
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:PABPC1-KHDRBS3 (FusionGDB2 ID:62190) |
Fusion Gene Summary for PABPC1-KHDRBS3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: PABPC1-KHDRBS3 | Fusion gene ID: 62190 | Hgene | Tgene | Gene symbol | PABPC1 | KHDRBS3 | Gene ID | 26986 | 10656 |
| Gene name | poly(A) binding protein cytoplasmic 1 | KH RNA binding domain containing, signal transduction associated 3 | |
| Synonyms | PAB1|PABP|PABP1|PABPC2|PABPL1 | Etle|SALP|SLM-2|SLM2|T-STAR|TSTAR|etoile | |
| Cytomap | 8q22.3 | 8q24.23 | |
| Type of gene | protein-coding | protein-coding | |
| Description | polyadenylate-binding protein 1poly(A) binding protein, cytoplasmic 2 | KH domain-containing, RNA-binding, signal transduction-associated protein 3KH domain containing, RNA binding, signal transduction associated 3RNA-binding protein T-StarSam68-like phosphotyrosine protein, T-STARsam68-like mammalian protein 2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | O75525 | |
| Ensembl transtripts involved in fusion gene | ENST00000318607, ENST00000519004, ENST00000522387, ENST00000519596, | ENST00000520981, ENST00000522578, ENST00000355849, | |
| Fusion gene scores | * DoF score | 31 X 28 X 11=9548 | 11 X 10 X 10=1100 |
| # samples | 37 | 14 | |
| ** MAII score | log2(37/9548*10)=-4.68960139056546 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(14/1100*10)=-2.97400479146706 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: PABPC1 [Title/Abstract] AND KHDRBS3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | PABPC1(101724880)-KHDRBS3(136554897), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | PABPC1 | GO:2000623 | negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 18447585 |
 Fusion gene breakpoints across PABPC1 (5'-gene) Fusion gene breakpoints across PABPC1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
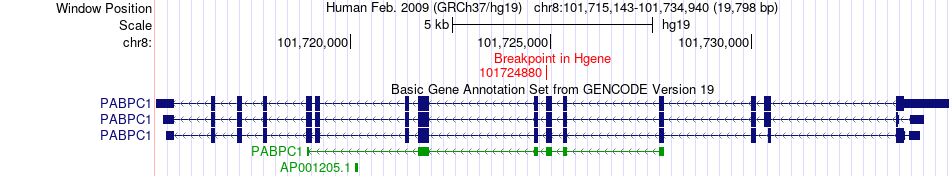 |
 Fusion gene breakpoints across KHDRBS3 (3'-gene) Fusion gene breakpoints across KHDRBS3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
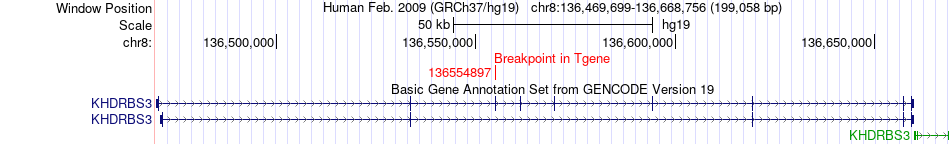 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LGG | TCGA-DH-A66G-01A | PABPC1 | chr8 | 101724880 | - | KHDRBS3 | chr8 | 136554897 | + |
| ChimerDB4 | LGG | TCGA-DH-A66G | PABPC1 | chr8 | 101724880 | - | KHDRBS3 | chr8 | 136554897 | + |
Top |
Fusion Gene ORF analysis for PABPC1-KHDRBS3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000318607 | ENST00000520981 | PABPC1 | chr8 | 101724880 | - | KHDRBS3 | chr8 | 136554897 | + |
