
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:NUF2-TIMM17A (FusionGDB2 ID:60877) |
Fusion Gene Summary for NUF2-TIMM17A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: NUF2-TIMM17A | Fusion gene ID: 60877 | Hgene | Tgene | Gene symbol | NUF2 | TIMM17A | Gene ID | 83540 | 10440 |
| Gene name | NUF2 component of NDC80 kinetochore complex | translocase of inner mitochondrial membrane 17A | |
| Synonyms | CDCA1|CT106|NUF2R | TIM17|TIM17A | |
| Cytomap | 1q23.3 | 1q32.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | kinetochore protein Nuf2NDC80 kinetochore complex component NUF2NUF2, NDC80 kinetochore complex component, homologcancer/testis antigen 106cell division cycle associated 1cell division cycle-associated protein 1hNuf2hNuf2RhsNuf2 | mitochondrial import inner membrane translocase subunit Tim17-Ainner membrane preprotein translocase Tim17amitochondrial inner membrane translocasepreprotein translocasetranslocase of inner mitochondrial membrane 17 homolog A | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000271452, ENST00000367900, ENST00000524800, ENST00000490881, | ENST00000482943, ENST00000367287, | |
| Fusion gene scores | * DoF score | 3 X 3 X 3=27 | 5 X 5 X 6=150 |
| # samples | 4 | 7 | |
| ** MAII score | log2(4/27*10)=0.567040592723894 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(7/150*10)=-1.09953567355091 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: NUF2 [Title/Abstract] AND TIMM17A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | NUF2(163317728)-TIMM17A(201926409), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across NUF2 (5'-gene) Fusion gene breakpoints across NUF2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
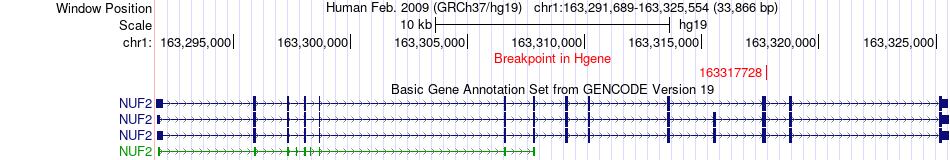 |
 Fusion gene breakpoints across TIMM17A (3'-gene) Fusion gene breakpoints across TIMM17A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
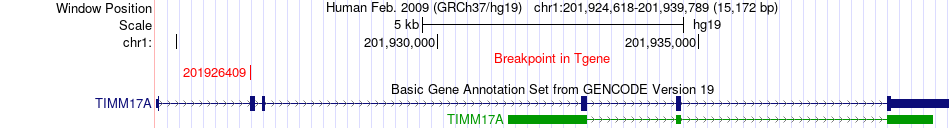 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-78-7159-01A | NUF2 | chr1 | 163317728 | - | TIMM17A | chr1 | 201926409 | + |
| ChimerDB4 | LUAD | TCGA-78-7159-01A | NUF2 | chr1 | 163317728 | + | TIMM17A | chr1 | 201926409 | + |
Top |
