
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:OGT-POLA1 (FusionGDB2 ID:61437) |
Fusion Gene Summary for OGT-POLA1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: OGT-POLA1 | Fusion gene ID: 61437 | Hgene | Tgene | Gene symbol | OGT | POLA1 | Gene ID | 8473 | 5422 |
| Gene name | O-linked N-acetylglucosamine (GlcNAc) transferase | DNA polymerase alpha 1, catalytic subunit | |
| Synonyms | HINCUT-1|HRNT1|MRX106|O-GLCNAC|OGT1 | NSX|POLA|VEODS|p180 | |
| Cytomap | Xq13.1 | Xp22.11-p21.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunitO-GlcNAc transferase p110 subunitO-GlcNAc transferase subunit p110O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminy | DNA polymerase alpha catalytic subunitDNA polymerase alpha catalytic subunit p180polymerase (DNA directed), alpha 1, catalytic subunitpolymerase (DNA) alpha 1, catalytic subunitpolymerase (DNA-directed), alpha (70kD) | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000498566, ENST00000373701, ENST00000373719, | ENST00000379059, ENST00000379068, ENST00000493342, | |
| Fusion gene scores | * DoF score | 9 X 9 X 4=324 | 10 X 10 X 7=700 |
| # samples | 10 | 12 | |
| ** MAII score | log2(10/324*10)=-1.6959938131099 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(12/700*10)=-2.54432051622381 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: OGT [Title/Abstract] AND POLA1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | OGT(70757922)-POLA1(24828015), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | OGT | GO:0006110 | regulation of glycolytic process | 22923583 |
| Hgene | OGT | GO:0006493 | protein O-linked glycosylation | 21240259|21285374|22923583|23222540|23352454|24474760 |
| Hgene | OGT | GO:0006915 | apoptotic process | 20824293 |
| Hgene | OGT | GO:0032868 | response to insulin | 18288188 |
| Hgene | OGT | GO:0035020 | regulation of Rac protein signal transduction | 18288188 |
| Hgene | OGT | GO:0043981 | histone H4-K5 acetylation | 20018852 |
| Hgene | OGT | GO:0043982 | histone H4-K8 acetylation | 20018852 |
| Hgene | OGT | GO:0043984 | histone H4-K16 acetylation | 20018852 |
| Hgene | OGT | GO:0045862 | positive regulation of proteolysis | 21285374 |
| Hgene | OGT | GO:0045944 | positive regulation of transcription by RNA polymerase II | 23222540|23353889 |
| Hgene | OGT | GO:0046626 | regulation of insulin receptor signaling pathway | 18288188 |
| Hgene | OGT | GO:0048015 | phosphatidylinositol-mediated signaling | 18288188 |
| Tgene | POLA1 | GO:0006269 | DNA replication, synthesis of RNA primer | 2175912 |
| Tgene | POLA1 | GO:0006270 | DNA replication initiation | 2175912 |
| Tgene | POLA1 | GO:0006272 | leading strand elongation | 2175912|7910375 |
| Tgene | POLA1 | GO:0006273 | lagging strand elongation | 2175912|7910375 |
| Tgene | POLA1 | GO:0006281 | DNA repair | 3335506 |
 Fusion gene breakpoints across OGT (5'-gene) Fusion gene breakpoints across OGT (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
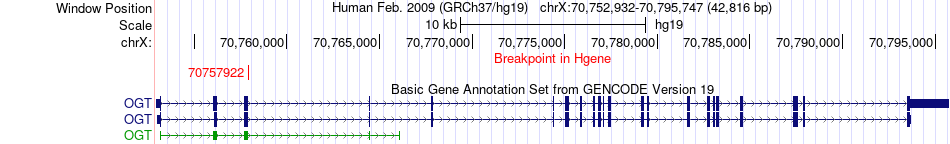 |
 Fusion gene breakpoints across POLA1 (3'-gene) Fusion gene breakpoints across POLA1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
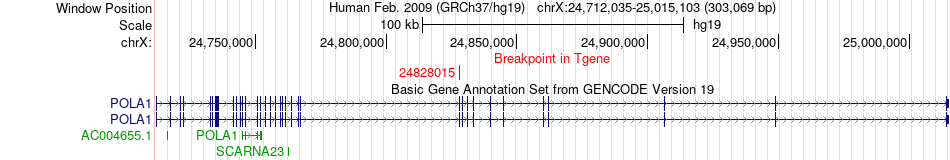 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-KK-A8IC-01A | OGT | chrX | 70757922 | - | POLA1 | chrX | 24828015 | + |
| ChimerDB4 | PRAD | TCGA-KK-A8IC-01A | OGT | chrX | 70757922 | + | POLA1 | chrX | 24828015 | + |
| ChimerDB4 | PRAD | TCGA-KK-A8IC | OGT | chrX | 70757922 | + | POLA1 | chrX | 24828015 | + |
Top |