| 5CDS-intron | ENST00000318607 | ENST00000522578 | PABPC1 | chr8 | 101724880 | - | KHDRBS3 | chr8 | 136554897 | + |
| 5CDS-intron | ENST00000519004 | ENST00000520981 | PABPC1 | chr8 | 101724880 | - | KHDRBS3 | chr8 | 136554897 | + |
| 5CDS-intron | ENST00000519004 | ENST00000522578 | PABPC1 | chr8 | 101724880 | - | KHDRBS3 | chr8 | 136554897 | + |
| 5CDS-intron | ENST00000522387 | ENST00000520981 | PABPC1 | chr8 | 101724880 | - | KHDRBS3 | chr8 | 136554897 | + |
| 5CDS-intron | ENST00000522387 | ENST00000522578 | PABPC1 | chr8 | 101724880 | - | KHDRBS3 | chr8 | 136554897 | + |
| 5UTR-3CDS | ENST00000519596 | ENST00000355849 | PABPC1 | chr8 | 101724880 | - | KHDRBS3 | chr8 | 136554897 | + |
| 5UTR-intron | ENST00000519596 | ENST00000520981 | PABPC1 | chr8 | 101724880 | - | KHDRBS3 | chr8 | 136554897 | + |
| 5UTR-intron | ENST00000519596 | ENST00000522578 | PABPC1 | chr8 | 101724880 | - | KHDRBS3 | chr8 | 136554897 | + |
| In-frame | ENST00000318607 | ENST00000355849 | PABPC1 | chr8 | 101724880 | - | KHDRBS3 | chr8 | 136554897 | + |
| In-frame | ENST00000519004 | ENST00000355849 | PABPC1 | chr8 | 101724880 | - | KHDRBS3 | chr8 | 136554897 | + |
| In-frame | ENST00000522387 | ENST00000355849 | PABPC1 | chr8 | 101724880 | - | KHDRBS3 | chr8 | 136554897 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000318607 | PABPC1 | chr8 | 101724880 | - | ENST00000355849 | KHDRBS3 | chr8 | 136554897 | + | 3365 | 2005 | 1129 | 2838 | 569 |
| ENST00000519004 | PABPC1 | chr8 | 101724880 | - | ENST00000355849 | KHDRBS3 | chr8 | 136554897 | + | 2464 | 1104 | 363 | 1937 | 524 |
| ENST00000522387 | PABPC1 | chr8 | 101724880 | - | ENST00000355849 | KHDRBS3 | chr8 | 136554897 | + | 2456 | 1096 | 316 | 1929 | 537 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000318607 | ENST00000355849 | PABPC1 | chr8 | 101724880 | - | KHDRBS3 | chr8 | 136554897 | + | 0.000732253 | 0.99926776 |
| ENST00000519004 | ENST00000355849 | PABPC1 | chr8 | 101724880 | - | KHDRBS3 | chr8 | 136554897 | + | 0.000523238 | 0.99947673 |
| ENST00000522387 | ENST00000355849 | PABPC1 | chr8 | 101724880 | - | KHDRBS3 | chr8 | 136554897 | + | 0.000205768 | 0.9997943 |
Top |
Fusion Genomic Features for PABPC1-KHDRBS3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| PABPC1 | chr8 | 101724879 | - | KHDRBS3 | chr8 | 136554896 | + | 3.91E-09 | 1 |
| PABPC1 | chr8 | 101724879 | - | KHDRBS3 | chr8 | 136554896 | + | 3.91E-09 | 1 |