Fusion Gene ORF analysis for NUF2-TIMM17A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000271452 | ENST00000482943 | NUF2 | chr1 | 163317728 | + | TIMM17A | chr1 | 201926409 | + |
| 5CDS-intron | ENST00000367900 | ENST00000482943 | NUF2 | chr1 | 163317728 | + | TIMM17A | chr1 | 201926409 | + |
| 5CDS-intron | ENST00000524800 | ENST00000482943 | NUF2 | chr1 | 163317728 | + | TIMM17A | chr1 | 201926409 | + |
| In-frame | ENST00000271452 | ENST00000367287 | NUF2 | chr1 | 163317728 | + | TIMM17A | chr1 | 201926409 | + |
| In-frame | ENST00000367900 | ENST00000367287 | NUF2 | chr1 | 163317728 | + | TIMM17A | chr1 | 201926409 | + |
| In-frame | ENST00000524800 | ENST00000367287 | NUF2 | chr1 | 163317728 | + | TIMM17A | chr1 | 201926409 | + |
| intron-3CDS | ENST00000490881 | ENST00000367287 | NUF2 | chr1 | 163317728 | + | TIMM17A | chr1 | 201926409 | + |
| intron-intron | ENST00000490881 | ENST00000482943 | NUF2 | chr1 | 163317728 | + | TIMM17A | chr1 | 201926409 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000524800 | NUF2 | chr1 | 163317728 | + | ENST00000367287 | TIMM17A | chr1 | 201926409 | + | 2913 | 1316 | 333 | 1805 | 490 |
| ENST00000367900 | NUF2 | chr1 | 163317728 | + | ENST00000367287 | TIMM17A | chr1 | 201926409 | + | 2873 | 1276 | 152 | 1765 | 537 |
| ENST00000271452 | NUF2 | chr1 | 163317728 | + | ENST00000367287 | TIMM17A | chr1 | 201926409 | + | 3000 | 1403 | 279 | 1892 | 537 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000524800 | ENST00000367287 | NUF2 | chr1 | 163317728 | + | TIMM17A | chr1 | 201926409 | + | 0.000238148 | 0.9997619 |
| ENST00000367900 | ENST00000367287 | NUF2 | chr1 | 163317728 | + | TIMM17A | chr1 | 201926409 | + | 0.000406476 | 0.99959356 |
| ENST00000271452 | ENST00000367287 | NUF2 | chr1 | 163317728 | + | TIMM17A | chr1 | 201926409 | + | 0.000424141 | 0.9995759 |
Top |
Fusion Genomic Features for NUF2-TIMM17A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| NUF2 | chr1 | 163317728 | + | TIMM17A | chr1 | 201926408 | + | 1.08E-05 | 0.99998915 |
| NUF2 | chr1 | 163317728 | + | TIMM17A | chr1 | 201926408 | + | 1.08E-05 | 0.99998915 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
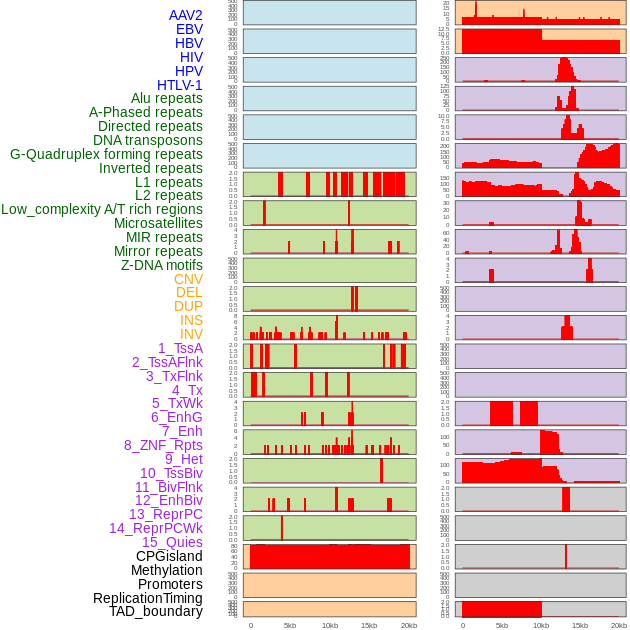 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
