATAGTGAGTAGTATGGTATCAGTTAATTCCAGTCTGAGCTTCTCTGTCAACTTCA GTTTCTCTCTCAGTTTAATGATTTAATAATAGTCCAGGTTTTTGTGTGTTTTTCTTTATACTGCAAATTAATAATGATTCACTTTATAGT TTGGGAGACAGAATCAGGTCTTGAATAAAATAATTGTAATGAGTGCTAAATGGGCACCATTATTCGAATCAGATACCTTTTATATTCTCT >60250_60250_11_NREP-COMMD10_NREP_chr5_111091467_ENST00000515855_COMMD10_chr5_115469765_ENST00000274458_length(amino acids)=105AA_BP= MIPYYSLFYISFLYYFRSKIYSSYSNKNAIVYLIVYGYFLNFIHVITKALYSTKVHILSCIHGRSLIHYGIIQQVKTKQDRQESNTMTVK -------------------------------------------------------------- >60250_60250_12_NREP-COMMD10_NREP_chr5_111091467_ENST00000515855_COMMD10_chr5_115469765_ENST00000515539_length(transcript)=498nt_BP=120nt ACATTTGTGTTTCAGGTGTAGAGAGGAGAGAGAGTGAACAGGGAGCGGGGCTTTTGTCTGTTGGTCTCCCTGGACTGAAGAGAGGGAGAA TAGAAGCCCAAGACTAAGATTCTCAAAATGCTAGAGACCGTTGGATGGCAGCTTAACCTTCAGATGGCTCACTCTGCTCAAGCAAAACTA AAATCTCCTCAAGCTGTGTTACAACTCGGAGTGAACAATGAAGATTCAAAGAGCCTGGAGAAAGTTCTTGTGGAATTCAGTCACAAGGAG TTGTTTGATTTCTATAACAAGCTAGAGACTATACAAGCACAGCTGGATTCCCTTACATGATGTTTTCGAAGACTGTTTTTTTCATCACGC TCCTGCCACCTCATTATTTTGCATTGAAGATACATTGCCAGGTTGTGTTTTCTGAAGGATTCAGTGACTTGCTTTCTGTAAATTATATGG >60250_60250_12_NREP-COMMD10_NREP_chr5_111091467_ENST00000515855_COMMD10_chr5_115469765_ENST00000515539_length(amino acids)=70AA_BP=1 -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for NREP-COMMD10 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for NREP-COMMD10 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for NREP-COMMD10 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