| PABPC1 | chr8 | 101724879 | - | KHDRBS3 | chr8 | 136554896 | + | 3.91E-09 | 1 |
| PABPC1 | chr8 | 101724879 | - | KHDRBS3 | chr8 | 136554896 | + | 3.91E-09 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for PABPC1-KHDRBS3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:101724880/chr8:136554897) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | KHDRBS3 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: RNA-binding protein that plays a role in the regulation of alternative splicing and influences mRNA splice site selection and exon inclusion. Binds preferentially to the 5'-[AU]UAAA-3' motif in vitro. Binds optimally to RNA containing 5'-[AU]UAA-3' as a bipartite motif spaced by more than 15 nucleotides. Binds poly(A). RNA-binding abilities are down-regulated by tyrosine kinase PTK6 (PubMed:10564820, PubMed:19561594, PubMed:26758068). Involved in splice site selection of vascular endothelial growth factor (PubMed:15901763). In vitro regulates CD44 alternative splicing by direct binding to purine-rich exonic enhancer (By similarity). Can regulate alternative splicing of neurexins NRXN1-3 in the laminin G-like domain 6 containing the evolutionary conserved neurexin alternative spliced segment 4 (AS4) involved in neurexin selective targeting to postsynaptic partners such as neuroligins and LRRTM family members (PubMed:26758068). Targeted, cell-type specific splicing regulation of NRXN1 at AS4 is involved in neuronal glutamatergic synapse function and plasticity (By similarity). May regulate expression of KHDRBS2/SLIM-1 in defined brain neuron populations by modifying its alternative splicing (By similarity). Can bind FABP9 mRNA (By similarity). May play a role as a negative regulator of cell growth. Inhibits cell proliferation. {ECO:0000250|UniProtKB:Q9JLP1, ECO:0000250|UniProtKB:Q9R226, ECO:0000269|PubMed:10564820, ECO:0000269|PubMed:15901763, ECO:0000269|PubMed:19561594, ECO:0000269|PubMed:26758068}.; FUNCTION: (Microbial infection) Involved in post-transcriptional regulation of HIV-1 gene expression. {ECO:0000269|PubMed:11741900}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PABPC1 | chr8:101724880 | chr8:136554897 | ENST00000318607 | - | 6 | 15 | 11_89 | 292 | 311.0 | Domain | RRM 1 |
| Hgene | PABPC1 | chr8:101724880 | chr8:136554897 | ENST00000318607 | - | 6 | 15 | 191_268 | 292 | 311.0 | Domain | RRM 3 |
| Hgene | PABPC1 | chr8:101724880 | chr8:136554897 | ENST00000318607 | - | 6 | 15 | 99_175 | 292 | 311.0 | Domain | RRM 2 |