Fusion Gene ORF analysis for OGT-POLA1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000498566 | ENST00000379059 | OGT | chrX | 70757922 | + | POLA1 | chrX | 24828015 | + |
| 3UTR-3CDS | ENST00000498566 | ENST00000379068 | OGT | chrX | 70757922 | + | POLA1 | chrX | 24828015 | + |
| 3UTR-intron | ENST00000498566 | ENST00000493342 | OGT | chrX | 70757922 | + | POLA1 | chrX | 24828015 | + |
| 5CDS-intron | ENST00000373701 | ENST00000493342 | OGT | chrX | 70757922 | + | POLA1 | chrX | 24828015 | + |
| 5CDS-intron | ENST00000373719 | ENST00000493342 | OGT | chrX | 70757922 | + | POLA1 | chrX | 24828015 | + |
| In-frame | ENST00000373701 | ENST00000379059 | OGT | chrX | 70757922 | + | POLA1 | chrX | 24828015 | + |
| In-frame | ENST00000373701 | ENST00000379068 | OGT | chrX | 70757922 | + | POLA1 | chrX | 24828015 | + |
| In-frame | ENST00000373719 | ENST00000379059 | OGT | chrX | 70757922 | + | POLA1 | chrX | 24828015 | + |
| In-frame | ENST00000373719 | ENST00000379068 | OGT | chrX | 70757922 | + | POLA1 | chrX | 24828015 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000373719 | OGT | chrX | 70757922 | + | ENST00000379068 | POLA1 | chrX | 24828015 | + | 3158 | 679 | 67 | 2121 | 684 |
| ENST00000373719 | OGT | chrX | 70757922 | + | ENST00000379059 | POLA1 | chrX | 24828015 | + | 3158 | 679 | 67 | 2121 | 684 |
| ENST00000373701 | OGT | chrX | 70757922 | + | ENST00000379068 | POLA1 | chrX | 24828015 | + | 3088 | 609 | 27 | 2051 | 674 |
| ENST00000373701 | OGT | chrX | 70757922 | + | ENST00000379059 | POLA1 | chrX | 24828015 | + | 3088 | 609 | 27 | 2051 | 674 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000373719 | ENST00000379068 | OGT | chrX | 70757922 | + | POLA1 | chrX | 24828015 | + | 0.001064185 | 0.99893576 |
| ENST00000373719 | ENST00000379059 | OGT | chrX | 70757922 | + | POLA1 | chrX | 24828015 | + | 0.001064185 | 0.99893576 |
| ENST00000373701 | ENST00000379068 | OGT | chrX | 70757922 | + | POLA1 | chrX | 24828015 | + | 0.001924151 | 0.99807584 |
| ENST00000373701 | ENST00000379059 | OGT | chrX | 70757922 | + | POLA1 | chrX | 24828015 | + | 0.001924151 | 0.99807584 |
Top |
Fusion Genomic Features for OGT-POLA1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| OGT | chrX | 70757922 | + | POLA1 | chrX | 24828014 | + | 4.27E-05 | 0.9999573 |
| OGT | chrX | 70757922 | + | POLA1 | chrX | 24828014 | + | 4.27E-05 | 0.9999573 |
| OGT | chrX | 70757922 | + | POLA1 | chrX | 24828014 | + | 4.27E-05 | 0.9999573 |
| OGT | chrX | 70757922 | + | POLA1 | chrX | 24828014 | + | 4.27E-05 | 0.9999573 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for OGT-POLA1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chrX:70757922/chrX:24828015) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373701 | + | 3 | 22 | 21_54 | 144 | 1037.0 | Repeat | Note=TPR 1 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373701 | + | 3 | 22 | 89_122 | 144 | 1037.0 | Repeat | Note=TPR 2 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373719 | + | 3 | 22 | 123_156 | 154 | 1047.0 | Repeat | Note=TPR 3 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373719 | + | 3 | 22 | 21_54 | 154 | 1047.0 | Repeat | Note=TPR 1 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373719 | + | 3 | 22 | 89_122 | 154 | 1047.0 | Repeat | Note=TPR 2 |
| Tgene | POLA1 | chrX:70757922 | chrX:24828015 | ENST00000379059 | 25 | 37 | 1047_1050 | 982 | 1463.0 | Compositional bias | Note=Poly-Leu | |
| Tgene | POLA1 | chrX:70757922 | chrX:24828015 | ENST00000379059 | 25 | 37 | 1051_1054 | 982 | 1463.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | POLA1 | chrX:70757922 | chrX:24828015 | ENST00000379059 | 25 | 37 | 1348_1374 | 982 | 1463.0 | Motif | CysB motif | |
