
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:NRG1-NRG1 (FusionGDB2 ID:60276) |
Fusion Gene Summary for NRG1-NRG1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: NRG1-NRG1 | Fusion gene ID: 60276 | Hgene | Tgene | Gene symbol | NRG1 | NRG1 | Gene ID | 3084 | 3084 |
| Gene name | neuregulin 1 | neuregulin 1 | |
| Synonyms | ARIA|GGF|GGF2|HGL|HRG|HRG1|HRGA|MST131|MSTP131|NDF|NRG1-IT2|SMDF | ARIA|GGF|GGF2|HGL|HRG|HRG1|HRGA|MST131|MSTP131|NDF|NRG1-IT2|SMDF | |
| Cytomap | 8p12 | 8p12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | pro-neuregulin-1, membrane-bound isoformacetylcholine receptor-inducing activityglial growth factor 2heregulin, alpha (45kD, ERBB2 p185-activator)neu differentiation factorpro-NRG1sensory and motor neuron derived factor | pro-neuregulin-1, membrane-bound isoformacetylcholine receptor-inducing activityglial growth factor 2heregulin, alpha (45kD, ERBB2 p185-activator)neu differentiation factorpro-NRG1sensory and motor neuron derived factor | |
| Modification date | 20200320 | 20200320 | |
| UniProtAcc | Q02297 | Q02297 | |
| Ensembl transtripts involved in fusion gene | ENST00000519301, ENST00000287842, ENST00000287845, ENST00000338921, ENST00000341377, ENST00000356819, ENST00000405005, ENST00000520407, ENST00000520502, ENST00000521670, ENST00000523079, ENST00000523681, ENST00000539990, | ENST00000520502, ENST00000523681, ENST00000539990, ENST00000287842, ENST00000287845, ENST00000338921, ENST00000341377, ENST00000356819, ENST00000405005, ENST00000519301, ENST00000520407, ENST00000521670, ENST00000523079, | |
| Fusion gene scores | * DoF score | 12 X 10 X 7=840 | 25 X 17 X 14=5950 |
| # samples | 12 | 27 | |
| ** MAII score | log2(12/840*10)=-2.8073549220576 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(27/5950*10)=-4.46185835603184 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: NRG1 [Title/Abstract] AND NRG1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | NRG1(31496947)-NRG1(32453344), # samples:13 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | NRG1 | GO:0003222 | ventricular trabecula myocardium morphogenesis | 17336907 |
| Hgene | NRG1 | GO:0031334 | positive regulation of protein complex assembly | 10559227 |
| Hgene | NRG1 | GO:0038127 | ERBB signaling pathway | 11389077 |
| Hgene | NRG1 | GO:0038129 | ERBB3 signaling pathway | 27353365 |
| Hgene | NRG1 | GO:0045892 | negative regulation of transcription, DNA-templated | 15073182 |
| Hgene | NRG1 | GO:0051048 | negative regulation of secretion | 10559227 |
| Hgene | NRG1 | GO:0060379 | cardiac muscle cell myoblast differentiation | 17336907 |
| Hgene | NRG1 | GO:0060956 | endocardial cell differentiation | 17336907 |
| Tgene | NRG1 | GO:0003222 | ventricular trabecula myocardium morphogenesis | 17336907 |
| Tgene | NRG1 | GO:0031334 | positive regulation of protein complex assembly | 10559227 |
| Tgene | NRG1 | GO:0038127 | ERBB signaling pathway | 11389077 |
| Tgene | NRG1 | GO:0038129 | ERBB3 signaling pathway | 27353365 |
| Tgene | NRG1 | GO:0045892 | negative regulation of transcription, DNA-templated | 15073182 |
| Tgene | NRG1 | GO:0051048 | negative regulation of secretion | 10559227 |
| Tgene | NRG1 | GO:0060379 | cardiac muscle cell myoblast differentiation | 17336907 |
| Tgene | NRG1 | GO:0060956 | endocardial cell differentiation | 17336907 |
 Fusion gene breakpoints across NRG1 (5'-gene) Fusion gene breakpoints across NRG1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
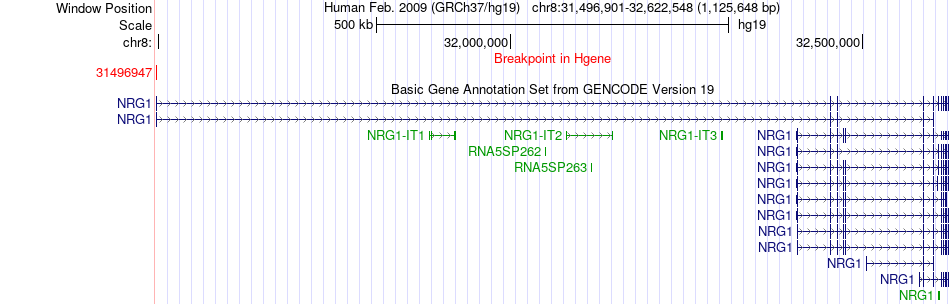 |
 Fusion gene breakpoints across NRG1 (3'-gene) Fusion gene breakpoints across NRG1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
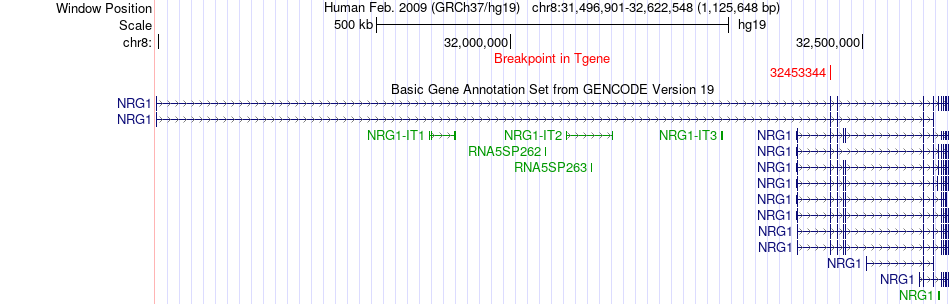 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChiTaRS5.0 | N/A | EF372273 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| ChiTaRS5.0 | N/A | EF372274 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| ChiTaRS5.0 | N/A | EF372275 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| ChiTaRS5.0 | N/A | EF372276 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| ChiTaRS5.0 | N/A | EF372277 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| ChiTaRS5.0 | N/A | EF517295 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| ChiTaRS5.0 | N/A | EF517296 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| ChiTaRS5.0 | N/A | EF517297 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| ChiTaRS5.0 | N/A | NM_001159995 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| ChiTaRS5.0 | N/A | NM_001159999 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| ChiTaRS5.0 | N/A | NM_001160001 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| ChiTaRS5.0 | N/A | NM_001322201 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| ChiTaRS5.0 | N/A | NM_001322202 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
Top |
Fusion Gene ORF analysis for NRG1-NRG1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000519301 | ENST00000520502 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| 5CDS-intron | ENST00000519301 | ENST00000523681 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| 5CDS-intron | ENST00000519301 | ENST00000539990 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| In-frame | ENST00000519301 | ENST00000287842 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| In-frame | ENST00000519301 | ENST00000287845 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| In-frame | ENST00000519301 | ENST00000338921 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| In-frame | ENST00000519301 | ENST00000341377 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| In-frame | ENST00000519301 | ENST00000356819 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| In-frame | ENST00000519301 | ENST00000405005 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| In-frame | ENST00000519301 | ENST00000519301 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| In-frame | ENST00000519301 | ENST00000520407 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| In-frame | ENST00000519301 | ENST00000521670 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| In-frame | ENST00000519301 | ENST00000523079 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287842 | ENST00000287842 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287842 | ENST00000287845 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287842 | ENST00000338921 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287842 | ENST00000341377 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287842 | ENST00000356819 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287842 | ENST00000405005 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287842 | ENST00000519301 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287842 | ENST00000520407 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287842 | ENST00000521670 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287842 | ENST00000523079 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287845 | ENST00000287842 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287845 | ENST00000287845 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287845 | ENST00000338921 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287845 | ENST00000341377 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287845 | ENST00000356819 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287845 | ENST00000405005 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287845 | ENST00000519301 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287845 | ENST00000520407 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287845 | ENST00000521670 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000287845 | ENST00000523079 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000338921 | ENST00000287842 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000338921 | ENST00000287845 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000338921 | ENST00000338921 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000338921 | ENST00000341377 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000338921 | ENST00000356819 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000338921 | ENST00000405005 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000338921 | ENST00000519301 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000338921 | ENST00000520407 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000338921 | ENST00000521670 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000338921 | ENST00000523079 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000341377 | ENST00000287842 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000341377 | ENST00000287845 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000341377 | ENST00000338921 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000341377 | ENST00000341377 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000341377 | ENST00000356819 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000341377 | ENST00000405005 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000341377 | ENST00000519301 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000341377 | ENST00000520407 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000341377 | ENST00000521670 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000341377 | ENST00000523079 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000356819 | ENST00000287842 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000356819 | ENST00000287845 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000356819 | ENST00000338921 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000356819 | ENST00000341377 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000356819 | ENST00000356819 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000356819 | ENST00000405005 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000356819 | ENST00000519301 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000356819 | ENST00000520407 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000356819 | ENST00000521670 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000356819 | ENST00000523079 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000405005 | ENST00000287842 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000405005 | ENST00000287845 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000405005 | ENST00000338921 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000405005 | ENST00000341377 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000405005 | ENST00000356819 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000405005 | ENST00000405005 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000405005 | ENST00000519301 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000405005 | ENST00000520407 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000405005 | ENST00000521670 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000405005 | ENST00000523079 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520407 | ENST00000287842 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520407 | ENST00000287845 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520407 | ENST00000338921 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520407 | ENST00000341377 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520407 | ENST00000356819 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520407 | ENST00000405005 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520407 | ENST00000519301 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520407 | ENST00000520407 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520407 | ENST00000521670 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520407 | ENST00000523079 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520502 | ENST00000287842 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520502 | ENST00000287845 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520502 | ENST00000338921 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520502 | ENST00000341377 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520502 | ENST00000356819 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520502 | ENST00000405005 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520502 | ENST00000519301 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520502 | ENST00000520407 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520502 | ENST00000521670 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000520502 | ENST00000523079 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000521670 | ENST00000287842 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000521670 | ENST00000287845 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000521670 | ENST00000338921 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000521670 | ENST00000341377 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000521670 | ENST00000356819 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000521670 | ENST00000405005 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000521670 | ENST00000519301 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000521670 | ENST00000520407 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000521670 | ENST00000521670 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000521670 | ENST00000523079 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523079 | ENST00000287842 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523079 | ENST00000287845 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523079 | ENST00000338921 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523079 | ENST00000341377 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523079 | ENST00000356819 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523079 | ENST00000405005 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523079 | ENST00000519301 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523079 | ENST00000520407 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523079 | ENST00000521670 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523079 | ENST00000523079 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523681 | ENST00000287842 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523681 | ENST00000287845 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523681 | ENST00000338921 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523681 | ENST00000341377 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523681 | ENST00000356819 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523681 | ENST00000405005 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523681 | ENST00000519301 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523681 | ENST00000520407 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523681 | ENST00000521670 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000523681 | ENST00000523079 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000539990 | ENST00000287842 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000539990 | ENST00000287845 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000539990 | ENST00000338921 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000539990 | ENST00000341377 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000539990 | ENST00000356819 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000539990 | ENST00000405005 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000539990 | ENST00000519301 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000539990 | ENST00000520407 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000539990 | ENST00000521670 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-3CDS | ENST00000539990 | ENST00000523079 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000287842 | ENST00000520502 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000287842 | ENST00000523681 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000287842 | ENST00000539990 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000287845 | ENST00000520502 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000287845 | ENST00000523681 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000287845 | ENST00000539990 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000338921 | ENST00000520502 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000338921 | ENST00000523681 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000338921 | ENST00000539990 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000341377 | ENST00000520502 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000341377 | ENST00000523681 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000341377 | ENST00000539990 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000356819 | ENST00000520502 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000356819 | ENST00000523681 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000356819 | ENST00000539990 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000405005 | ENST00000520502 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000405005 | ENST00000523681 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000405005 | ENST00000539990 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000520407 | ENST00000520502 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000520407 | ENST00000523681 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000520407 | ENST00000539990 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000520502 | ENST00000520502 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000520502 | ENST00000523681 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000520502 | ENST00000539990 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000521670 | ENST00000520502 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000521670 | ENST00000523681 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000521670 | ENST00000539990 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000523079 | ENST00000520502 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000523079 | ENST00000523681 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000523079 | ENST00000539990 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000523681 | ENST00000520502 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000523681 | ENST00000523681 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000523681 | ENST00000539990 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000539990 | ENST00000520502 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000539990 | ENST00000523681 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
| intron-intron | ENST00000539990 | ENST00000539990 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000519301 | NRG1 | chr8 | 31496947 | + | ENST00000519301 | NRG1 | chr8 | 32453344 | + | 1785 | 46 | 9 | 1781 | 590 |
| ENST00000519301 | NRG1 | chr8 | 31496947 | + | ENST00000520407 | NRG1 | chr8 | 32453344 | + | 1026 | 46 | 9 | 569 | 186 |
| ENST00000519301 | NRG1 | chr8 | 31496947 | + | ENST00000523079 | NRG1 | chr8 | 32453344 | + | 1788 | 46 | 9 | 1208 | 399 |
| ENST00000519301 | NRG1 | chr8 | 31496947 | + | ENST00000338921 | NRG1 | chr8 | 32453344 | + | 2521 | 46 | 9 | 1892 | 627 |
| ENST00000519301 | NRG1 | chr8 | 31496947 | + | ENST00000356819 | NRG1 | chr8 | 32453344 | + | 2512 | 46 | 9 | 1883 | 624 |
| ENST00000519301 | NRG1 | chr8 | 31496947 | + | ENST00000287845 | NRG1 | chr8 | 32453344 | + | 2410 | 46 | 9 | 1781 | 590 |
| ENST00000519301 | NRG1 | chr8 | 31496947 | + | ENST00000341377 | NRG1 | chr8 | 32453344 | + | 2556 | 46 | 689 | 1927 | 412 |
| ENST00000519301 | NRG1 | chr8 | 31496947 | + | ENST00000287842 | NRG1 | chr8 | 32453344 | + | 1969 | 46 | 9 | 1859 | 616 |
| ENST00000519301 | NRG1 | chr8 | 31496947 | + | ENST00000521670 | NRG1 | chr8 | 32453344 | + | 1607 | 46 | 9 | 1334 | 441 |
| ENST00000519301 | NRG1 | chr8 | 31496947 | + | ENST00000405005 | NRG1 | chr8 | 32453344 | + | 1938 | 46 | 9 | 1868 | 619 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000519301 | ENST00000519301 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + | 0.012085608 | 0.9879144 |
| ENST00000519301 | ENST00000520407 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + | 0.004736614 | 0.99526334 |
| ENST00000519301 | ENST00000523079 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + | 0.006751682 | 0.99324834 |
| ENST00000519301 | ENST00000338921 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + | 0.004683498 | 0.9953165 |
| ENST00000519301 | ENST00000356819 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + | 0.003776387 | 0.99622357 |
| ENST00000519301 | ENST00000287845 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + | 0.00392518 | 0.9960748 |
| ENST00000519301 | ENST00000341377 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + | 0.004885315 | 0.9951147 |
| ENST00000519301 | ENST00000287842 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + | 0.007732549 | 0.99226743 |
| ENST00000519301 | ENST00000521670 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + | 0.031561658 | 0.9684383 |
| ENST00000519301 | ENST00000405005 | NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453344 | + | 0.010651933 | 0.98934805 |
Top |
Fusion Genomic Features for NRG1-NRG1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453345 | + | 0.9971923 | 0.002807661 |
| NRG1 | chr8 | 31496947 | + | NRG1 | chr8 | 32453345 | + | 0.9971923 | 0.002807661 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
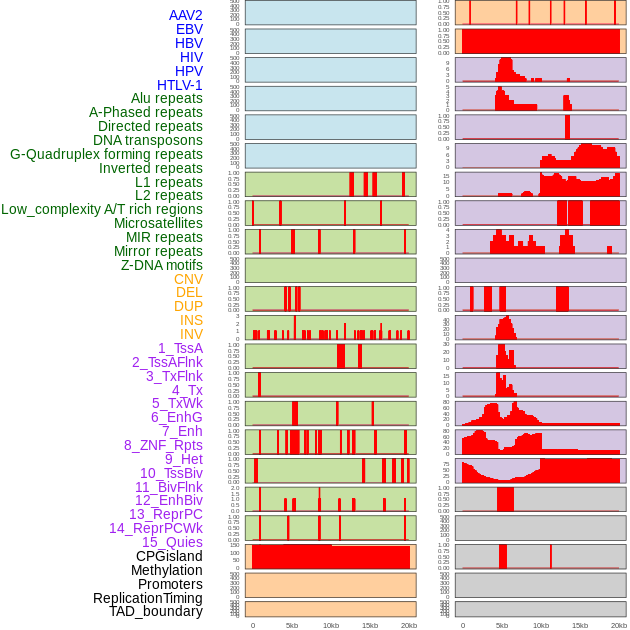 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
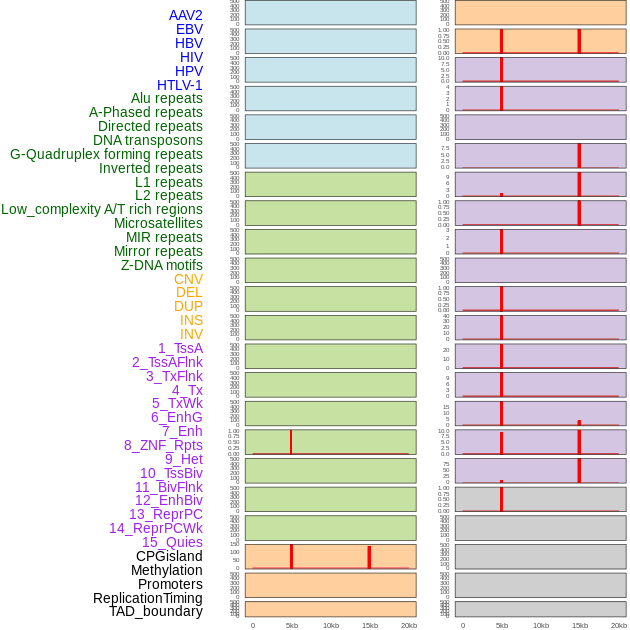 |
Top |
Fusion Protein Features for NRG1-NRG1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:31496947/chr8:32453344) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NRG1 | NRG1 |
| FUNCTION: Direct ligand for ERBB3 and ERBB4 tyrosine kinase receptors. Concomitantly recruits ERBB1 and ERBB2 coreceptors, resulting in ligand-stimulated tyrosine phosphorylation and activation of the ERBB receptors. The multiple isoforms perform diverse functions such as inducing growth and differentiation of epithelial, glial, neuronal, and skeletal muscle cells; inducing expression of acetylcholine receptor in synaptic vesicles during the formation of the neuromuscular junction; stimulating lobuloalveolar budding and milk production in the mammary gland and inducing differentiation of mammary tumor cells; stimulating Schwann cell proliferation; implication in the development of the myocardium such as trabeculation of the developing heart. Isoform 10 may play a role in motor and sensory neuron development. Binds to ERBB4 (PubMed:10867024, PubMed:7902537). Binds to ERBB3 (PubMed:20682778). Acts as a ligand for integrins and binds (via EGF domain) to integrins ITGAV:ITGB3 or ITGA6:ITGB4. Its binding to integrins and subsequent ternary complex formation with integrins and ERRB3 are essential for NRG1-ERBB signaling. Induces the phosphorylation and activation of MAPK3/ERK1, MAPK1/ERK2 and AKT1 (PubMed:20682778). Ligand-dependent ERBB4 endocytosis is essential for the NRG1-mediated activation of these kinases in neurons (By similarity). {ECO:0000250|UniProtKB:P43322, ECO:0000269|PubMed:10867024, ECO:0000269|PubMed:1348215, ECO:0000269|PubMed:20682778, ECO:0000269|PubMed:7902537}. | FUNCTION: Direct ligand for ERBB3 and ERBB4 tyrosine kinase receptors. Concomitantly recruits ERBB1 and ERBB2 coreceptors, resulting in ligand-stimulated tyrosine phosphorylation and activation of the ERBB receptors. The multiple isoforms perform diverse functions such as inducing growth and differentiation of epithelial, glial, neuronal, and