| Hgene | PABPC1 | chr8:101724880 | chr8:136554897 | ENST00000318607 | - | 6 | 15 | 166_289 | 292 | 311.0 | Region | Note=CSDE1-binding |
| Tgene | KHDRBS3 | chr8:101724880 | chr8:136554897 | ENST00000355849 | 1 | 9 | 250_261 | 69 | 347.0 | Compositional bias | Note=Pro-rich | |
| Tgene | KHDRBS3 | chr8:101724880 | chr8:136554897 | ENST00000355849 | 1 | 9 | 266_316 | 69 | 347.0 | Compositional bias | Note=Tyr-rich |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PABPC1 | chr8:101724880 | chr8:136554897 | ENST00000318607 | - | 6 | 15 | 495_501 | 292 | 311.0 | Compositional bias | Note=Poly-Ala |
| Hgene | PABPC1 | chr8:101724880 | chr8:136554897 | ENST00000318607 | - | 6 | 15 | 294_370 | 292 | 311.0 | Domain | RRM 4 |
| Hgene | PABPC1 | chr8:101724880 | chr8:136554897 | ENST00000318607 | - | 6 | 15 | 542_619 | 292 | 311.0 | Domain | PABC |
| Hgene | PABPC1 | chr8:101724880 | chr8:136554897 | ENST00000318607 | - | 6 | 15 | 541_636 | 292 | 311.0 | Region | (Microbial infection) Binding to HRSV M2-1 protein |
| Tgene | KHDRBS3 | chr8:101724880 | chr8:136554897 | ENST00000355849 | 1 | 9 | 61_127 | 69 | 347.0 | Domain | Note=KH | |
| Tgene | KHDRBS3 | chr8:101724880 | chr8:136554897 | ENST00000355849 | 1 | 9 | 1_160 | 69 | 347.0 | Region | Involved in homodimerization |
Top |
Fusion Gene Sequence for PABPC1-KHDRBS3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >62190_62190_1_PABPC1-KHDRBS3_PABPC1_chr8_101724880_ENST00000318607_KHDRBS3_chr8_136554897_ENST00000355849_length(transcript)=3365nt_BP=2005nt CGTAGTGCGGTCCGGGTAGGAGAGAGCAGGGGGCCACGCTGGCTCCATCTACGCACGTTTCGGAAACCGGGCCCGGAGGGGACCACGCCC GCTCACCCGTGCCCGGCGTCCCCCGGAGCTGGGGCTAGAGGAGCCGGGCCGGCCGCAGCGGGAAGAGGCCACAGGCGGCCGCGCACGTGG GGGGTGGTTAGCGCGGAGGAACCCGGAAGCCCTTTGCACCGCCCACCGCCGCGGTGACGGGTTAACGCTCCTCCTGGTGGAGGCAGGGGC GGGCGGGGCGGAAGGCGGGGACGCCTCAGCCAACACCCCCGAGGACCGCCCGCGAGCCGTCCTCCCCACCCCCACGGACGCGACGCAGCG CGGGCTCCGCTTCCCCGGCCGCGGCGCCTGCGCAAAAGCCCCGCCCCTTCGCCCGAGCCCCACCCCCACACCGCAGCCTCCACCGCCGCC ACCTCTCCCTTCCTCTCTGCTCTTTCCTCCTGTTTTCTCTTCCCTCCTCCCCCTGGCCTCCGCGTCTCCTCCTACTCCGGCGCTGACGCT CGCGTAGGGGCCCTGGCGTCAGACGCGCGGGGGCGGGGCGAGTGCGGCGCGGGGTATAAGTAGAGGGTGCAGGAGGCGGTGCTTCCCCTT CTCCCCGGCGGTTAGTGCTGAGAGTGCGGAGTGTGTGCTCCGGGCTCGGAACACACATTTATTATTAAAAAATCCAAAAAAAATCTAAAA AAATCTTTTAAAAAACCCCAAAAAAATTTACAAAAAATCCGCGTCTCCCCCGCCGGAGACTTTTATTTTTTTTCTTCCTCTTTTATAAAA TAACCCGGTGAAGCAGCCGAGACCGACCCGCCCGCCCGCGGCCCCGCAGCAGCTCCAAGAAGGAACCAAGAGACCGAGGCCTTCCCGCTG CCCGGACCCGACACCGCCACCCTCGCTCCCCGCCGGCAGCCGGCAGCCAGCGGCAGTGGATCGACCCCGTTCTGCGGCCGTTGAGTAGTT