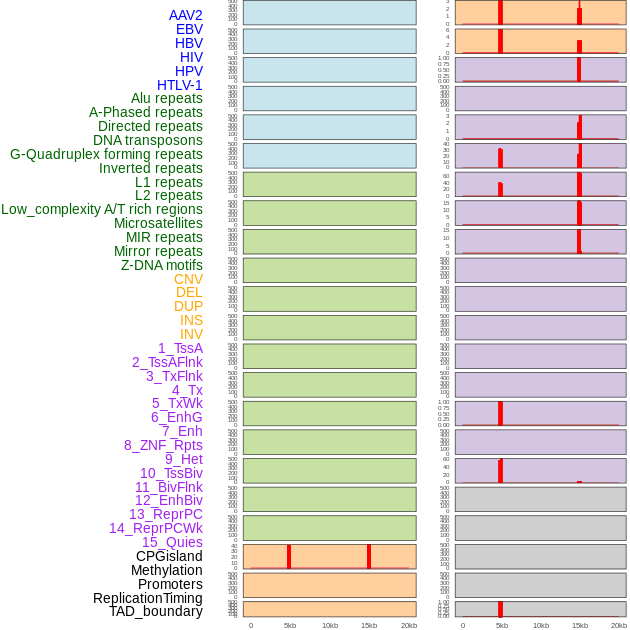 |
Top |
Fusion Protein Features for NUF2-TIMM17A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:163317728/chr1:201926409) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NUF2 | chr1:163317728 | chr1:201926409 | ENST00000271452 | + | 12 | 14 | 147_345 | 374 | 465.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NUF2 | chr1:163317728 | chr1:201926409 | ENST00000367900 | + | 12 | 14 | 147_345 | 374 | 465.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Tgene | TIMM17A | chr1:163317728 | chr1:201926409 | ENST00000367287 | 0 | 6 | 113_133 | 8 | 172.0 | Transmembrane | Helical | |
| Tgene | TIMM17A | chr1:163317728 | chr1:201926409 | ENST00000367287 | 0 | 6 | 17_37 | 8 | 172.0 | Transmembrane | Helical | |
| Tgene | TIMM17A | chr1:163317728 | chr1:201926409 | ENST00000367287 | 0 | 6 | 63_77 | 8 | 172.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NUF2 | chr1:163317728 | chr1:201926409 | ENST00000271452 | + | 12 | 14 | 389_459 | 374 | 465.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | NUF2 | chr1:163317728 | chr1:201926409 | ENST00000367900 | + | 12 | 14 | 389_459 | 374 | 465.0 | Coiled coil | Ontology_term=ECO:0000255 |
Top |
Fusion Gene Sequence for NUF2-TIMM17A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >60877_60877_1_NUF2-TIMM17A_NUF2_chr1_163317728_ENST00000271452_TIMM17A_chr1_201926409_ENST00000367287_length(transcript)=3000nt_BP=1403nt AGTAGGAGGCGGCAAGTTTGAAAAGTGATGACGGTTGACGTTTGCTGATTTTTGACTTTGCTTGTAGCTGCTCCCCGAACTCGCCGTCTT CCTGTCGGCGGCCGGCACTGTAGGTGAGCGCGAGAGGACGGAGGAAGGAAGCCTGCAGACAGACGCCTTCTCCATCCCAAGGCGCGGGCA GGTGCCGGGACGCTGGGCCTGGCGGTGTTTTCGTCGTGCTCAGCGGTGGGAGGAGGCGGAAGAAACCAGAGCCTGGGAGATTAACAGGAA ACTTCCAAGATGGAAACTTTGTCTTTCCCCAGATATAATGTAGCTGAGATTGTGATTCATATTCGCAATAAGATCTTAACAGGAGCTGAT GGTAAAAACCTCACCAAGAATGATCTTTATCCAAATCCAAAGCCTGAAGTCTTGCACATGATCTACATGAGAGCCTTACAAATAGTATAT GGAATTCGACTGGAACATTTTTACATGATGCCAGTGAACTCTGAAGTCATGTATCCACATTTAATGGAAGGCTTCTTACCATTCAGCAAT TTAGTTACTCATCTGGACTCATTTTTGCCTATCTGCCGGGTGAATGACTTTGAGACTGCTGATATTCTATGTCCAAAAGCAAAACGGACA AGTCGGTTTTTAAGTGGCATTATCAACTTTATTCACTTCAGAGAAGCATGCCGTGAAACGTATATGGAATTTCTTTGGCAATATAAATCC