| Tgene | POLA1 | chrX:70757922 | chrX:24828015 | ENST00000379059 | 25 | 37 | 1245_1376 | 982 | 1463.0 | Region | DNA-binding | |
| Tgene | POLA1 | chrX:70757922 | chrX:24828015 | ENST00000379059 | 25 | 37 | 1283_1318 | 982 | 1463.0 | Zinc finger | CysA-type |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373701 | + | 3 | 22 | 487_503 | 144 | 1037.0 | Motif | Nuclear localization signal |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373719 | + | 3 | 22 | 487_503 | 154 | 1047.0 | Motif | Nuclear localization signal |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373701 | + | 3 | 22 | 906_908 | 144 | 1037.0 | Nucleotide binding | UDP |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373701 | + | 3 | 22 | 911_914 | 144 | 1037.0 | Nucleotide binding | UDP |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373701 | + | 3 | 22 | 930_932 | 144 | 1037.0 | Nucleotide binding | UDP |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373719 | + | 3 | 22 | 906_908 | 154 | 1047.0 | Nucleotide binding | UDP |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373719 | + | 3 | 22 | 911_914 | 154 | 1047.0 | Nucleotide binding | UDP |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373719 | + | 3 | 22 | 930_932 | 154 | 1047.0 | Nucleotide binding | UDP |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373701 | + | 3 | 22 | 991_1010 | 144 | 1037.0 | Region | Note=Required for phosphatidylinositol 3%2C4%2C5-triphosphate binding |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373719 | + | 3 | 22 | 991_1010 | 154 | 1047.0 | Region | Note=Required for phosphatidylinositol 3%2C4%2C5-triphosphate binding |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373701 | + | 3 | 22 | 123_156 | 144 | 1037.0 | Repeat | Note=TPR 3 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373701 | + | 3 | 22 | 157_190 | 144 | 1037.0 | Repeat | Note=TPR 4 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373701 | + | 3 | 22 | 191_224 | 144 | 1037.0 | Repeat | Note=TPR 5 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373701 | + | 3 | 22 | 225_258 | 144 | 1037.0 | Repeat | Note=TPR 6 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373701 | + | 3 | 22 | 259_292 | 144 | 1037.0 | Repeat | Note=TPR 7 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373701 | + | 3 | 22 | 293_326 | 144 | 1037.0 | Repeat | Note=TPR 8 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373701 | + | 3 | 22 | 327_360 | 144 | 1037.0 | Repeat | Note=TPR 9 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373701 | + | 3 | 22 | 361_394 | 144 | 1037.0 | Repeat | Note=TPR 10 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373701 | + | 3 | 22 | 395_428 | 144 | 1037.0 | Repeat | Note=TPR 11 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373701 | + | 3 | 22 | 429_462 | 144 | 1037.0 | Repeat | Note=TPR 12 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373701 | + | 3 | 22 | 463_473 | 144 | 1037.0 | Repeat | Note=TPR 13%3B truncated |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373719 | + | 3 | 22 | 157_190 | 154 | 1047.0 | Repeat | Note=TPR 4 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373719 | + | 3 | 22 | 191_224 | 154 | 1047.0 | Repeat | Note=TPR 5 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373719 | + | 3 | 22 | 225_258 | 154 | 1047.0 | Repeat | Note=TPR 6 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373719 | + | 