skeletal muscle cells; inducing expression of acetylcholine receptor in synaptic vesicles during the formation of the neuromuscular junction; stimulating lobuloalveolar budding and milk production in the mammary gland and inducing differentiation of mammary tumor cells; stimulating Schwann cell proliferation; implication in the development of the myocardium such as trabeculation of the developing heart. Isoform 10 may play a role in motor and sensory neuron development. Binds to ERBB4 (PubMed:10867024, PubMed:7902537). Binds to ERBB3 (PubMed:20682778). Acts as a ligand for integrins and binds (via EGF domain) to integrins ITGAV:ITGB3 or ITGA6:ITGB4. Its binding to integrins and subsequent ternary complex formation with integrins and ERRB3 are essential for NRG1-ERBB signaling. Induces the phosphorylation and activation of MAPK3/ERK1, MAPK1/ERK2 and AKT1 (PubMed:20682778). Ligand-dependent ERBB4 endocytosis is essential for the NRG1-mediated activation of these kinases in neurons (By similarity). {ECO:0000250|UniProtKB:P43322, ECO:0000269|PubMed:10867024, ECO:0000269|PubMed:1348215, ECO:0000269|PubMed:20682778, ECO:0000269|PubMed:7902537}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000287842 | 0 | 12 | 165_177 | 33 | 638.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000341377 | 0 | 13 | 165_177 | 33 | 525.3333333333334 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000356819 | 0 | 13 | 165_177 | 33 | 646.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000405005 | 0 | 12 | 165_177 | 33 | 641.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000519301 | 0 | 11 | 165_177 | 12 | 591.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520502 | 0 | 3 | 165_177 | 0 | 297.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000521670 | 0 | 13 | 165_177 | 33 | 477.3333333333333 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000523079 | 0 | 11 | 165_177 | 33 | 421.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000287842 | 0 | 12 | 178_222 | 33 | 638.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000287842 | 0 | 12 | 37_128 | 33 | 638.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000341377 | 0 | 13 | 178_222 | 33 | 525.3333333333334 | Domain | EGF-like | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000341377 | 0 | 13 | 37_128 | 33 | 525.3333333333334 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000356819 | 0 | 13 | 178_222 | 33 | 646.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000356819 | 0 | 13 | 37_128 | 33 | 646.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000405005 | 0 | 12 | 178_222 | 33 | 641.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000405005 | 0 | 12 | 37_128 | 33 | 641.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000519301 | 0 | 11 | 178_222 | 12 | 591.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000519301 | 0 | 11 | 37_128 | 12 | 591.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520502 | 0 | 3 | 178_222 | 0 | 297.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520502 | 0 | 3 | 37_128 | 0 | 297.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000521670 | 0 | 13 | 178_222 | 33 | 477.3333333333333 | Domain | EGF-like | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000521670 | 0 | 13 | 37_128 | 33 | 477.3333333333333 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000523079 | 0 | 11 | 178_222 | 33 | 421.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000523079 | 0 | 11 | 37_128 | 33 | 421.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000287842 | 0 | 12 | 266_640 | 33 | 638.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000341377 | 0 | 13 | 266_640 | 33 | 525.3333333333334 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000356819 | 0 | 13 | 266_640 | 33 | 646.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000405005 | 0 | 12 | 266_640 | 33 | 641.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000519301 | 0 | 11 | 20_242 | 12 | 591.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000519301 | 0 | 11 | 266_640 | 12 | 591.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520407 | 0 | 5 | 266_640 | 248 | 423.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520502 | 0 | 3 | 20_242 | 0 | 297.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520502 | 0 | 3 | 266_640 | 0 | 297.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000521670 | 0 | 13 | 266_640 | 33 | 477.3333333333333 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000523079 | 0 | 11 | 266_640 | 33 | 421.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000287842 | 0 | 12 | 243_265 | 33 | 638.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000341377 | 0 | 13 | 243_265 | 33 | 525.3333333333334 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000356819 | 0 | 13 | 243_265 | 33 | 646.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000405005 | 0 | 12 | 243_265 | 33 | 641.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000519301 | 0 | 11 | 243_265 | 12 | 591.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520502 | 0 | 3 | 243_265 | 0 | 297.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000521670 | 0 | 13 | 243_265 | 33 | 477.3333333333333 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000523079 | 0 | 11 | 243_265 | 33 | 421.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000287842 | + | 1 | 12 | 165_177 | 0 | 638.0 | Compositional bias | Note=Ser/Thr-rich |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000341377 | + | 1 | 13 | 165_177 | 0 | 525.3333333333334 | Compositional bias | Note=Ser/Thr-rich |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000356819 | + | 1 | 13 | 165_177 | 0 | 646.0 | Compositional bias | Note=Ser/Thr-rich |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000405005 | + | 1 | 12 | 165_177 | 0 | 641.0 | Compositional bias | Note=Ser/Thr-rich |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000519301 | + | 1 | 11 | 165_177 | 12 | 591.0 | Compositional bias | Note=Ser/Thr-rich |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520407 | + | 1 | 5 | 165_177 | 0 | 423.0 | Compositional bias | Note=Ser/Thr-rich |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520502 | + | 1 | 3 | 165_177 | 0 | 297.0 | Compositional bias | Note=Ser/Thr-rich |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000521670 | + | 1 | 13 | 165_177 | 0 | 477.3333333333333 | Compositional bias | Note=Ser/Thr-rich |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000523079 | + | 1 | 11 | 165_177 | 0 | 421.0 | Compositional bias | Note=Ser/Thr-rich |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000287842 | + | 1 | 12 | 178_222 | 0 | 638.0 | Domain | EGF-like |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000287842 | + | 1 | 12 | 37_128 | 0 | 638.0 | Domain | Note=Ig-like C2-type |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000341377 | + | 1 | 13 | 178_222 | 0 | 525.3333333333334 | Domain | EGF-like |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000341377 | + | 1 | 13 | 37_128 | 0 | 525.3333333333334 | Domain | Note=Ig-like C2-type |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000356819 | + | 1 | 13 | 178_222 | 0 | 646.0 | Domain | EGF-like |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000356819 | + | 1 | 13 | 37_128 | 0 | 646.0 | Domain | Note=Ig-like C2-type |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000405005 | + | 1 | 12 | 178_222 | 0 | 641.0 | Domain | EGF-like |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000405005 | + | 1 | 12 | 37_128 | 0 | 641.0 | Domain | Note=Ig-like C2-type |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000519301 | + | 1 | 11 | 178_222 | 12 | 591.0 | Domain | EGF-like |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000519301 | + | 1 | 11 | 37_128 | 12 | 591.0 | Domain | Note=Ig-like C2-type |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520407 | + | 1 | 5 | 178_222 | 0 | 423.0 | Domain | EGF-like |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520407 | + | 1 | 5 | 37_128 | 0 | 423.0 | Domain | Note=Ig-like C2-type |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520502 | + | 1 | 3 | 178_222 | 0 | 297.0 | Domain | EGF-like |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520502 | + | 1 | 3 | 37_128 | 0 | 297.0 | Domain | Note=Ig-like C2-type |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000521670 | + | 1 | 13 | 178_222 | 0 | 477.3333333333333 | Domain | EGF-like |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000521670 | + | 1 | 13 | 37_128 | 0 | 477.3333333333333 | Domain | Note=Ig-like C2-type |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000523079 | + | 1 | 11 | 178_222 | 0 | 421.0 | Domain | EGF-like |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000523079 | + | 1 | 11 | 37_128 | 0 | 421.0 | Domain | Note=Ig-like C2-type |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000287842 | + | 1 | 12 | 20_242 | 0 | 638.0 | Topological domain | Extracellular |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000287842 | + | 1 | 12 | 266_640 | 0 | 638.0 | Topological domain | Cytoplasmic |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000341377 | + | 1 | 13 | 20_242 | 0 | 525.3333333333334 | Topological domain | Extracellular |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000341377 | + | 1 | 13 | 266_640 | 0 | 525.3333333333334 | Topological domain | Cytoplasmic |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000356819 | + | 1 | 13 | 20_242 | 0 | 646.0 | Topological domain | Extracellular |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000356819 | + | 1 | 13 | 266_640 | 0 | 646.0 | Topological domain | Cytoplasmic |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000405005 | + | 1 | 12 | 20_242 | 0 | 641.0 | Topological domain | Extracellular |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000405005 | + | 1 | 12 | 266_640 | 0 | 641.0 | Topological domain | Cytoplasmic |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000519301 | + | 1 | 11 | 20_242 | 12 | 591.0 | Topological domain | Extracellular |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000519301 | + | 1 | 11 | 266_640 | 12 | 591.0 | Topological domain | Cytoplasmic |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520407 | + | 1 | 5 | 20_242 | 0 | 423.0 | Topological domain | Extracellular |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520407 | + | 1 | 5 | 266_640 | 0 | 423.0 | Topological domain | Cytoplasmic |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520502 | + | 1 | 3 | 20_242 | 0 | 297.0 | Topological domain | Extracellular |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520502 | + | 1 | 3 | 266_640 | 0 | 297.0 | Topological domain | Cytoplasmic |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000521670 | + | 1 | 13 | 20_242 | 0 | 477.3333333333333 | Topological domain | Extracellular |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000521670 | + | 1 | 13 | 266_640 | 0 | 477.3333333333333 | Topological domain | Cytoplasmic |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000523079 | + | 1 | 11 | 20_242 | 0 | 421.0 | Topological domain | Extracellular |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000523079 | + | 1 | 11 | 266_640 | 0 | 421.0 | Topological domain | Cytoplasmic |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000287842 | + | 1 | 12 | 243_265 | 0 | 638.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000341377 | + | 1 | 13 | 243_265 | 0 | 525.3333333333334 | Transmembrane | Helical%3B Note%3DInternal signal sequence |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000356819 | + | 1 | 13 | 243_265 | 0 | 646.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000405005 | + | 1 | 12 | 243_265 | 0 | 641.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000519301 | + | 1 | 11 | 243_265 | 12 | 591.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520407 | + | 1 | 5 | 243_265 | 0 | 423.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520502 | + | 1 | 3 | 243_265 | 0 | 297.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000521670 | + | 1 | 13 | 243_265 | 0 | 477.3333333333333 | Transmembrane | Helical%3B Note%3DInternal signal sequence |
| Hgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000523079 | + | 1 | 11 | 243_265 | 0 | 421.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520407 | 0 | 5 | 165_177 | 248 | 423.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520407 | 0 | 5 | 178_222 | 248 | 423.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520407 | 0 | 5 | 37_128 | 248 | 423.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000287842 | 0 | 12 | 20_242 | 33 | 638.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000341377 | 0 | 13 | 20_242 | 33 | 525.3333333333334 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000356819 | 0 | 13 | 20_242 | 33 | 646.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000405005 | 0 | 12 | 20_242 | 33 | 641.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520407 | 0 | 5 | 20_242 | 248 | 423.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000521670 | 0 | 13 | 20_242 | 33 | 477.3333333333333 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000523079 | 0 | 11 | 20_242 | 33 | 421.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr8:31496947 | chr8:32453344 | ENST00000520407 | 0 | 5 | 243_265 | 248 | 423.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence |
Top |
Fusion Gene Sequence for NRG1-NRG1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >60276_60276_1_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000287842_length(transcript)=1969nt_BP=46nt GGCAGCAGCATGGGGAAAGGACGCGCGGGCCGAGTTGGCACCACAGCCTTGCCTCCCCGATTGAAAGAGATGAAAAGCCAGGAATCGGCT GCAGGTTCCAAACTAGTCCTTCGGTGTGAAACCAGTTCTGAATACTCCTCTCTCAGATTCAAGTGGTTCAAGAATGGGAATGAATTGAAT CGAAAAAACAAACCACAAAATATCAAGATACAAAAAAAGCCAGGGAAGTCAGAACTTCGCATTAACAAAGCATCACTGGCTGATTCTGGA GAGTATATGTGCAAAGTGATCAGCAAATTAGGAAATGACAGTGCCTCTGCCAATATCACCATCGTGGAATCAAACGAGATCATCACTGGT ATGCCAGCCTCAACTGAAGGAGCATATGTGTCTTCAGAGTCTCCCATTAGAATATCAGTATCCACAGAAGGAGCAAATACTTCTTCATCT ACATCTACATCCACCACTGGGACAAGCCATCTTGTAAAATGTGCGGAGAAGGAGAAAACTTTCTGTGTGAATGGAGGGGAGTGCTTCATG GTGAAAGACCTTTCAAACCCCTCGAGATACTTGTGCAAGTGCCCAAATGAGTTTACTGGTGATCGCTGCCAAAACTACGTAATGGCCAGC TTCTACAAGGCGGAGGAGCTGTACCAGAAGAGAGTGCTGACCATAACCGGCATCTGCATCGCCCTCCTTGTGGTCGGCATCATGTGTGTG GTGGCCTACTGCAAAACCAAGAAACAGCGGAAAAAGCTGCATGACCGTCTTCGGCAGAGCCTTCGGTCTGAACGAAACAATATGATGAAC ATTGCCAATGGGCCTCACCATCCTAACCCACCCCCCGAGAATGTCCAGCTGGTGAATCAATACGTATCTAAAAACGTCATCTCCAGTGAG CATATTGTTGAGAGAGAAGCAGAGACATCCTTTTCCACCAGTCACTATACTTCCACAGCCCATCACTCCACTACTGTCACCCAGACTCCT AGCCACAGCTGGAGCAACGGACACACTGAAAGCATCCTTTCCGAAAGCCACTCTGTAATCGTGATGTCATCCGTAGAAAACAGTAGGCAC AGCAGCCCAACTGGGGGCCCAAGAGGACGTCTTAATGGCACAGGAGGCCCTCGTGAATGTAACAGCTTCCTCAGGCATGCCAGAGAAACC CCTGATTCCTACCGAGACTCTCCTCATAGTGAAAGGTATGTGTCAGCCATGACCACCCCGGCTCGTATGTCACCTGTAGATTTCCACACG CCAAGCTCCCCCAAATCGCCCCCTTCGGAAATGTCTCCACCCGTGTCCAGCATGACGGTGTCCATGCCTTCCATGGCGGTCAGCCCCTTC ATGGAAGAAGAGAGACCTCTACTTCTCGTGACACCACCAAGGCTGCGGGAGAAGAAGTTTGACCATCACCCTCAGCAGTTCAGCTCCTTC CACCACAACCCCGCGCATGACAGTAACAGCCTCCCTGCTAGCCCCTTGAGGATAGTGGAGGATGAGGAGTATGAAACGACCCAAGAGTAC GAGCCAGCCCAAGAGCCTGTTAAGAAACTCGCCAATAGCCGGCGGGCCAAAAGAACCAAGCCCAATGGCCACATTGCTAACAGATTGGAA GTGGACAGCAACACAAGCTCCCAGAGCAGTAACTCAGAGAGTGAAACAGAAGATGAAAGAGTAGGTGAAGATACGCCTTTCCTGGGCATA CAGAACCCCCTGGCAGCCAGTCTTGAGGCAACACCTGCCTTCCGCCTGGCTGACAGCAGGACTAACCCAGCAGGCCGCTTCTCGACACAG GAAGAAATCCAGGCCAGGCTGTCTAGTGTAATTGCTAACCAAGACCCTATTGCTGTATAAAACCTAAATAAACACATAGATTCACCTGTA >60276_60276_1_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000287842_length(amino acids)=616AA_BP=12 MGKGRAGRVGTTALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYM CKVISKLGNDSASANITIVESNEIITGMPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKD LSNPSRYLCKCPNEFTGDRCQNYVMASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIAN GPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSP TGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEE ERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDS -------------------------------------------------------------- >60276_60276_2_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000287845_length(transcript)=2410nt_BP=46nt GGCAGCAGCATGGGGAAAGGACGCGCGGGCCGAGTTGGCACCACAGCCTTGCCTCCCCGATTGAAAGAGATGAAAAGCCAGGAATCGGCT GCAGGTTCCAAACTAGTCCTTCGGTGTGAAACCAGTTCTGAATACTCCTCTCTCAGATTCAAGTGGTTCAAGAATGGGAATGAATTGAAT CGAAAAAACAAACCACAAAATATCAAGATACAAAAAAAGCCAGGGAAGTCAGAACTTCGCATTAACAAAGCATCACTGGCTGATTCTGGA GAGTATATGTGCAAAGTGATCAGCAAATTAGGAAATGACAGTGCCTCTGCCAATATCACCATCGTGGAATCAAACGCTACATCTACATCC ACCACTGGGACAAGCCATCTTGTAAAATGTGCGGAGAAGGAGAAAACTTTCTGTGTGAATGGAGGGGAGTGCTTCATGGTGAAAGACCTT TCAAACCCCTCGAGATACTTGTGCAAGTGCCCAAATGAGTTTACTGGTGATCGCTGCCAAAACTACGTAATGGCCAGCTTCTACAAGCAT CTTGGGATTGAATTTATGGAGGCGGAGGAGCTGTACCAGAAGAGAGTGCTGACCATAACCGGCATCTGCATCGCCCTCCTTGTGGTCGGC ATCATGTGTGTGGTGGCCTACTGCAAAACCAAGAAACAGCGGAAAAAGCTGCATGACCGTCTTCGGCAGAGCCTTCGGTCTGAACGAAAC AATATGATGAACATTGCCAATGGGCCTCACCATCCTAACCCACCCCCCGAGAATGTCCAGCTGGTGAATCAATACGTATCTAAAAACGTC ATCTCCAGTGAGCATATTGTTGAGAGAGAAGCAGAGACATCCTTTTCCACCAGTCACTATACTTCCACAGCCCATCACTCCACTACTGTC ACCCAGACTCCTAGCCACAGCTGGAGCAACGGACACACTGAAAGCATCCTTTCCGAAAGCCACTCTGTAATCGTGATGTCATCCGTAGAA AACAGTAGGCACAGCAGCCCAACTGGGGGCCCAAGAGGACGTCTTAATGGCACAGGAGGCCCTCGTGAATGTAACAGCTTCCTCAGGCAT GCCAGAGAAACCCCTGATTCCTACCGAGACTCTCCTCATAGTGAAAGGTATGTGTCAGCCATGACCACCCCGGCTCGTATGTCACCTGTA GATTTCCACACGCCAAGCTCCCCCAAATCGCCCCCTTCGGAAATGTCTCCACCCGTGTCCAGCATGACGGTGTCCATGCCTTCCATGGCG GTCAGCCCCTTCATGGAAGAAGAGAGACCTCTACTTCTCGTGACACCACCAAGGCTGCGGGAGAAGAAGTTTGACCATCACCCTCAGCAG TTCAGCTCCTTCCACCACAACCCCGCGCATGACAGTAACAGCCTCCCTGCTAGCCCCTTGAGGATAGTGGAGGATGAGGAGTATGAAACG ACCCAAGAGTACGAGCCAGCCCAAGAGCCTGTTAAGAAACTCGCCAATAGCCGGCGGGCCAAAAGAACCAAGCCCAATGGCCACATTGCT AACAGATTGGAAGTGGACAGCAACACAAGCTCCCAGAGCAGTAACTCAGAGAGTGAAACAGAAGATGAAAGAGTAGGTGAAGATACGCCT TTCCTGGGCATACAGAACCCCCTGGCAGCCAGTCTTGAGGCAACACCTGCCTTCCGCCTGGCTGACAGCAGGACTAACCCAGCAGGCCGC TTCTCGACACAGGAAGAAATCCAGGCCAGGCTGTCTAGTGTAATTGCTAACCAAGACCCTATTGCTGTATAAAACCTAAATAAACACATA GATTCACCTGTAAAACTTTATTTTATATAATAAAGTATTCCACCTTAAATTAAACAATTTATTTTATTTTAGCAGTTCTGCAAATAGAAA ACAGGAAAAAAACTTTTATAAATTAAATATATGTATGTAAAAATGTGTTATGTGCCATATGTAGCAATTTTTTACAGTATTTCAAAACGA GAAAGATATCAATGGTGCCTTTATGTTATGTTATGTCGAGAGCAAGTTTTGTACAGTTACAGTGATTGCTTTTCCACAGTATTTCTGCAA AACCTCTCATAGATTCAGTTTTTGCTGGCTTCTTGTGCATTGCATTATGATGTTGACTGGATGTATGATTTGCAAGACTTGCAACTGTCC CTCTGTTTGCTTGTAGTAGCACCCGATCAGTATGTCTTGTAATGGCACATCCATCCAGATATGCCTCTCTTGTGTATGAAGTTTTCTTTG CTTTCAGAATATGAAATGAGTTGTGTCTACTCTGCCAGCCAAAGGTTTGCCTCATTGGGCTCTGAGATAATAGTAGATCCAACAGCATGC >60276_60276_2_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000287845_length(amino acids)=590AA_BP=12 MGKGRAGRVGTTALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYM CKVISKLGNDSASANITIVESNATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGI EFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISS EHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARE TPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSS FHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLG -------------------------------------------------------------- >60276_60276_3_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000338921_length(transcript)=2521nt_BP=46nt GGCAGCAGCATGGGGAAAGGACGCGCGGGCCGAGTTGGCACCACAGCCTTGCCTCCCCGATTGAAAGAGATGAAAAGCCAGGAATCGGCT GCAGGTTCCAAACTAGTCCTTCGGTGTGAAACCAGTTCTGAATACTCCTCTCTCAGATTCAAGTGGTTCAAGAATGGGAATGAATTGAAT CGAAAAAACAAACCACAAAATATCAAGATACAAAAAAAGCCAGGGAAGTCAGAACTTCGCATTAACAAAGCATCACTGGCTGATTCTGGA GAGTATATGTGCAAAGTGATCAGCAAATTAGGAAATGACAGTGCCTCTGCCAATATCACCATCGTGGAATCAAACGAGATCATCACTGGT ATGCCAGCCTCAACTGAAGGAGCATATGTGTCTTCAGAGTCTCCCATTAGAATATCAGTATCCACAGAAGGAGCAAATACTTCTTCATCT ACATCTACATCCACCACTGGGACAAGCCATCTTGTAAAATGTGCGGAGAAGGAGAAAACTTTCTGTGTGAATGGAGGGGAGTGCTTCATG GTGAAAGACCTTTCAAACCCCTCGAGATACTTGTGCAAGTGCCAACCTGGATTCACTGGAGCAAGATGTACTGAGAATGTGCCCATGAAA GTCCAAAACCAAGAAAAGCATTCTGGAGAACCCTTTCCAGAGGCGGAGGAGCTGTACCAGAAGAGAGTGCTGACCATAACCGGCATCTGC ATCGCCCTCCTTGTGGTCGGCATCATGTGTGTGGTGGCCTACTGCAAAACCAAGAAACAGCGGAAAAAGCTGCATGACCGTCTTCGGCAG AGCCTTCGGTCTGAACGAAACAATATGATGAACATTGCCAATGGGCCTCACCATCCTAACCCACCCCCCGAGAATGTCCAGCTGGTGAAT CAATACGTATCTAAAAACGTCATCTCCAGTGAGCATATTGTTGAGAGAGAAGCAGAGACATCCTTTTCCACCAGTCACTATACTTCCACA GCCCATCACTCCACTACTGTCACCCAGACTCCTAGCCACAGCTGGAGCAACGGACACACTGAAAGCATCCTTTCCGAAAGCCACTCTGTA ATCGTGATGTCATCCGTAGAAAACAGTAGGCACAGCAGCCCAACTGGGGGCCCAAGAGGACGTCTTAATGGCACAGGAGGCCCTCGTGAA TGTAACAGCTTCCTCAGGCATGCCAGAGAAACCCCTGATTCCTACCGAGACTCTCCTCATAGTGAAAGGTATGTGTCAGCCATGACCACC CCGGCTCGTATGTCACCTGTAGATTTCCACACGCCAAGCTCCCCCAAATCGCCCCCTTCGGAAATGTCTCCACCCGTGTCCAGCATGACG GTGTCCATGCCTTCCATGGCGGTCAGCCCCTTCATGGAAGAAGAGAGACCTCTACTTCTCGTGACACCACCAAGGCTGCGGGAGAAGAAG TTTGACCATCACCCTCAGCAGTTCAGCTCCTTCCACCACAACCCCGCGCATGACAGTAACAGCCTCCCTGCTAGCCCCTTGAGGATAGTG GAGGATGAGGAGTATGAAACGACCCAAGAGTACGAGCCAGCCCAAGAGCCTGTTAAGAAACTCGCCAATAGCCGGCGGGCCAAAAGAACC AAGCCCAATGGCCACATTGCTAACAGATTGGAAGTGGACAGCAACACAAGCTCCCAGAGCAGTAACTCAGAGAGTGAAACAGAAGATGAA AGAGTAGGTGAAGATACGCCTTTCCTGGGCATACAGAACCCCCTGGCAGCCAGTCTTGAGGCAACACCTGCCTTCCGCCTGGCTGACAGC AGGACTAACCCAGCAGGCCGCTTCTCGACACAGGAAGAAATCCAGGCCAGGCTGTCTAGTGTAATTGCTAACCAAGACCCTATTGCTGTA TAAAACCTAAATAAACACATAGATTCACCTGTAAAACTTTATTTTATATAATAAAGTATTCCACCTTAAATTAAACAATTTATTTTATTT TAGCAGTTCTGCAAATAGAAAACAGGAAAAAAACTTTTATAAATTAAATATATGTATGTAAAAATGTGTTATGTGCCATATGTAGCAATT TTTTACAGTATTTCAAAACGAGAAAGATATCAATGGTGCCTTTATGTTATGTTATGTCGAGAGCAAGTTTTGTACAGTTACAGTGATTGC TTTTCCACAGTATTTCTGCAAAACCTCTCATAGATTCAGTTTTTGCTGGCTTCTTGTGCATTGCATTATGATGTTGACTGGATGTATGAT TTGCAAGACTTGCAACTGTCCCTCTGTTTGCTTGTAGTAGCACCCGATCAGTATGTCTTGTAATGGCACATCCATCCAGATATGCCTCTC TTGTGTATGAAGTTTTCTTTGCTTTCAGAATATGAAATGAGTTGTGTCTACTCTGCCAGCCAAAGGTTTGCCTCATTGGGCTCTGAGATA ATAGTAGATCCAACAGCATGCTACTATTAAATACAGCAAGAAACTGCATTAAGTAATGTTAAATATTAGGAAGAAAGTAATACTGTGATT >60276_60276_3_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000338921_length(amino acids)=627AA_BP=12 MGKGRAGRVGTTALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYM CKVISKLGNDSASANITIVESNEIITGMPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKD LSNPSRYLCKCQPGFTGARCTENVPMKVQNQEKHSGEPFPEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLR SERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVM SSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSM PSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPN -------------------------------------------------------------- >60276_60276_4_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000341377_length(transcript)=2556nt_BP=46nt GGCAGCAGCATGGGGAAAGGACGCGCGGGCCGAGTTGGCACCACAGCCTTGCCTCCCCGATTGAAAGAGATGAAAAGCCAGGAATCGGCT GCAGGTTCCAAACTAGTCCTTCGGTGTGAAACCAGTTCTGAATACTCCTCTCTCAGATTCAAGTGGTTCAAGAATGGGAATGAATTGAAT CGAAAAAACAAACCACAAAATATCAAGATACAAAAAAAGCCAGGGAAGTCAGAACTTCGCATTAACAAAGCATCACTGGCTGATTCTGGA GAGTATATGTGCAAAGTGATCAGCAAATTAGGAAATGACAGTGCCTCTGCCAATATCACCATCGTGGAATCAAACGAGATCATCACTGGT ATGCCAGCCTCAACTGAAGGAGCATATGTGTCTTCAGAGTCTCCCATTAGAATATCAGTATCCACAGAAGGAGCAAATACTTCTTCATCT ACATCTACATCCACCACTGGGACAAGCCATCTTGTAAAATGTGCGGAGAAGGAGAAAACTTTCTGTGTGAATGGAGGGGAGTGCTTCATG GTGAAAGACCTTTCAAACCCCTCGAGATACTTGTGCAAGTGCCAACCTGGATTCACTGGAGCAAGATGTACTGAGAATGTGCCCATGAAA GTCCAAAACCAAGAAAGTGCCCAAATGAGTTTACTGGTGATCGCTGCCAAAACTACGTAATGGCCAGCTTCTACAAGGCGGAGGAGCTGT ACCAGAAGAGAGTGCTGACCATAACCGGCATCTGCATCGCCCTCCTTGTGGTCGGCATCATGTGTGTGGTGGCCTACTGCAAAACCAAGA AACAGCGGAAAAAGCTGCATGACCGTCTTCGGCAGAGCCTTCGGTCTGAACGAAACAATATGATGAACATTGCCAATGGGCCTCACCATC CTAACCCACCCCCCGAGAATGTCCAGCTGGTGAATCAATACGTATCTAAAAACGTCATCTCCAGTGAGCATATTGTTGAGAGAGAAGCAG AGACATCCTTTTCCACCAGTCACTATACTTCCACAGCCCATCACTCCACTACTGTCACCCAGACTCCTAGCCACAGCTGGAGCAACGGAC