TCTGCGGACAAAATGCAACAGTTAAACGCCGCACACCAGGAGGCATTAATGAAACTGGAGAGACTTGATTCTGTTCCAGTTGAAGAGCAA GAAGAGTTCAAGCAGCTTTCAGATGGAATTCAGGAGCTACAACAATCACTAAATCAGGATTTTCATCAAAAAACGATAGTGCTGCAAGAG GGAAATTCCCAAAAGAAGTCAAATATTTCAGAGAAAACCAAGCGTTTGAATGAACTAAAATTGTCGGTGGTTTCTTTGAAAGAAATACAA GAGAGTTTGAAAACAAAAATTGTGGATTCTCCAGAGAAGTTAAAGAATTATAAAGAAAAAATGAAAGATACGGTCCAGAAGCTTAAAAAT GCCAGACAAGAAGTGGTGGAGAAATATGAAATCTATGGAGACTCAGTTGACTGCCTGCCTTCATGTCAGTTGGAAGTGCAGTTATATCAA AAGAAAATACAGGACCTTTCAGATAATAGGGAAAAATTAGCCAGTATCTTAAAGGAGAGCCTGAACTTGGAGGACCAAATTGAGAGTGAT GAGTCAGAACTGAAGAAATTGAAGACTGAAGAAAATTCGTTCAAAAGACTGATGATTGTGAAGAAGGAAAAACTTGCCACAGCACAATTC AAAATAAATAAGAAGCATGAAGATGTTAAGCAATACAAACGCACAGTAATTGACCCATGGCGAATTGTGGATGACTGTGGTGGGGCCTTT ACGATGGGTACCATTGGTGGTGGTATCTTTCAAGCAATCAAAGGTTTTCGCAATTCTCCAGTGGGAGTAAACCACAGACTACGAGGGAGT TTGACAGCTATTAAAACCAGGGCTCCACAGTTAGGAGGTAGCTTTGCAGTTTGGGGAGGGCTGTTTTCCATGATTGACTGTAGTATGGTT CAAGTCAGAGGAAAGGAAGATCCCTGGAACTCCATCACAAGTGGTGCCTTAACGGGAGCCATACTGGCAGCAAGAAATGGACCAGTGGCC ATGGTTGGGTCAGCCGCAATGGGTGGCATTCTCCTAGCTTTAATTGAAGGAGCTGGTATCTTGTTGACAAGATTTGCCTCTGCACAGTTT CCCAATGGTCCTCAGTTTGCAGAAGACCCCTCCCAGTTGCCTTCAACTCAGTTACCTTCCTCACCTTTTGGAGACTATCGACAATATCAG TAGGACTTCTTTCCTAGGATTTCTTTAACAGAACGAGTTGTGGTTCGAGAAGGATTTCAGAAGATCAAGTTACAGTCTGTTTTTAAAACC ATAGGTGGGACAGCTATGGCCAATAGGCTATAAAGAGACATTTAGCACTTTTTTCTATTTAAAGGAACAAGCGGGGAAGGGTGCTAAAAG ATAATACGTTTATTTATTCACACTTGAATTGCATTTGTGATCAAAATAAATGTTTAAATCGCTAAAGGAAAATACAGTAAGTGCTTGAAA GATGAAGGACCAAAAGGCCAAAAAACAGTGAAATATGATCATCATCTCCTTGCGGACTTCTCTGCCTGGTTTTGTGTGTTCTGTTATTCA AACAATAAAAAGCTGGTGGAACTTACTCTTTCTTTTAAGATAAGTTGTAGACTTCGATGTTTCATGCTCATGTACTTCAAATAATGCATG TTTTATAGTTAGTCCCTCATCACTTGAAGTGACTTCTGAGAATTATGCAGAGTCAACATGGATCATTTCACAGTGAGATGCTTTATGGAT TGAAGGATATGGTAAAATGTTTATAGTTTACTTTGAAAGTAAAATATACTATGTCTTGGTTTTGAGGATATTGGATACAAAACTCTCTTC CTTTAGGGCTACTGAGTCTTGATTCCTGATCATCAGAAATTTCACCAGAAACAACTTGCTTCCAATATACCCAATTCTATATGAAGAATT CATGGAGAGTGTACTGGCACTGGAAGAGTTTAGTGTTTCTTGTATGCTTGAAAATAAAGTATGTACTGTTTTGAATGTGTTCCAAGTCCT CTGCATAAACGATGTATTTTGGGGTCTGGTTGGGCCTGGAAAATGGATGAGCACTTCAGAACAGGTCATTTTCCTGATATTGGAAGTGAC ATGTGGCCCTATAGGAGGCATGATGTTAGTTAATTACACATTTGCCTACATCTGTGGGAAATGGAGAACAAAGCCATGTGGGTACTGTAA ACACACGTTTATCTTTTGGCCCAATGCCATACATATGGTAGGCATTTAATTACTGATTGTGTTTGGATAATTTGGGAATTTTCGACTGTG >60877_60877_1_NUF2-TIMM17A_NUF2_chr1_163317728_ENST00000271452_TIMM17A_chr1_201926409_ENST00000367287_length(amino acids)=537AA_BP=375 METLSFPRYNVAEIVIHIRNKILTGADGKNLTKNDLYPNPKPEVLHMIYMRALQIVYGIRLEHFYMMPVNSEVMYPHLMEGFLPFSNLVT HLDSFLPICRVNDFETADILCPKAKRTSRFLSGIINFIHFREACRETYMEFLWQYKSSADKMQQLNAAHQEALMKLERLDSVPVEEQEEF KQLSDGIQELQQSLNQDFHQKTIVLQEGNSQKKSNISEKTKRLNELKLSVVSLKEIQESLKTKIVDSPEKLKNYKEKMKDTVQKLKNARQ EVVEKYEIYGDSVDCLPSCQLEVQLYQKKIQDLSDNREKLASILKESLNLEDQIESDESELKKLKTEENSFKRLMIVKKEKLATAQFKIN KKHEDVKQYKRTVIDPWRIVDDCGGAFTMGTIGGGIFQAIKGFRNSPVGVNHRLRGSLTAIKTRAPQLGGSFAVWGGLFSMIDCSMVQVR -------------------------------------------------------------- >60877_60877_2_NUF2-TIMM17A_NUF2_chr1_163317728_ENST00000367900_TIMM17A_chr1_201926409_ENST00000367287_length(transcript)=2873nt_BP=1276nt GGAATGGGGCGGGACTTCCAGTAGGAGGCGGCAAGTTTGAAAAGTGATGACGGTTGACGTTTGCTGATTTTTGACTTTGCTTGTAGCTGC TCCCCGAACTCGCCGTCTTCCTGTCGGCGGCCGGCACTGTAGATTAACAGGAAACTTCCAAGATGGAAACTTTGTCTTTCCCCAGATATA ATGTAGCTGAGATTGTGATTCATATTCGCAATAAGATCTTAACAGGAGCTGATGGTAAAAACCTCACCAAGAATGATCTTTATCCAAATC CAAAGCCTGAAGTCTTGCACATGATCTACATGAGAGCCTTACAAATAGTATATGGAATTCGACTGGAACATTTTTACATGATGCCAGTGA ACTCTGAAGTCATGTATCCACATTTAATGGAAGGCTTCTTACCATTCAGCAATTTAGTTACTCATCTGGACTCATTTTTGCCTATCTGCC GGGTGAATGACTTTGAGACTGCTGATATTCTATGTCCAAAAGCAAAACGGACAAGTCGGTTTTTAAGTGGCATTATCAACTTTATTCACT TCAGAGAAGCATGCCGTGAAACGTATATGGAATTTCTTTGGCAATATAAATCCTCTGCGGACAAAATGCAACAGTTAAACGCCGCACACC AGGAGGCATTAATGAAACTGGAGAGACTTGATTCTGTTCCAGTTGAAGAGCAAGAAGAGTTCAAGCAGCTTTCAGATGGAATTCAGGAGC TACAACAATCACTAAATCAGGATTTTCATCAAAAAACGATAGTGCTGCAAGAGGGAAATTCCCAAAAGAAGTCAAATATTTCAGAGAAAA CCAAGCGTTTGAATGAACTAAAATTGTCGGTGGTTTCTTTGAAAGAAATACAAGAGAGTTTGAAAACAAAAATTGTGGATTCTCCAGAGA AGTTAAAGAATTATAAAGAAAAAATGAAAGATACGGTCCAGAAGCTTAAAAATGCCAGACAAGAAGTGGTGGAGAAATATGAAATCTATG GAGACTCAGTTGACTGCCTGCCTTCATGTCAGTTGGAAGTGCAGTTATATCAAAAGAAAATACAGGACCTTTCAGATAATAGGGAAAAAT TAGCCAGTATCTTAAAGGAGAGCCTGAACTTGGAGGACCAAATTGAGAGTGATGAGTCAGAACTGAAGAAATTGAAGACTGAAGAAAATT CGTTCAAAAGACTGATGATTGTGAAGAAGGAAAAACTTGCCACAGCACAATTCAAAATAAATAAGAAGCATGAAGATGTTAAGCAATACA AACGCACAGTAATTGACCCATGGCGAATTGTGGATGACTGTGGTGGGGCCTTTACGATGGGTACCATTGGTGGTGGTATCTTTCAAGCAA