3 | 22 | 259_292 | 154 | 1047.0 | Repeat | Note=TPR 7 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373719 | + | 3 | 22 | 293_326 | 154 | 1047.0 | Repeat | Note=TPR 8 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373719 | + | 3 | 22 | 327_360 | 154 | 1047.0 | Repeat | Note=TPR 9 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373719 | + | 3 | 22 | 361_394 | 154 | 1047.0 | Repeat | Note=TPR 10 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373719 | + | 3 | 22 | 395_428 | 154 | 1047.0 | Repeat | Note=TPR 11 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373719 | + | 3 | 22 | 429_462 | 154 | 1047.0 | Repeat | Note=TPR 12 |
| Hgene | OGT | chrX:70757922 | chrX:24828015 | ENST00000373719 | + | 3 | 22 | 463_473 | 154 | 1047.0 | Repeat | Note=TPR 13%3B truncated |
| Tgene | POLA1 | chrX:70757922 | chrX:24828015 | ENST00000379059 | 25 | 37 | 75_112 | 982 | 1463.0 | Compositional bias | Note=Asp-rich | |
| Tgene | POLA1 | chrX:70757922 | chrX:24828015 | ENST00000379059 | 25 | 37 | 650_715 | 982 | 1463.0 | Region | DNA-binding |
Top |
Fusion Gene Sequence for OGT-POLA1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >61437_61437_1_OGT-POLA1_OGT_chrX_70757922_ENST00000373701_POLA1_chrX_24828015_ENST00000379059_length(transcript)=3088nt_BP=609nt TAGATGGTCAATTAGAGTTCCCAGGGTTTGAAGCCTGTAACTGCTGCCGCCGCTCAAGCCCTCCAGAGCATTGCTACGGCTGCTGCCCTT GTACTACTACCTCCAAATACGTTCTTGCTGGTAGTGGCGGCAGCAGGACCAATTACCTCTTTTTTGCTCTCCCTCGAGAAGCTCCAGATG GCGTCTTCCGTGGGCAACGTGGCCGACAGCACAGGGTTAGCTGAGTTGGCACATCGAGAATATCAGGCAGGAGATTTTGAGGCAGCTGAG AGACACTGCATGCAGCTCTGGAGACAAGAGCCAGACAATACTGGTGTGCTTTTATTACTTTCATCTATACACTTCCAGTGTCGAAGGCTG GACAGATCTGCTCACTTTAGCACTCTGGCAATTAAACAGAACCCCCTTCTGGCAGAAGCTTATTCGAATTTGGGGAATGTGTACAAGGAA AGAGGGCAGTTGCAGGAGGCAATTGAGCATTATCGACATGCATTGCGTCTCAAACCTGATTTCATCGATGGTTATATTAACCTGGCAGCC GCCTTGGTAGCAGCGGGTGACATGGAAGGGGCAGTACAAGCTTACGTCTCTGCTCTTCAGTACAATCCTATTTTGATGCATACGAAAGAG ATGGTACAAAAGATGAATCTTGAAGTTATTTATGGAGATACAGATTCAATTATGATAAACACCAATAGCACCAATCTGGAAGAAGTATTT AAGTTGGGAAACAAGGTAAAAAGTGAAGTGAATAAGTTGTACAAACTGCTTGAAATAGACATTGATGGGGTTTTCAAGTCTCTGCTACTG CTGAAAAAAAAGAAGTACGCTGCTCTGGTTGTTGAGCCAACGTCGGATGGGAATTATGTCACCAAACAGGAGCTCAAAGGATTAGATATA GTTAGAAGAGATTGGTGTGATCTTGCTAAAGACACTGGAAACTTTGTGATTGGCCAGATTCTTTCTGATCAAAGCCGGGACACTATAGTG GAAAACATTCAGAAGAGGCTGATAGAAATTGGAGAAAATGTGCTAAATGGCAGTGTCCCAGTGAGCCAGTTTGAAATTAACAAGGCATTG ACAAAGGATCCCCAGGATTACCCTGATAAAAAAAGCCTACCTCATGTACATGTTGCCCTCTGGATAAATTCTCAAGGAGGCAGAAAGGTG AAAGCTGGAGATACTGTGTCATATGTCATCTGTCAGGATGGATCAAACCTCACTGCAAGTCAGAGGGCCTATGCGCCTGAGCAGCTGCAG AAACAGGATAATCTAACCATTGACACCCAGTACTACCTGGCCCAGCAGATCCACCCAGTCGTGGCTCGGATCTGTGAACCAATAGACGGA ATTGATGCTGTCCTCATTGCAACGTGGTTGGGACTTGACCCCACCCAATTTAGAGTTCATCATTATCATAAAGATGAAGAGAATGATGCT CTACTTGGTGGCCCAGCACAGCTCACTGATGAAGAGAAATACAGGGACTGTGAAAGATTCAAATGTCCATGCCCTACATGTGGAACTGAG AATATTTATGATAATGTCTTTGATGGTTCGGGAACAGATATGGAGCCCAGCTTGTATCGTTGCAGTAACATCGATTGTAAGGCTTCACCT CTGACCTTTACAGTACAACTGAGCAACAAATTGATCATGGACATTAGACGTTTCATTAAAAAGTACTATGATGGCTGGTTGATATGTGAA GAGCCAACCTGTCGCAATCGAACTCGTCACCTTCCCCTTCAATTCTCCCGAACTGGGCCTCTTTGCCCAGCCTGCATGAAAGCTACACTT CAACCAGAGTATTCTGACAAGTCCCTGTACACCCAGCTGTGCTTTTACCGGTACATTTTTGATGCGGAGTGTGCACTGGAGAAACTTACT ACCGATCATGAGAAAGATAAATTGAAGAAGCAATTTTTTACCCCCAAAGTTCTGCAGGACTACAGAAAACTCAAGAACACAGCAGAGCAA TTCTTGTCCCGAAGTGGCTACTCCGAAGTGAATCTGAGCAAACTCTTCGCTGGTTGTGCCGTGAAATCCTAAGGGAATCCCAGGAGTAAC CAAGGAGGGGGTAGTTGAAAAATCCCAGCTTCCTCTGTGCCTCCACTCTGGCCCTAAATGCTCCTCCAGCATCTGTTTCTCCCTTGGGAC TGTGTCTCATGTTTGTGTGAATGTAGACCAGGAAAGGGGGCTGCAAAAATGTTGAGTCTAATGTTCGTAAGCATCATAGAAATTCCTGTC