ACACTGAAAGCATCCTTTCCGAAAGCCACTCTGTAATCGTGATGTCATCCGTAGAAAACAGTAGGCACAGCAGCCCAACTGGGGGCCCAA GAGGACGTCTTAATGGCACAGGAGGCCCTCGTGAATGTAACAGCTTCCTCAGGCATGCCAGAGAAACCCCTGATTCCTACCGAGACTCTC CTCATAGTGAAAGGTATGTGTCAGCCATGACCACCCCGGCTCGTATGTCACCTGTAGATTTCCACACGCCAAGCTCCCCCAAATCGCCCC CTTCGGAAATGTCTCCACCCGTGTCCAGCATGACGGTGTCCATGCCTTCCATGGCGGTCAGCCCCTTCATGGAAGAAGAGAGACCTCTAC TTCTCGTGACACCACCAAGGCTGCGGGAGAAGAAGTTTGACCATCACCCTCAGCAGTTCAGCTCCTTCCACCACAACCCCGCGCATGACA GTAACAGCCTCCCTGCTAGCCCCTTGAGGATAGTGGAGGATGAGGAGTATGAAACGACCCAAGAGTACGAGCCAGCCCAAGAGCCTGTTA AGAAACTCGCCAATAGCCGGCGGGCCAAAAGAACCAAGCCCAATGGCCACATTGCTAACAGATTGGAAGTGGACAGCAACACAAGCTCCC AGAGCAGTAACTCAGAGAGTGAAACAGAAGATGAAAGAGTAGGTGAAGATACGCCTTTCCTGGGCATACAGAACCCCCTGGCAGCCAGTC TTGAGGCAACACCTGCCTTCCGCCTGGCTGACAGCAGGACTAACCCAGCAGGCCGCTTCTCGACACAGGAAGAAATCCAGGCCAGGCTGT CTAGTGTAATTGCTAACCAAGACCCTATTGCTGTATAAAACCTAAATAAACACATAGATTCACCTGTAAAACTTTATTTTATATAATAAA GTATTCCACCTTAAATTAAACAATTTATTTTATTTTAGCAGTTCTGCAAATAGAAAACAGGAAAAAAACTTTTATAAATTAAATATATGT ATGTAAAAATGTGTTATGTGCCATATGTAGCAATTTTTTACAGTATTTCAAAACGAGAAAGATATCAATGGTGCCTTTATGTTATGTTAT GTCGAGAGCAAGTTTTGTACAGTTACAGTGATTGCTTTTCCACAGTATTTCTGCAAAACCTCTCATAGATTCAGTTTTTGCTGGCTTCTT GTGCATTGCATTATGATGTTGACTGGATGTATGATTTGCAAGACTTGCAACTGTCCCTCTGTTTGCTTGTAGTAGCACCCGATCAGTATG TCTTGTAATGGCACATCCATCCAGATATGCCTCTCTTGTGTATGAAGTTTTCTTTGCTTTCAGAATATGAAATGAGTTGTGTCTACTCTG CCAGCCAAAGGTTTGCCTCATTGGGCTCTGAGATAATAGTAGATCCAACAGCATGCTACTATTAAATACAGCAAGAAACTGCATTAAGTA >60276_60276_4_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000341377_length(amino acids)=412AA_BP= MASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >60276_60276_5_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000356819_length(transcript)=2512nt_BP=46nt GGCAGCAGCATGGGGAAAGGACGCGCGGGCCGAGTTGGCACCACAGCCTTGCCTCCCCGATTGAAAGAGATGAAAAGCCAGGAATCGGCT GCAGGTTCCAAACTAGTCCTTCGGTGTGAAACCAGTTCTGAATACTCCTCTCTCAGATTCAAGTGGTTCAAGAATGGGAATGAATTGAAT CGAAAAAACAAACCACAAAATATCAAGATACAAAAAAAGCCAGGGAAGTCAGAACTTCGCATTAACAAAGCATCACTGGCTGATTCTGGA GAGTATATGTGCAAAGTGATCAGCAAATTAGGAAATGACAGTGCCTCTGCCAATATCACCATCGTGGAATCAAACGAGATCATCACTGGT ATGCCAGCCTCAACTGAAGGAGCATATGTGTCTTCAGAGTCTCCCATTAGAATATCAGTATCCACAGAAGGAGCAAATACTTCTTCATCT ACATCTACATCCACCACTGGGACAAGCCATCTTGTAAAATGTGCGGAGAAGGAGAAAACTTTCTGTGTGAATGGAGGGGAGTGCTTCATG GTGAAAGACCTTTCAAACCCCTCGAGATACTTGTGCAAGTGCCCAAATGAGTTTACTGGTGATCGCTGCCAAAACTACGTAATGGCCAGC TTCTACAAGCATCTTGGGATTGAATTTATGGAGGCGGAGGAGCTGTACCAGAAGAGAGTGCTGACCATAACCGGCATCTGCATCGCCCTC CTTGTGGTCGGCATCATGTGTGTGGTGGCCTACTGCAAAACCAAGAAACAGCGGAAAAAGCTGCATGACCGTCTTCGGCAGAGCCTTCGG TCTGAACGAAACAATATGATGAACATTGCCAATGGGCCTCACCATCCTAACCCACCCCCCGAGAATGTCCAGCTGGTGAATCAATACGTA TCTAAAAACGTCATCTCCAGTGAGCATATTGTTGAGAGAGAAGCAGAGACATCCTTTTCCACCAGTCACTATACTTCCACAGCCCATCAC TCCACTACTGTCACCCAGACTCCTAGCCACAGCTGGAGCAACGGACACACTGAAAGCATCCTTTCCGAAAGCCACTCTGTAATCGTGATG TCATCCGTAGAAAACAGTAGGCACAGCAGCCCAACTGGGGGCCCAAGAGGACGTCTTAATGGCACAGGAGGCCCTCGTGAATGTAACAGC TTCCTCAGGCATGCCAGAGAAACCCCTGATTCCTACCGAGACTCTCCTCATAGTGAAAGGTATGTGTCAGCCATGACCACCCCGGCTCGT ATGTCACCTGTAGATTTCCACACGCCAAGCTCCCCCAAATCGCCCCCTTCGGAAATGTCTCCACCCGTGTCCAGCATGACGGTGTCCATG CCTTCCATGGCGGTCAGCCCCTTCATGGAAGAAGAGAGACCTCTACTTCTCGTGACACCACCAAGGCTGCGGGAGAAGAAGTTTGACCAT CACCCTCAGCAGTTCAGCTCCTTCCACCACAACCCCGCGCATGACAGTAACAGCCTCCCTGCTAGCCCCTTGAGGATAGTGGAGGATGAG GAGTATGAAACGACCCAAGAGTACGAGCCAGCCCAAGAGCCTGTTAAGAAACTCGCCAATAGCCGGCGGGCCAAAAGAACCAAGCCCAAT GGCCACATTGCTAACAGATTGGAAGTGGACAGCAACACAAGCTCCCAGAGCAGTAACTCAGAGAGTGAAACAGAAGATGAAAGAGTAGGT GAAGATACGCCTTTCCTGGGCATACAGAACCCCCTGGCAGCCAGTCTTGAGGCAACACCTGCCTTCCGCCTGGCTGACAGCAGGACTAAC CCAGCAGGCCGCTTCTCGACACAGGAAGAAATCCAGGCCAGGCTGTCTAGTGTAATTGCTAACCAAGACCCTATTGCTGTATAAAACCTA AATAAACACATAGATTCACCTGTAAAACTTTATTTTATATAATAAAGTATTCCACCTTAAATTAAACAATTTATTTTATTTTAGCAGTTC TGCAAATAGAAAACAGGAAAAAAACTTTTATAAATTAAATATATGTATGTAAAAATGTGTTATGTGCCATATGTAGCAATTTTTTACAGT ATTTCAAAACGAGAAAGATATCAATGGTGCCTTTATGTTATGTTATGTCGAGAGCAAGTTTTGTACAGTTACAGTGATTGCTTTTCCACA GTATTTCTGCAAAACCTCTCATAGATTCAGTTTTTGCTGGCTTCTTGTGCATTGCATTATGATGTTGACTGGATGTATGATTTGCAAGAC TTGCAACTGTCCCTCTGTTTGCTTGTAGTAGCACCCGATCAGTATGTCTTGTAATGGCACATCCATCCAGATATGCCTCTCTTGTGTATG AAGTTTTCTTTGCTTTCAGAATATGAAATGAGTTGTGTCTACTCTGCCAGCCAAAGGTTTGCCTCATTGGGCTCTGAGATAATAGTAGAT >60276_60276_5_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000356819_length(amino acids)=624AA_BP=12 MGKGRAGRVGTTALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYM CKVISKLGNDSASANITIVESNEIITGMPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKD LSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSER NNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSV ENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSM AVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHI -------------------------------------------------------------- >60276_60276_6_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000405005_length(transcript)=1938nt_BP=46nt GGCAGCAGCATGGGGAAAGGACGCGCGGGCCGAGTTGGCACCACAGCCTTGCCTCCCCGATTGAAAGAGATGAAAAGCCAGGAATCGGCT GCAGGTTCCAAACTAGTCCTTCGGTGTGAAACCAGTTCTGAATACTCCTCTCTCAGATTCAAGTGGTTCAAGAATGGGAATGAATTGAAT CGAAAAAACAAACCACAAAATATCAAGATACAAAAAAAGCCAGGGAAGTCAGAACTTCGCATTAACAAAGCATCACTGGCTGATTCTGGA GAGTATATGTGCAAAGTGATCAGCAAATTAGGAAATGACAGTGCCTCTGCCAATATCACCATCGTGGAATCAAACGAGATCATCACTGGT ATGCCAGCCTCAACTGAAGGAGCATATGTGTCTTCAGAGTCTCCCATTAGAATATCAGTATCCACAGAAGGAGCAAATACTTCTTCATCT ACATCTACATCCACCACTGGGACAAGCCATCTTGTAAAATGTGCGGAGAAGGAGAAAACTTTCTGTGTGAATGGAGGGGAGTGCTTCATG GTGAAAGACCTTTCAAACCCCTCGAGATACTTGTGCAAGTGCCAACCTGGATTCACTGGAGCAAGATGTACTGAGAATGTGCCCATGAAA GTCCAAAACCAAGAAAAGGCGGAGGAGCTGTACCAGAAGAGAGTGCTGACCATAACCGGCATCTGCATCGCCCTCCTTGTGGTCGGCATC ATGTGTGTGGTGGCCTACTGCAAAACCAAGAAACAGCGGAAAAAGCTGCATGACCGTCTTCGGCAGAGCCTTCGGTCTGAACGAAACAAT ATGATGAACATTGCCAATGGGCCTCACCATCCTAACCCACCCCCCGAGAATGTCCAGCTGGTGAATCAATACGTATCTAAAAACGTCATC TCCAGTGAGCATATTGTTGAGAGAGAAGCAGAGACATCCTTTTCCACCAGTCACTATACTTCCACAGCCCATCACTCCACTACTGTCACC CAGACTCCTAGCCACAGCTGGAGCAACGGACACACTGAAAGCATCCTTTCCGAAAGCCACTCTGTAATCGTGATGTCATCCGTAGAAAAC AGTAGGCACAGCAGCCCAACTGGGGGCCCAAGAGGACGTCTTAATGGCACAGGAGGCCCTCGTGAATGTAACAGCTTCCTCAGGCATGCC AGAGAAACCCCTGATTCCTACCGAGACTCTCCTCATAGTGAAAGGTATGTGTCAGCCATGACCACCCCGGCTCGTATGTCACCTGTAGAT TTCCACACGCCAAGCTCCCCCAAATCGCCCCCTTCGGAAATGTCTCCACCCGTGTCCAGCATGACGGTGTCCATGCCTTCCATGGCGGTC AGCCCCTTCATGGAAGAAGAGAGACCTCTACTTCTCGTGACACCACCAAGGCTGCGGGAGAAGAAGTTTGACCATCACCCTCAGCAGTTC AGCTCCTTCCACCACAACCCCGCGCATGACAGTAACAGCCTCCCTGCTAGCCCCTTGAGGATAGTGGAGGATGAGGAGTATGAAACGACC CAAGAGTACGAGCCAGCCCAAGAGCCTGTTAAGAAACTCGCCAATAGCCGGCGGGCCAAAAGAACCAAGCCCAATGGCCACATTGCTAAC AGATTGGAAGTGGACAGCAACACAAGCTCCCAGAGCAGTAACTCAGAGAGTGAAACAGAAGATGAAAGAGTAGGTGAAGATACGCCTTTC CTGGGCATACAGAACCCCCTGGCAGCCAGTCTTGAGGCAACACCTGCCTTCCGCCTGGCTGACAGCAGGACTAACCCAGCAGGCCGCTTC TCGACACAGGAAGAAATCCAGGCCAGGCTGTCTAGTGTAATTGCTAACCAAGACCCTATTGCTGTATAAAACCTAAATAAACACATAGAT >60276_60276_6_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000405005_length(amino acids)=619AA_BP=12 MGKGRAGRVGTTALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYM CKVISKLGNDSASANITIVESNEIITGMPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKD LSNPSRYLCKCQPGFTGARCTENVPMKVQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMN IANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRH SSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPF MEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLE -------------------------------------------------------------- >60276_60276_7_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000519301_length(transcript)=1785nt_BP=46nt GGCAGCAGCATGGGGAAAGGACGCGCGGGCCGAGTTGGCACCACAGCCTTGCCTCCCCGATTGAAAGAGATGAAAAGCCAGGAATCGGCT GCAGGTTCCAAACTAGTCCTTCGGTGTGAAACCAGTTCTGAATACTCCTCTCTCAGATTCAAGTGGTTCAAGAATGGGAATGAATTGAAT CGAAAAAACAAACCACAAAATATCAAGATACAAAAAAAGCCAGGGAAGTCAGAACTTCGCATTAACAAAGCATCACTGGCTGATTCTGGA GAGTATATGTGCAAAGTGATCAGCAAATTAGGAAATGACAGTGCCTCTGCCAATATCACCATCGTGGAATCAAACGCTACATCTACATCC ACCACTGGGACAAGCCATCTTGTAAAATGTGCGGAGAAGGAGAAAACTTTCTGTGTGAATGGAGGGGAGTGCTTCATGGTGAAAGACCTT