TCAAAGGTTTTCGCAATTCTCCAGTGGGAGTAAACCACAGACTACGAGGGAGTTTGACAGCTATTAAAACCAGGGCTCCACAGTTAGGAG GTAGCTTTGCAGTTTGGGGAGGGCTGTTTTCCATGATTGACTGTAGTATGGTTCAAGTCAGAGGAAAGGAAGATCCCTGGAACTCCATCA CAAGTGGTGCCTTAACGGGAGCCATACTGGCAGCAAGAAATGGACCAGTGGCCATGGTTGGGTCAGCCGCAATGGGTGGCATTCTCCTAG CTTTAATTGAAGGAGCTGGTATCTTGTTGACAAGATTTGCCTCTGCACAGTTTCCCAATGGTCCTCAGTTTGCAGAAGACCCCTCCCAGT TGCCTTCAACTCAGTTACCTTCCTCACCTTTTGGAGACTATCGACAATATCAGTAGGACTTCTTTCCTAGGATTTCTTTAACAGAACGAG TTGTGGTTCGAGAAGGATTTCAGAAGATCAAGTTACAGTCTGTTTTTAAAACCATAGGTGGGACAGCTATGGCCAATAGGCTATAAAGAG ACATTTAGCACTTTTTTCTATTTAAAGGAACAAGCGGGGAAGGGTGCTAAAAGATAATACGTTTATTTATTCACACTTGAATTGCATTTG TGATCAAAATAAATGTTTAAATCGCTAAAGGAAAATACAGTAAGTGCTTGAAAGATGAAGGACCAAAAGGCCAAAAAACAGTGAAATATG ATCATCATCTCCTTGCGGACTTCTCTGCCTGGTTTTGTGTGTTCTGTTATTCAAACAATAAAAAGCTGGTGGAACTTACTCTTTCTTTTA AGATAAGTTGTAGACTTCGATGTTTCATGCTCATGTACTTCAAATAATGCATGTTTTATAGTTAGTCCCTCATCACTTGAAGTGACTTCT GAGAATTATGCAGAGTCAACATGGATCATTTCACAGTGAGATGCTTTATGGATTGAAGGATATGGTAAAATGTTTATAGTTTACTTTGAA AGTAAAATATACTATGTCTTGGTTTTGAGGATATTGGATACAAAACTCTCTTCCTTTAGGGCTACTGAGTCTTGATTCCTGATCATCAGA AATTTCACCAGAAACAACTTGCTTCCAATATACCCAATTCTATATGAAGAATTCATGGAGAGTGTACTGGCACTGGAAGAGTTTAGTGTT TCTTGTATGCTTGAAAATAAAGTATGTACTGTTTTGAATGTGTTCCAAGTCCTCTGCATAAACGATGTATTTTGGGGTCTGGTTGGGCCT GGAAAATGGATGAGCACTTCAGAACAGGTCATTTTCCTGATATTGGAAGTGACATGTGGCCCTATAGGAGGCATGATGTTAGTTAATTAC ACATTTGCCTACATCTGTGGGAAATGGAGAACAAAGCCATGTGGGTACTGTAAACACACGTTTATCTTTTGGCCCAATGCCATACATATG >60877_60877_2_NUF2-TIMM17A_NUF2_chr1_163317728_ENST00000367900_TIMM17A_chr1_201926409_ENST00000367287_length(amino acids)=537AA_BP=375 METLSFPRYNVAEIVIHIRNKILTGADGKNLTKNDLYPNPKPEVLHMIYMRALQIVYGIRLEHFYMMPVNSEVMYPHLMEGFLPFSNLVT HLDSFLPICRVNDFETADILCPKAKRTSRFLSGIINFIHFREACRETYMEFLWQYKSSADKMQQLNAAHQEALMKLERLDSVPVEEQEEF KQLSDGIQELQQSLNQDFHQKTIVLQEGNSQKKSNISEKTKRLNELKLSVVSLKEIQESLKTKIVDSPEKLKNYKEKMKDTVQKLKNARQ EVVEKYEIYGDSVDCLPSCQLEVQLYQKKIQDLSDNREKLASILKESLNLEDQIESDESELKKLKTEENSFKRLMIVKKEKLATAQFKIN KKHEDVKQYKRTVIDPWRIVDDCGGAFTMGTIGGGIFQAIKGFRNSPVGVNHRLRGSLTAIKTRAPQLGGSFAVWGGLFSMIDCSMVQVR -------------------------------------------------------------- >60877_60877_3_NUF2-TIMM17A_NUF2_chr1_163317728_ENST00000524800_TIMM17A_chr1_201926409_ENST00000367287_length(transcript)=2913nt_BP=1316nt