TTCATATTAAGATGTACTGCTTTAAAACACAACTCCAGAGCCCCTCCCCAAGCTCCCCTCCCCAAGCTCCTGAAGACCCGGTTTCTGAGG GAGGGAAATTGCTACTTGGATTGAGAGTAGCTGGAATGTAAGTGACCCCAGGCTTTGCCTCAGGGCCTTTAGCCTATGTCCCCCCCACAT AAAGAGAGCTTCTCAGAGCCTGACTGAAGAGCTGACGTTTTGCTTTTTCATATGCCAATTAAACCCGGTCTAAATCCAAATGCTTCTCCA GCCATCCAGGAGTGGCTGTCCTTTTCAGTCTTGTCTTTTATATAGGTAGCTGAGGGGGAAGATTTAGAAGCCTTGCACTCACTAAATAGA TTAAACAGAGCAGGCTTGTTTGTTGAATTGCTCCAAAGTCCAACAGACACACACTGAGCAGGTGTTTTACACTCACATTCCCTTTTTGCC CCTTAAATAGAAAGTGCAGGTAAAGGTTTATACAACAAGAAAGCACATTGAAAATAATTTGATACTCTAACAATCCATTAACATGTGTAG GGGTTACGGTGAGGATCACTGTGTTGTATTCAGAAAAACGGGGAGAGGGATGCTTAATTGGCCCTGGCGCTTGCTATTTTTTTCTCATTT CTTCACAATAGGACCGTCTTTGGCAGCAGCAAAATGTATTTCAGTATGGCAGTCTTTCCTCTCTTACATTATTGGTAAGATTATACTAAC AAAATGTTTCCCCTTGTACAATTATGCTGTGTTTTTAAAAAACATTGACCTGTGTGTTTTTATAAAAGAAAAAGTATGTTGTGCCTTCTT >61437_61437_1_OGT-POLA1_OGT_chrX_70757922_ENST00000373701_POLA1_chrX_24828015_ENST00000379059_length(amino acids)=674AA_BP=194 MKPVTAAAAQALQSIATAAALVLLPPNTFLLVVAAAGPITSFLLSLEKLQMASSVGNVADSTGLAELAHREYQAGDFEAAERHCMQLWRQ EPDNTGVLLLLSSIHFQCRRLDRSAHFSTLAIKQNPLLAEAYSNLGNVYKERGQLQEAIEHYRHALRLKPDFIDGYINLAAALVAAGDME GAVQAYVSALQYNPILMHTKEMVQKMNLEVIYGDTDSIMINTNSTNLEEVFKLGNKVKSEVNKLYKLLEIDIDGVFKSLLLLKKKKYAAL VVEPTSDGNYVTKQELKGLDIVRRDWCDLAKDTGNFVIGQILSDQSRDTIVENIQKRLIEIGENVLNGSVPVSQFEINKALTKDPQDYPD KKSLPHVHVALWINSQGGRKVKAGDTVSYVICQDGSNLTASQRAYAPEQLQKQDNLTIDTQYYLAQQIHPVVARICEPIDGIDAVLIATW LGLDPTQFRVHHYHKDEENDALLGGPAQLTDEEKYRDCERFKCPCPTCGTENIYDNVFDGSGTDMEPSLYRCSNIDCKASPLTFTVQLSN KLIMDIRRFIKKYYDGWLICEEPTCRNRTRHLPLQFSRTGPLCPACMKATLQPEYSDKSLYTQLCFYRYIFDAECALEKLTTDHEKDKLK -------------------------------------------------------------- >61437_61437_2_OGT-POLA1_OGT_chrX_70757922_ENST00000373701_POLA1_chrX_24828015_ENST00000379068_length(transcript)=3088nt_BP=609nt TAGATGGTCAATTAGAGTTCCCAGGGTTTGAAGCCTGTAACTGCTGCCGCCGCTCAAGCCCTCCAGAGCATTGCTACGGCTGCTGCCCTT GTACTACTACCTCCAAATACGTTCTTGCTGGTAGTGGCGGCAGCAGGACCAATTACCTCTTTTTTGCTCTCCCTCGAGAAGCTCCAGATG GCGTCTTCCGTGGGCAACGTGGCCGACAGCACAGGGTTAGCTGAGTTGGCACATCGAGAATATCAGGCAGGAGATTTTGAGGCAGCTGAG AGACACTGCATGCAGCTCTGGAGACAAGAGCCAGACAATACTGGTGTGCTTTTATTACTTTCATCTATACACTTCCAGTGTCGAAGGCTG GACAGATCTGCTCACTTTAGCACTCTGGCAATTAAACAGAACCCCCTTCTGGCAGAAGCTTATTCGAATTTGGGGAATGTGTACAAGGAA AGAGGGCAGTTGCAGGAGGCAATTGAGCATTATCGACATGCATTGCGTCTCAAACCTGATTTCATCGATGGTTATATTAACCTGGCAGCC GCCTTGGTAGCAGCGGGTGACATGGAAGGGGCAGTACAAGCTTACGTCTCTGCTCTTCAGTACAATCCTATTTTGATGCATACGAAAGAG ATGGTACAAAAGATGAATCTTGAAGTTATTTATGGAGATACAGATTCAATTATGATAAACACCAATAGCACCAATCTGGAAGAAGTATTT AAGTTGGGAAACAAGGTAAAAAGTGAAGTGAATAAGTTGTACAAACTGCTTGAAATAGACATTGATGGGGTTTTCAAGTCTCTGCTACTG CTGAAAAAAAAGAAGTACGCTGCTCTGGTTGTTGAGCCAACGTCGGATGGGAATTATGTCACCAAACAGGAGCTCAAAGGATTAGATATA GTTAGAAGAGATTGGTGTGATCTTGCTAAAGACACTGGAAACTTTGTGATTGGCCAGATTCTTTCTGATCAAAGCCGGGACACTATAGTG GAAAACATTCAGAAGAGGCTGATAGAAATTGGAGAAAATGTGCTAAATGGCAGTGTCCCAGTGAGCCAGTTTGAAATTAACAAGGCATTG ACAAAGGATCCCCAGGATTACCCTGATAAAAAAAGCCTACCTCATGTACATGTTGCCCTCTGGATAAATTCTCAAGGAGGCAGAAAGGTG AAAGCTGGAGATACTGTGTCATATGTCATCTGTCAGGATGGATCAAACCTCACTGCAAGTCAGAGGGCCTATGCGCCTGAGCAGCTGCAG AAACAGGATAATCTAACCATTGACACCCAGTACTACCTGGCCCAGCAGATCCACCCAGTCGTGGCTCGGATCTGTGAACCAATAGACGGA ATTGATGCTGTCCTCATTGCAACGTGGTTGGGACTTGACCCCACCCAATTTAGAGTTCATCATTATCATAAAGATGAAGAGAATGATGCT CTACTTGGTGGCCCAGCACAGCTCACTGATGAAGAGAAATACAGGGACTGTGAAAGATTCAAATGTCCATGCCCTACATGTGGAACTGAG AATATTTATGATAATGTCTTTGATGGTTCGGGAACAGATATGGAGCCCAGCTTGTATCGTTGCAGTAACATCGATTGTAAGGCTTCACCT CTGACCTTTACAGTACAACTGAGCAACAAATTGATCATGGACATTAGACGTTTCATTAAAAAGTACTATGATGGCTGGTTGATATGTGAA