TCAAACCCCTCGAGATACTTGTGCAAGTGCCCAAATGAGTTTACTGGTGATCGCTGCCAAAACTACGTAATGGCCAGCTTCTACAAGCAT CTTGGGATTGAATTTATGGAGGCGGAGGAGCTGTACCAGAAGAGAGTGCTGACCATAACCGGCATCTGCATCGCCCTCCTTGTGGTCGGC ATCATGTGTGTGGTGGCCTACTGCAAAACCAAGAAACAGCGGAAAAAGCTGCATGACCGTCTTCGGCAGAGCCTTCGGTCTGAACGAAAC AATATGATGAACATTGCCAATGGGCCTCACCATCCTAACCCACCCCCCGAGAATGTCCAGCTGGTGAATCAATACGTATCTAAAAACGTC ATCTCCAGTGAGCATATTGTTGAGAGAGAAGCAGAGACATCCTTTTCCACCAGTCACTATACTTCCACAGCCCATCACTCCACTACTGTC ACCCAGACTCCTAGCCACAGCTGGAGCAACGGACACACTGAAAGCATCCTTTCCGAAAGCCACTCTGTAATCGTGATGTCATCCGTAGAA AACAGTAGGCACAGCAGCCCAACTGGGGGCCCAAGAGGACGTCTTAATGGCACAGGAGGCCCTCGTGAATGTAACAGCTTCCTCAGGCAT GCCAGAGAAACCCCTGATTCCTACCGAGACTCTCCTCATAGTGAAAGGTATGTGTCAGCCATGACCACCCCGGCTCGTATGTCACCTGTA GATTTCCACACGCCAAGCTCCCCCAAATCGCCCCCTTCGGAAATGTCTCCACCCGTGTCCAGCATGACGGTGTCCATGCCTTCCATGGCG GTCAGCCCCTTCATGGAAGAAGAGAGACCTCTACTTCTCGTGACACCACCAAGGCTGCGGGAGAAGAAGTTTGACCATCACCCTCAGCAG TTCAGCTCCTTCCACCACAACCCCGCGCATGACAGTAACAGCCTCCCTGCTAGCCCCTTGAGGATAGTGGAGGATGAGGAGTATGAAACG ACCCAAGAGTACGAGCCAGCCCAAGAGCCTGTTAAGAAACTCGCCAATAGCCGGCGGGCCAAAAGAACCAAGCCCAATGGCCACATTGCT AACAGATTGGAAGTGGACAGCAACACAAGCTCCCAGAGCAGTAACTCAGAGAGTGAAACAGAAGATGAAAGAGTAGGTGAAGATACGCCT TTCCTGGGCATACAGAACCCCCTGGCAGCCAGTCTTGAGGCAACACCTGCCTTCCGCCTGGCTGACAGCAGGACTAACCCAGCAGGCCGC >60276_60276_7_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000519301_length(amino acids)=590AA_BP=12 MGKGRAGRVGTTALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYM CKVISKLGNDSASANITIVESNATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGI EFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISS EHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARE TPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSS FHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLG -------------------------------------------------------------- >60276_60276_8_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000520407_length(transcript)=1026nt_BP=46nt GGCAGCAGCATGGGGAAAGGACGCGCGGGCCGAGTTGGCACCACAGCCTTGCCTCCCCGATTGAAAGAGATGAAAAGCCAGGAATCGGCT GCAGGTTCCAAACTAGTCCTTCGGTGTGAAACCAGTTCTGAATACTCCTCTCTCAGATTCAAGTGGTTCAAGAATGGGAATGAATTGAAT CGAAAAAACAAACCACAAAATATCAAGATACAAAAAAAGCCAGGGAAGTCAGAACTTCGCATTAACAAAGCATCACTGGCTGATTCTGGA GAGTATATGTGCAAAGTGATCAGCAAATTAGGAAATGACAGTGCCTCTGCCAATATCACCATCGTGGAATCAAACGCTACATCTACATCC ACCACTGGGACAAGCCATCTTGTAAAATGTGCGGAGAAGGAGAAAACTTTCTGTGTGAATGGAGGGGAGTGCTTCATGGTGAAAGACCTT TCAAACCCCTCGAGATACTTGTGCAAGTGCCCAAATGAGTTTACTGGTGATCGCTGCCAAAACTACGTAATGGCCAGCTTCTACAGTACG TCCACTCCCTTTCTGTCTCTGCCTGAATAGGAGCATGCTCAGTTGGTGCTGCTTTCTTGTTGCTGCATCTCCCCTCAGATTCCACCTAGA GCTAGATGTGTCTTACCAGATCTAATATTGACTGCCTCTGCCTGTCGCATGAGAACATTAACAAAAGCAATTGTATTACTTCCTCTGTTC GCGACTAGTTGGCTCTGAGATACTAATAGGTGTGTGAGGCTCCGGATGTTTCTGGAATTGATATTGAATGATGTGATACAAATTGATAGT CAATATCAAGCAGTGAAATATGATAATAAAGGCATTTCAAAGTCTCACTTTTATTGATAAAATAAAAATCATTCTACTGAACAGTCCATC TTCTTTATACAATGACCACATCCTGAAAAGGGTGTTGCTAAGCTGTAACCGATATGCACTTGAAATGATGGTAAGTTAATTTTGATTCAG >60276_60276_8_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000520407_length(amino acids)=186AA_BP=12 MGKGRAGRVGTTALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYM CKVISKLGNDSASANITIVESNATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYSTSTP -------------------------------------------------------------- >60276_60276_9_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000521670_length(transcript)=1607nt_BP=46nt GGCAGCAGCATGGGGAAAGGACGCGCGGGCCGAGTTGGCACCACAGCCTTGCCTCCCCGATTGAAAGAGATGAAAAGCCAGGAATCGGCT GCAGGTTCCAAACTAGTCCTTCGGTGTGAAACCAGTTCTGAATACTCCTCTCTCAGATTCAAGTGGTTCAAGAATGGGAATGAATTGAAT CGAAAAAACAAACCACAAAATATCAAGATACAAAAAAAGCCAGGGAAGTCAGAACTTCGCATTAACAAAGCATCACTGGCTGATTCTGGA GAGTATATGTGCAAAGTGATCAGCAAATTAGGAAATGACAGTGCCTCTGCCAATATCACCATCGTGGAATCAAACGAGATCATCACTGGT ATGCCAGCCTCAACTGAAGGAGCATATGTGTCTTCAGAGTCTCCCATTAGAATATCAGTATCCACAGAAGGAGCAAATACTTCTTCATCT ACATCTACATCCACCACTGGGACAAGCCATCTTGTAAAATGTGCGGAGAAGGAGAAAACTTTCTGTGTGAATGGAGGGGAGTGCTTCATG GTGAAAGACCTTTCAAACCCCTCGAGATACTTGTGCAAGTGCCAACCTGGATTCACTGGAGCAAGATGTACTGAGAATGTGCCCATGAAA GTCCAAAACCAAGAAAAGGCGGAGGAGCTGTACCAGAAGAGAGTGCTGACCATAACCGGCATCTGCATCGCCCTCCTTGTGGTCGGCATC ATGTGTGTGGTGGCCTACTGCAAAACCAAGAAACAGCGGAAAAAGCTGCATGACCGTCTTCGGCAGAGCCTTCGGTCTGAACGAAACAAT ATGATGAACATTGCCAATGGGCCTCACCATCCTAACCCACCCCCCGAGAATGTCCAGCTGGTGAATCAATACGTATCTAAAAACGTCATC TCCAGTGAGCATATTGTTGAGAGAGAAGCAGAGACATCCTTTTCCACCAGTCACTATACTTCCACAGCCCATCACTCCACTACTGTCACC CAGACTCCTAGCCACAGCTGGAGCAACGGACACACTGAAAGCATCCTTTCCGAAAGCCACTCTGTAATCGTGATGTCATCCGTAGAAAAC AGTAGGCACAGCAGCCCAACTGGGGGCCCAAGAGGACGTCTTAATGGCACAGGAGGCCCTCGTGAATGTAACAGCTTCCTCAGGCATGCC AGAGAAACCCCTGATTCCTACCGAGACTCTCCTCATAGTGAAAGACATAACCTTATAGCTGAGCTAAGGAGAAACAAGGCACACAGATCC AAATGCATGCAGATCCAGCTATCAGCAACTCATCTTAGATCTTCTTCCATTCCCCATTTGGGCTTCATTCTCTAAGACCCCTTGGCCTTT AGGAAGGTATGTGTCAGCCATGACCACCCCGGCTCGTATGTCACCTGTAGATTTCCACACGCCAAGCTCCCCCAAATCGCCCCCTTCGGA AATGTCTCCACCCGTGTCCAGCATGACGGTGTCCATGCCTTCCATGGCGGTCAGCCCCTTCATGGAAGAAGAGAGACCTCTACTTCTCGT >60276_60276_9_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000521670_length(amino acids)=441AA_BP=12 MGKGRAGRVGTTALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYM CKVISKLGNDSASANITIVESNEIITGMPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKD LSNPSRYLCKCQPGFTGARCTENVPMKVQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMN IANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRH -------------------------------------------------------------- >60276_60276_10_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000523079_length(transcript)=1788nt_BP=46nt GGCAGCAGCATGGGGAAAGGACGCGCGGGCCGAGTTGGCACCACAGCCTTGCCTCCCCGATTGAAAGAGATGAAAAGCCAGGAATCGGCT GCAGGTTCCAAACTAGTCCTTCGGTGTGAAACCAGTTCTGAATACTCCTCTCTCAGATTCAAGTGGTTCAAGAATGGGAATGAATTGAAT CGAAAAAACAAACCACAAAATATCAAGATACAAAAAAAGCCAGGGAAGTCAGAACTTCGCATTAACAAAGCATCACTGGCTGATTCTGGA GAGTATATGTGCAAAGTGATCAGCAAATTAGGAAATGACAGTGCCTCTGCCAATATCACCATCGTGGAATCAAACGAGATCATCACTGGT ATGCCAGCCTCAACTGAAGGAGCATATGTGTCTTCAGAGTCTCCCATTAGAATATCAGTATCCACAGAAGGAGCAAATACTTCTTCATCT ACATCTACATCCACCACTGGGACAAGCCATCTTGTAAAATGTGCGGAGAAGGAGAAAACTTTCTGTGTGAATGGAGGGGAGTGCTTCATG GTGAAAGACCTTTCAAACCCCTCGAGATACTTGTGCAAGTGCCCAAATGAGTTTACTGGTGATCGCTGCCAAAACTACGTAATGGCCAGC TTCTACAAGGCGGAGGAGCTGTACCAGAAGAGAGTGCTGACCATAACCGGCATCTGCATCGCCCTCCTTGTGGTCGGCATCATGTGTGTG GTGGCCTACTGCAAAACCAAGAAACAGCGGAAAAAGCTGCATGACCGTCTTCGGCAGAGCCTTCGGTCTGAACGAAACAATATGATGAAC ATTGCCAATGGGCCTCACCATCCTAACCCACCCCCCGAGAATGTCCAGCTGGTGAATCAATACGTATCTAAAAACGTCATCTCCAGTGAG CATATTGTTGAGAGAGAAGCAGAGACATCCTTTTCCACCAGTCACTATACTTCCACAGCCCATCACTCCACTACTGTCACCCAGACTCCT AGCCACAGCTGGAGCAACGGACACACTGAAAGCATCCTTTCCGAAAGCCACTCTGTAATCGTGATGTCATCCGTAGAAAACAGTAGGCAC AGCAGCCCAACTGGGGGCCCAAGAGGACGTCTTAATGGCACAGGAGGCCCTCGTGAATGTAACAGCTTCCTCAGGCATGCCAGAGAAACC CCTGATTCCTACCGAGACTCTCCTCATAGTGAAAGGTAAAACCGAAGGGCAAAGCTACTGCAGAGGAGAAACTCAGTCAGAGAATCCCTG TGAGCACCTGCGGTCTCACCTCAGGAAATCTACTCTAATCAGAATAAGGGGCGGCAGTTACCTGTTCTAGGAGTGCTCCTAGTTGATGAA GTCATCTCTTTGTTTGACGGAACTTATTTCTTCTGAGCTTCTCTCGTCGTCCCAGTGACTGACAGGCAACAGACTCTTAAAGAGCTGGGA TGCTTTGATGCGGAAGGTGCAGCACATGGAGTTTCCAGCTCTGGCCATGGGCTCAGACCCACTCGGGGTCTCAGTGTCCTCAGTTGTAAC ATTAGAGAGATGGCATCAATGCTTGATAAGGACCCTTCTATAATTCCAATTGCCAGTTATCCAAACTCTGATTCGGTGGTCGAGCTGGCC TCGTGTTCTTATCTGCTAACCCTGTCTTACCTTCCAGCCTCAGTTAAGTCAAATCAAGGGCTATGTCATTGCTGAATGTCATGGGGGGCA >60276_60276_10_NRG1-NRG1_NRG1_chr8_31496947_ENST00000519301_NRG1_chr8_32453344_ENST00000523079_length(amino acids)=399AA_BP=12 MGKGRAGRVGTTALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYM CKVISKLGNDSASANITIVESNEIITGMPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKD LSNPSRYLCKCPNEFTGDRCQNYVMASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIAN GPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for NRG1-NRG1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for NRG1-NRG1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for NRG1-NRG1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