AACCGGCATCCGTCGCGGTTGGACGTGGGCTTGGCGGAATGGGGCGGGACTTCCAGTAGGAGGCGGCAAGTTTGAAAAGTGATGACGGTT GACGTTTGCTGATTTTTGACTTTGCTTGTAGCTGCTCCCCGAACTCGCCGTCTTCCTGTCGGCGGCCGGCACTGTAGGTGAGCGCGAGAG GACGGAGGAAGGAAGCCTGCAGACAGACGCCTTCTCCATCCCAAGGCGCGGGCAGGTGCCGGGACGCTGGGCCTGGCGGTGTTTTCGTCG TGCTCAGCGGTGGGAGGAGGCGGAAGAAACCAGAGCCTGGGAGATTAACAGGAAACTTCCAAGATGGAAACTTTGTCTTTCCCCAGATAT AATGTAGCTGAGATTGTGATTCATATTCGCAATAAGATCTTAACAGGAGCTGATGGTAAAAACCTCACCAAGAATGATCTTTATCCAAAT CCAAAGCCTGAAGTCTTGCACATGATCTACATGAGAGCCTTACAAATAGTATATGGAATTCGACTGGAACATTTTTACATGATGCCAGTG AACTCTGAAGTCATGTATCCACATTTAATGGAAGGCTTCTTACCATTCAGCAATTTAGTTACTCATCTGGACTCATTTTTGCCTATCTGC CGGGTGAATGACTTTGAGACTGCTGATATTCTATGTCCAAAAGCAAAACGGACAAGTCGGTTTTTAAGTGGCATTATCAACTTTATTCAC TTCAGAGAAGCATGCCGTGAAACGTATATGGAATTTCTTTGGCAATATAAATCCTCTGCGGACAAAATGCAACAGTTAAACGCCGCACAC CAGGAGGCATTAATGAAACTGGAGAGACTTGATTCTGTTCCAGTTGAAGAGCAAGAAGAGTTCAAGCAGCTTTCAGATGGAATTCAGGAG CTACAACAATCACTAAATCAGGATTTTCATCAAAAAACGATAGTGCTGCAAGAGGGAAATTCCCAAAAGAAGTCAAATATTTCAGAGAAA ACCAAGCGTTTGAATGAACTAAAATTGTCGGTGGTTTCTTTGAAAGAAATACAAGAGAGTTTGAAAACAAAAATTGTGGATTCTCCAGAG AAGTTAAAGAATTATAAAGAAAAAATGAAAGATACGGTCCAGAAGCTTAAAAATGCCAGAAGCCTGAACTTGGAGGACCAAATTGAGAGT GATGAGTCAGAACTGAAGAAATTGAAGACTGAAGAAAATTCGTTCAAAAGACTGATGATTGTGAAGAAGGAAAAACTTGCCACAGCACAA TTCAAAATAAATAAGAAGCATGAAGATGTTAAGCAATACAAACGCACAGTAATTGACCCATGGCGAATTGTGGATGACTGTGGTGGGGCC TTTACGATGGGTACCATTGGTGGTGGTATCTTTCAAGCAATCAAAGGTTTTCGCAATTCTCCAGTGGGAGTAAACCACAGACTACGAGGG AGTTTGACAGCTATTAAAACCAGGGCTCCACAGTTAGGAGGTAGCTTTGCAGTTTGGGGAGGGCTGTTTTCCATGATTGACTGTAGTATG GTTCAAGTCAGAGGAAAGGAAGATCCCTGGAACTCCATCACAAGTGGTGCCTTAACGGGAGCCATACTGGCAGCAAGAAATGGACCAGTG GCCATGGTTGGGTCAGCCGCAATGGGTGGCATTCTCCTAGCTTTAATTGAAGGAGCTGGTATCTTGTTGACAAGATTTGCCTCTGCACAG TTTCCCAATGGTCCTCAGTTTGCAGAAGACCCCTCCCAGTTGCCTTCAACTCAGTTACCTTCCTCACCTTTTGGAGACTATCGACAATAT CAGTAGGACTTCTTTCCTAGGATTTCTTTAACAGAACGAGTTGTGGTTCGAGAAGGATTTCAGAAGATCAAGTTACAGTCTGTTTTTAAA ACCATAGGTGGGACAGCTATGGCCAATAGGCTATAAAGAGACATTTAGCACTTTTTTCTATTTAAAGGAACAAGCGGGGAAGGGTGCTAA AAGATAATACGTTTATTTATTCACACTTGAATTGCATTTGTGATCAAAATAAATGTTTAAATCGCTAAAGGAAAATACAGTAAGTGCTTG AAAGATGAAGGACCAAAAGGCCAAAAAACAGTGAAATATGATCATCATCTCCTTGCGGACTTCTCTGCCTGGTTTTGTGTGTTCTGTTAT TCAAACAATAAAAAGCTGGTGGAACTTACTCTTTCTTTTAAGATAAGTTGTAGACTTCGATGTTTCATGCTCATGTACTTCAAATAATGC