GAGCCAACCTGTCGCAATCGAACTCGTCACCTTCCCCTTCAATTCTCCCGAACTGGGCCTCTTTGCCCAGCCTGCATGAAAGCTACACTT CAACCAGAGTATTCTGACAAGTCCCTGTACACCCAGCTGTGCTTTTACCGGTACATTTTTGATGCGGAGTGTGCACTGGAGAAACTTACT ACCGATCATGAGAAAGATAAATTGAAGAAGCAATTTTTTACCCCCAAAGTTCTGCAGGACTACAGAAAACTCAAGAACACAGCAGAGCAA TTCTTGTCCCGAAGTGGCTACTCCGAAGTGAATCTGAGCAAACTCTTCGCTGGTTGTGCCGTGAAATCCTAAGGGAATCCCAGGAGTAAC CAAGGAGGGGGTAGTTGAAAAATCCCAGCTTCCTCTGTGCCTCCACTCTGGCCCTAAATGCTCCTCCAGCATCTGTTTCTCCCTTGGGAC TGTGTCTCATGTTTGTGTGAATGTAGACCAGGAAAGGGGGCTGCAAAAATGTTGAGTCTAATGTTCGTAAGCATCATAGAAATTCCTGTC TTCATATTAAGATGTACTGCTTTAAAACACAACTCCAGAGCCCCTCCCCAAGCTCCCCTCCCCAAGCTCCTGAAGACCCGGTTTCTGAGG GAGGGAAATTGCTACTTGGATTGAGAGTAGCTGGAATGTAAGTGACCCCAGGCTTTGCCTCAGGGCCTTTAGCCTATGTCCCCCCCACAT AAAGAGAGCTTCTCAGAGCCTGACTGAAGAGCTGACGTTTTGCTTTTTCATATGCCAATTAAACCCGGTCTAAATCCAAATGCTTCTCCA GCCATCCAGGAGTGGCTGTCCTTTTCAGTCTTGTCTTTTATATAGGTAGCTGAGGGGGAAGATTTAGAAGCCTTGCACTCACTAAATAGA TTAAACAGAGCAGGCTTGTTTGTTGAATTGCTCCAAAGTCCAACAGACACACACTGAGCAGGTGTTTTACACTCACATTCCCTTTTTGCC CCTTAAATAGAAAGTGCAGGTAAAGGTTTATACAACAAGAAAGCACATTGAAAATAATTTGATACTCTAACAATCCATTAACATGTGTAG GGGTTACGGTGAGGATCACTGTGTTGTATTCAGAAAAACGGGGAGAGGGATGCTTAATTGGCCCTGGCGCTTGCTATTTTTTTCTCATTT CTTCACAATAGGACCGTCTTTGGCAGCAGCAAAATGTATTTCAGTATGGCAGTCTTTCCTCTCTTACATTATTGGTAAGATTATACTAAC AAAATGTTTCCCCTTGTACAATTATGCTGTGTTTTTAAAAAACATTGACCTGTGTGTTTTTATAAAAGAAAAAGTATGTTGTGCCTTCTT >61437_61437_2_OGT-POLA1_OGT_chrX_70757922_ENST00000373701_POLA1_chrX_24828015_ENST00000379068_length(amino acids)=674AA_BP=194 MKPVTAAAAQALQSIATAAALVLLPPNTFLLVVAAAGPITSFLLSLEKLQMASSVGNVADSTGLAELAHREYQAGDFEAAERHCMQLWRQ EPDNTGVLLLLSSIHFQCRRLDRSAHFSTLAIKQNPLLAEAYSNLGNVYKERGQLQEAIEHYRHALRLKPDFIDGYINLAAALVAAGDME GAVQAYVSALQYNPILMHTKEMVQKMNLEVIYGDTDSIMINTNSTNLEEVFKLGNKVKSEVNKLYKLLEIDIDGVFKSLLLLKKKKYAAL VVEPTSDGNYVTKQELKGLDIVRRDWCDLAKDTGNFVIGQILSDQSRDTIVENIQKRLIEIGENVLNGSVPVSQFEINKALTKDPQDYPD KKSLPHVHVALWINSQGGRKVKAGDTVSYVICQDGSNLTASQRAYAPEQLQKQDNLTIDTQYYLAQQIHPVVARICEPIDGIDAVLIATW LGLDPTQFRVHHYHKDEENDALLGGPAQLTDEEKYRDCERFKCPCPTCGTENIYDNVFDGSGTDMEPSLYRCSNIDCKASPLTFTVQLSN KLIMDIRRFIKKYYDGWLICEEPTCRNRTRHLPLQFSRTGPLCPACMKATLQPEYSDKSLYTQLCFYRYIFDAECALEKLTTDHEKDKLK -------------------------------------------------------------- >61437_61437_3_OGT-POLA1_OGT_chrX_70757922_ENST00000373719_POLA1_chrX_24828015_ENST00000379059_length(transcript)=3158nt_BP=679nt GGCCGAGGAGCCGTCGCCGCCATTTCAAGACCGTACTAGGTAGATGGTCAATTAGAGTTCCCAGGGTTTGAAGCCTGTAACTGCTGCCGC CGCTCAAGCCCTCCAGAGCATTGCTACGGCTGCTGCCCTTGTACTACTACCTCCAAATACGTTCTTGCTGGTAGTGGCGGCAGCAGGACC AATTACCTCTTTTTTGCTCTCCCTCGAGAAGCTCCAGATGGCGTCTTCCGTGGGCAACGTGGCCGACAGCACAGAACCAACGAAACGTAT GCTTTCCTTCCAAGGGTTAGCTGAGTTGGCACATCGAGAATATCAGGCAGGAGATTTTGAGGCAGCTGAGAGACACTGCATGCAGCTCTG GAGACAAGAGCCAGACAATACTGGTGTGCTTTTATTACTTTCATCTATACACTTCCAGTGTCGAAGGCTGGACAGATCTGCTCACTTTAG CACTCTGGCAATTAAACAGAACCCCCTTCTGGCAGAAGCTTATTCGAATTTGGGGAATGTGTACAAGGAAAGAGGGCAGTTGCAGGAGGC AATTGAGCATTATCGACATGCATTGCGTCTCAAACCTGATTTCATCGATGGTTATATTAACCTGGCAGCCGCCTTGGTAGCAGCGGGTGA CATGGAAGGGGCAGTACAAGCTTACGTCTCTGCTCTTCAGTACAATCCTATTTTGATGCATACGAAAGAGATGGTACAAAAGATGAATCT TGAAGTTATTTATGGAGATACAGATTCAATTATGATAAACACCAATAGCACCAATCTGGAAGAAGTATTTAAGTTGGGAAACAAGGTAAA AAGTGAAGTGAATAAGTTGTACAAACTGCTTGAAATAGACATTGATGGGGTTTTCAAGTCTCTGCTACTGCTGAAAAAAAAGAAGTACGC TGCTCTGGTTGTTGAGCCAACGTCGGATGGGAATTATGTCACCAAACAGGAGCTCAAAGGATTAGATATAGTTAGAAGAGATTGGTGTGA TCTTGCTAAAGACACTGGAAACTTTGTGATTGGCCAGATTCTTTCTGATCAAAGCCGGGACACTATAGTGGAAAACATTCAGAAGAGGCT GATAGAAATTGGAGAAAATGTGCTAAATGGCAGTGTCCCAGTGAGCCAGTTTGAAATTAACAAGGCATTGACAAAGGATCCCCAGGATTA CCCTGATAAAAAAAGCCTACCTCATGTACATGTTGCCCTCTGGATAAATTCTCAAGGAGGCAGAAAGGTGAAAGCTGGAGATACTGTGTC ATATGTCATCTGTCAGGATGGATCAAACCTCACTGCAAGTCAGAGGGCCTATGCGCCTGAGCAGCTGCAGAAACAGGATAATCTAACCAT TGACACCCAGTACTACCTGGCCCAGCAGATCCACCCAGTCGTGGCTCGGATCTGTGAACCAATAGACGGAATTGATGCTGTCCTCATTGC AACGTGGTTGGGACTTGACCCCACCCAATTTAGAGTTCATCATTATCATAAAGATGAAGAGAATGATGCTCTACTTGGTGGCCCAGCACA GCTCACTGATGAAGAGAAATACAGGGACTGTGAAAGATTCAAATGTCCATGCCCTACATGTGGAACTGAGAATATTTATGATAATGTCTT TGATGGTTCGGGAACAGATATGGAGCCCAGCTTGTATCGTTGCAGTAACATCGATTGTAAGGCTTCACCTCTGACCTTTACAGTACAACT GAGCAACAAATTGATCATGGACATTAGACGTTTCATTAAAAAGTACTATGATGGCTGGTTGATATGTGAAGAGCCAACCTGTCGCAATCG AACTCGTCACCTTCCCCTTCAATTCTCCCGAACTGGGCCTCTTTGCCCAGCCTGCATGAAAGCTACACTTCAACCAGAGTATTCTGACAA GTCCCTGTACACCCAGCTGTGCTTTTACCGGTACATTTTTGATGCGGAGTGTGCACTGGAGAAACTTACTACCGATCATGAGAAAGATAA ATTGAAGAAGCAATTTTTTACCCCCAAAGTTCTGCAGGACTACAGAAAACTCAAGAACACAGCAGAGCAATTCTTGTCCCGAAGTGGCTA CTCCGAAGTGAATCTGAGCAAACTCTTCGCTGGTTGTGCCGTGAAATCCTAAGGGAATCCCAGGAGTAACCAAGGAGGGGGTAGTTGAAA AATCCCAGCTTCCTCTGTGCCTCCACTCTGGCCCTAAATGCTCCTCCAGCATCTGTTTCTCCCTTGGGACTGTGTCTCATGTTTGTGTGA ATGTAGACCAGGAAAGGGGGCTGCAAAAATGTTGAGTCTAATGTTCGTAAGCATCATAGAAATTCCTGTCTTCATATTAAGATGTACTGC TTTAAAACACAACTCCAGAGCCCCTCCCCAAGCTCCCCTCCCCAAGCTCCTGAAGACCCGGTTTCTGAGGGAGGGAAATTGCTACTTGGA TTGAGAGTAGCTGGAATGTAAGTGACCCCAGGCTTTGCCTCAGGGCCTTTAGCCTATGTCCCCCCCACATAAAGAGAGCTTCTCAGAGCC TGACTGAAGAGCTGACGTTTTGCTTTTTCATATGCCAATTAAACCCGGTCTAAATCCAAATGCTTCTCCAGCCATCCAGGAGTGGCTGTC CTTTTCAGTCTTGTCTTTTATATAGGTAGCTGAGGGGGAAGATTTAGAAGCCTTGCACTCACTAAATAGATTAAACAGAGCAGGCTTGTT TGTTGAATTGCTCCAAAGTCCAACAGACACACACTGAGCAGGTGTTTTACACTCACATTCCCTTTTTGCCCCTTAAATAGAAAGTGCAGG TAAAGGTTTATACAACAAGAAAGCACATTGAAAATAATTTGATACTCTAACAATCCATTAACATGTGTAGGGGTTACGGTGAGGATCACT GTGTTGTATTCAGAAAAACGGGGAGAGGGATGCTTAATTGGCCCTGGCGCTTGCTATTTTTTTCTCATTTCTTCACAATAGGACCGTCTT TGGCAGCAGCAAAATGTATTTCAGTATGGCAGTCTTTCCTCTCTTACATTATTGGTAAGATTATACTAACAAAATGTTTCCCCTTGTACA ATTATGCTGTGTTTTTAAAAAACATTGACCTGTGTGTTTTTATAAAAGAAAAAGTATGTTGTGCCTTCTTCTTAAGAATAAAGTTTTCTA >61437_61437_3_OGT-POLA1_OGT_chrX_70757922_ENST00000373719_POLA1_chrX_24828015_ENST00000379059_length(amino acids)=684AA_BP=204 MKPVTAAAAQALQSIATAAALVLLPPNTFLLVVAAAGPITSFLLSLEKLQMASSVGNVADSTEPTKRMLSFQGLAELAHREYQAGDFEAA ERHCMQLWRQEPDNTGVLLLLSSIHFQCRRLDRSAHFSTLAIKQNPLLAEAYSNLGNVYKERGQLQEAIEHYRHALRLKPDFIDGYINLA AALVAAGDMEGAVQAYVSALQYNPILMHTKEMVQKMNLEVIYGDTDSIMINTNSTNLEEVFKLGNKVKSEVNKLYKLLEIDIDGVFKSLL LLKKKKYAALVVEPTSDGNYVTKQELKGLDIVRRDWCDLAKDTGNFVIGQILSDQSRDTIVENIQKRLIEIGENVLNGSVPVSQFEINKA LTKDPQDYPDKKSLPHVHVALWINSQGGRKVKAGDTVSYVICQDGSNLTASQRAYAPEQLQKQDNLTIDTQYYLAQQIHPVVARICEPID GIDAVLIATWLGLDPTQFRVHHYHKDEENDALLGGPAQLTDEEKYRDCERFKCPCPTCGTENIYDNVFDGSGTDMEPSLYRCSNIDCKAS PLTFTVQLSNKLIMDIRRFIKKYYDGWLICEEPTCRNRTRHLPLQFSRTGPLCPACMKATLQPEYSDKSLYTQLCFYRYIFDAECALEKL -------------------------------------------------------------- >61437_61437_4_OGT-POLA1_OGT_chrX_70757922_ENST00000373719_POLA1_chrX_24828015_ENST00000379068_length(transcript)=3158nt_BP=679nt GGCCGAGGAGCCGTCGCCGCCATTTCAAGACCGTACTAGGTAGATGGTCAATTAGAGTTCCCAGGGTTTGAAGCCTGTAACTGCTGCCGC CGCTCAAGCCCTCCAGAGCATTGCTACGGCTGCTGCCCTTGTACTACTACCTCCAAATACGTTCTTGCTGGTAGTGGCGGCAGCAGGACC AATTACCTCTTTTTTGCTCTCCCTCGAGAAGCTCCAGATGGCGTCTTCCGTGGGCAACGTGGCCGACAGCACAGAACCAACGAAACGTAT GCTTTCCTTCCAAGGGTTAGCTGAGTTGGCACATCGAGAATATCAGGCAGGAGATTTTGAGGCAGCTGAGAGACACTGCATGCAGCTCTG GAGACAAGAGCCAGACAATACTGGTGTGCTTTTATTACTTTCATCTATACACTTCCAGTGTCGAAGGCTGGACAGATCTGCTCACTTTAG CACTCTGGCAATTAAACAGAACCCCCTTCTGGCAGAAGCTTATTCGAATTTGGGGAATGTGTACAAGGAAAGAGGGCAGTTGCAGGAGGC AATTGAGCATTATCGACATGCATTGCGTCTCAAACCTGATTTCATCGATGGTTATATTAACCTGGCAGCCGCCTTGGTAGCAGCGGGTGA