ATGTTTTATAGTTAGTCCCTCATCACTTGAAGTGACTTCTGAGAATTATGCAGAGTCAACATGGATCATTTCACAGTGAGATGCTTTATG GATTGAAGGATATGGTAAAATGTTTATAGTTTACTTTGAAAGTAAAATATACTATGTCTTGGTTTTGAGGATATTGGATACAAAACTCTC TTCCTTTAGGGCTACTGAGTCTTGATTCCTGATCATCAGAAATTTCACCAGAAACAACTTGCTTCCAATATACCCAATTCTATATGAAGA ATTCATGGAGAGTGTACTGGCACTGGAAGAGTTTAGTGTTTCTTGTATGCTTGAAAATAAAGTATGTACTGTTTTGAATGTGTTCCAAGT CCTCTGCATAAACGATGTATTTTGGGGTCTGGTTGGGCCTGGAAAATGGATGAGCACTTCAGAACAGGTCATTTTCCTGATATTGGAAGT GACATGTGGCCCTATAGGAGGCATGATGTTAGTTAATTACACATTTGCCTACATCTGTGGGAAATGGAGAACAAAGCCATGTGGGTACTG TAAACACACGTTTATCTTTTGGCCCAATGCCATACATATGGTAGGCATTTAATTACTGATTGTGTTTGGATAATTTGGGAATTTTCGACT >60877_60877_3_NUF2-TIMM17A_NUF2_chr1_163317728_ENST00000524800_TIMM17A_chr1_201926409_ENST00000367287_length(amino acids)=490AA_BP=328 METLSFPRYNVAEIVIHIRNKILTGADGKNLTKNDLYPNPKPEVLHMIYMRALQIVYGIRLEHFYMMPVNSEVMYPHLMEGFLPFSNLVT HLDSFLPICRVNDFETADILCPKAKRTSRFLSGIINFIHFREACRETYMEFLWQYKSSADKMQQLNAAHQEALMKLERLDSVPVEEQEEF KQLSDGIQELQQSLNQDFHQKTIVLQEGNSQKKSNISEKTKRLNELKLSVVSLKEIQESLKTKIVDSPEKLKNYKEKMKDTVQKLKNARS LNLEDQIESDESELKKLKTEENSFKRLMIVKKEKLATAQFKINKKHEDVKQYKRTVIDPWRIVDDCGGAFTMGTIGGGIFQAIKGFRNSP VGVNHRLRGSLTAIKTRAPQLGGSFAVWGGLFSMIDCSMVQVRGKEDPWNSITSGALTGAILAARNGPVAMVGSAAMGGILLALIEGAGI -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for NUF2-TIMM17A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | NUF2 | chr1:163317728 | chr1:201926409 | ENST00000271452 | + | 12 | 14 | 386_464 | 374.6666666666667 | 465.0 | the C-terminus of NDC80 and the SPBC24-SPBC25 subcomplex |
| Hgene | NUF2 | chr1:163317728 | chr1:201926409 | ENST00000367900 | + | 12 | 14 | 386_464 | 374.6666666666667 | 465.0 | the C-terminus of NDC80 and the SPBC24-SPBC25 subcomplex |
| Hgene | NUF2 | chr1:163317728 | chr1:201926409 | ENST00000271452 | + | 12 | 14 | 1_385 | 374.6666666666667 | 465.0 | the N-terminus of NDC80 |
| Hgene | NUF2 | chr1:163317728 | chr1:201926409 | ENST00000367900 | + | 12 | 14 | 1_385 | 374.6666666666667 | 465.0 | the N-terminus of NDC80 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for NUF2-TIMM17A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for NUF2-TIMM17A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