CATGGAAGGGGCAGTACAAGCTTACGTCTCTGCTCTTCAGTACAATCCTATTTTGATGCATACGAAAGAGATGGTACAAAAGATGAATCT TGAAGTTATTTATGGAGATACAGATTCAATTATGATAAACACCAATAGCACCAATCTGGAAGAAGTATTTAAGTTGGGAAACAAGGTAAA AAGTGAAGTGAATAAGTTGTACAAACTGCTTGAAATAGACATTGATGGGGTTTTCAAGTCTCTGCTACTGCTGAAAAAAAAGAAGTACGC TGCTCTGGTTGTTGAGCCAACGTCGGATGGGAATTATGTCACCAAACAGGAGCTCAAAGGATTAGATATAGTTAGAAGAGATTGGTGTGA TCTTGCTAAAGACACTGGAAACTTTGTGATTGGCCAGATTCTTTCTGATCAAAGCCGGGACACTATAGTGGAAAACATTCAGAAGAGGCT GATAGAAATTGGAGAAAATGTGCTAAATGGCAGTGTCCCAGTGAGCCAGTTTGAAATTAACAAGGCATTGACAAAGGATCCCCAGGATTA CCCTGATAAAAAAAGCCTACCTCATGTACATGTTGCCCTCTGGATAAATTCTCAAGGAGGCAGAAAGGTGAAAGCTGGAGATACTGTGTC ATATGTCATCTGTCAGGATGGATCAAACCTCACTGCAAGTCAGAGGGCCTATGCGCCTGAGCAGCTGCAGAAACAGGATAATCTAACCAT TGACACCCAGTACTACCTGGCCCAGCAGATCCACCCAGTCGTGGCTCGGATCTGTGAACCAATAGACGGAATTGATGCTGTCCTCATTGC AACGTGGTTGGGACTTGACCCCACCCAATTTAGAGTTCATCATTATCATAAAGATGAAGAGAATGATGCTCTACTTGGTGGCCCAGCACA GCTCACTGATGAAGAGAAATACAGGGACTGTGAAAGATTCAAATGTCCATGCCCTACATGTGGAACTGAGAATATTTATGATAATGTCTT TGATGGTTCGGGAACAGATATGGAGCCCAGCTTGTATCGTTGCAGTAACATCGATTGTAAGGCTTCACCTCTGACCTTTACAGTACAACT GAGCAACAAATTGATCATGGACATTAGACGTTTCATTAAAAAGTACTATGATGGCTGGTTGATATGTGAAGAGCCAACCTGTCGCAATCG AACTCGTCACCTTCCCCTTCAATTCTCCCGAACTGGGCCTCTTTGCCCAGCCTGCATGAAAGCTACACTTCAACCAGAGTATTCTGACAA GTCCCTGTACACCCAGCTGTGCTTTTACCGGTACATTTTTGATGCGGAGTGTGCACTGGAGAAACTTACTACCGATCATGAGAAAGATAA ATTGAAGAAGCAATTTTTTACCCCCAAAGTTCTGCAGGACTACAGAAAACTCAAGAACACAGCAGAGCAATTCTTGTCCCGAAGTGGCTA CTCCGAAGTGAATCTGAGCAAACTCTTCGCTGGTTGTGCCGTGAAATCCTAAGGGAATCCCAGGAGTAACCAAGGAGGGGGTAGTTGAAA AATCCCAGCTTCCTCTGTGCCTCCACTCTGGCCCTAAATGCTCCTCCAGCATCTGTTTCTCCCTTGGGACTGTGTCTCATGTTTGTGTGA ATGTAGACCAGGAAAGGGGGCTGCAAAAATGTTGAGTCTAATGTTCGTAAGCATCATAGAAATTCCTGTCTTCATATTAAGATGTACTGC TTTAAAACACAACTCCAGAGCCCCTCCCCAAGCTCCCCTCCCCAAGCTCCTGAAGACCCGGTTTCTGAGGGAGGGAAATTGCTACTTGGA TTGAGAGTAGCTGGAATGTAAGTGACCCCAGGCTTTGCCTCAGGGCCTTTAGCCTATGTCCCCCCCACATAAAGAGAGCTTCTCAGAGCC TGACTGAAGAGCTGACGTTTTGCTTTTTCATATGCCAATTAAACCCGGTCTAAATCCAAATGCTTCTCCAGCCATCCAGGAGTGGCTGTC CTTTTCAGTCTTGTCTTTTATATAGGTAGCTGAGGGGGAAGATTTAGAAGCCTTGCACTCACTAAATAGATTAAACAGAGCAGGCTTGTT TGTTGAATTGCTCCAAAGTCCAACAGACACACACTGAGCAGGTGTTTTACACTCACATTCCCTTTTTGCCCCTTAAATAGAAAGTGCAGG TAAAGGTTTATACAACAAGAAAGCACATTGAAAATAATTTGATACTCTAACAATCCATTAACATGTGTAGGGGTTACGGTGAGGATCACT GTGTTGTATTCAGAAAAACGGGGAGAGGGATGCTTAATTGGCCCTGGCGCTTGCTATTTTTTTCTCATTTCTTCACAATAGGACCGTCTT TGGCAGCAGCAAAATGTATTTCAGTATGGCAGTCTTTCCTCTCTTACATTATTGGTAAGATTATACTAACAAAATGTTTCCCCTTGTACA ATTATGCTGTGTTTTTAAAAAACATTGACCTGTGTGTTTTTATAAAAGAAAAAGTATGTTGTGCCTTCTTCTTAAGAATAAAGTTTTCTA >61437_61437_4_OGT-POLA1_OGT_chrX_70757922_ENST00000373719_POLA1_chrX_24828015_ENST00000379068_length(amino acids)=684AA_BP=204 MKPVTAAAAQALQSIATAAALVLLPPNTFLLVVAAAGPITSFLLSLEKLQMASSVGNVADSTEPTKRMLSFQGLAELAHREYQAGDFEAA ERHCMQLWRQEPDNTGVLLLLSSIHFQCRRLDRSAHFSTLAIKQNPLLAEAYSNLGNVYKERGQLQEAIEHYRHALRLKPDFIDGYINLA AALVAAGDMEGAVQAYVSALQYNPILMHTKEMVQKMNLEVIYGDTDSIMINTNSTNLEEVFKLGNKVKSEVNKLYKLLEIDIDGVFKSLL LLKKKKYAALVVEPTSDGNYVTKQELKGLDIVRRDWCDLAKDTGNFVIGQILSDQSRDTIVENIQKRLIEIGENVLNGSVPVSQFEINKA LTKDPQDYPDKKSLPHVHVALWINSQGGRKVKAGDTVSYVICQDGSNLTASQRAYAPEQLQKQDNLTIDTQYYLAQQIHPVVARICEPID GIDAVLIATWLGLDPTQFRVHHYHKDEENDALLGGPAQLTDEEKYRDCERFKCPCPTCGTENIYDNVFDGSGTDMEPSLYRCSNIDCKAS PLTFTVQLSNKLIMDIRRFIKKYYDGWLICEEPTCRNRTRHLPLQFSRTGPLCPACMKATLQPEYSDKSLYTQLCFYRYIFDAECALEKL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for OGT-POLA1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for OGT-POLA1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for OGT-POLA1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