TTCAATTCCGGTTGATTTTTGTCCCTCTGCGCTTGCTCCCCGCTCCCCTCCCCCCGGCTCCGGCCCCCAGCCCCGGCACTCGCTCTCCTC CTCTCACGGAAAGGTCGCGGCCTGTGGCCCTGCGGGCAGCCGTGCCGAGATGAACCCCAGTGCCCCCAGCTACCCCATGGCCTCGCTCTA CGTGGGGGACCTCCACCCCGACGTGACCGAGGCGATGCTCTACGAGAAGTTCAGCCCGGCCGGGCCCATCCTCTCCATCCGGGTCTGCAG GGACATGATCACCCGCCGCTCCTTGGGCTACGCGTATGTGAACTTCCAGCAGCCGGCGGACGCGGAGCGTGCTTTGGACACCATGAATTT TGATGTTATAAAGGGCAAGCCAGTACGCATCATGTGGTCTCAGCGTGATCCATCACTTCGCAAAAGTGGAGTAGGCAACATATTCATTAA AAATCTGGACAAATCCATTGATAATAAAGCACTGTATGATACATTTTCTGCTTTTGGTAACATCCTTTCATGTAAGGTGGTTTGTGATGA AAATGGTTCCAAGGGCTATGGATTTGTACACTTTGAGACGCAGGAAGCAGCTGAAAGAGCTATTGAAAAAATGAATGGAATGCTCCTAAA TGATCGCAAAGTATTTGTTGGACGATTTAAGTCTCGTAAAGAACGAGAAGCTGAACTTGGAGCTAGGGCAAAAGAATTCACCAATGTTTA CATCAAGAATTTTGGAGAAGACATGGATGATGAGCGCCTTAAGGATCTCTTTGGCAAGTTTGGGCCTGCCTTAAGTGTGAAAGTAATGAC TGATGAAAGTGGAAAATCCAAAGGATTTGGATTTGTAAGCTTTGAAAGGCATGAAGATGCACAGAAAGCTGTGGATGAGATGAACGGAAA GGAGCTCAATGGAAAACAAATTTATGTTGGTCGAGCTCAGAAAAAGGTGGAACGGCAGACGGAACTTAAGCGCAAATTTGAACAGATGAA ACAAGATAGGATCACCAGATACCAGTTCAACTTTGTGGGGAAACTTTTGGGTCCACGTGGCAATTCTCTGAAGCGTTTACAAGAAGAAAC CTTGACAAAAATGTCCATCCTTGGGAAAGGTTCCATGAGAGACAAGGCCAAGGAAGAAGAGTTGAGGAAAAGTGGAGAAGCGAAGTACTT CCATCTCAATGATGATCTCCATGTTCTCATTGAAGTGTTTGCCCCACCTGCAGAAGCTTATGCCAGGATGGGACATGCTTTGGAAGAAAT CAAAAAGTTCCTCATCCCTGATTATAATGATGAGATCAGGCAAGCACAGCTCCAGGAGTTAACATATTTGAATGGTGGTTCAGAAAATGC AGATGTTCCAGTGGTTCGAGGGAAACCCACCTTGCGTACAAGAGGTGTACCAGCCCCAGCAATAACCAGGGGAAGGGGAGGAGTTACAGC CCGGCCAGTTGGAGTTGTAGTACCACGAGGGACGCCAACTCCCAGAGGAGTCCTGTCCACCCGAGGGCCAGTGAGTCGGGGAAGAGGACT TCTCACTCCCAGAGCAAGAGGAGTCCCCCCAACTGGGTACAGACCTCCACCGCCACCCCCGACACAAGAGACTTATGGAGAATATGACTA TGATGATGGATATGGCACTGCTTATGATGAACAGAGTTATGATTCCTATGATAACAGCTATAGCACCCCAGCCCAAAGTGGTGCTGATTA CTATGATTACGGACATGGACTCAGTGAGGAGACTTATGATTCCTACGGGCAAGAAGAGTGGACTAACTCAAGACACAAGGCACCTTCAGC GAGGACAGCAAAGGGCGTCTACAGAGACCAGCCATATGGCAGATACTGATTGTACTGTCTGATGTTGTGAAATAGCCAATCTCCACCAGT CCTGTATACTGTTCAAAGTAATTTTTTTCTATGAACAATCCCTTTTTAAATAAATCAGAATGCTTAAAATCTGAATGGATGGAACTTAAA ACTACTTTGTTGAAACATCAACCTGGGCAGAAAAAAAAAAAAAAAGACATGTAAAATTTTGTTATTTCCAGTCTGTATATGAAAAAATAG GTCATCAAAAGGAAAAAAAATAACTTTGATTAACTAGTGTTAAACAAAAAATAGGTTTACTAAATATGTTAATCTATTCTTTTAACATAA GCCTCACCTTTCATTTTAAAGGTTTCCATAGAATTTAGTTATTTTATCTTTCAGCCATATGCTAGTTTTTTTTTCTCTTGCCAACATGCG TAAAAAGGGAAGCCAATTACAAGTGCAAATAATGTGGTATTCTTTTGTAACTCAAGTCTTGAAATGTTCTGTAGTGTTAAGCAAAGTCTC >62190_62190_1_PABPC1-KHDRBS3_PABPC1_chr8_101724880_ENST00000318607_KHDRBS3_chr8_136554897_ENST00000355849_length(amino acids)=569AA_BP=292 MNPSAPSYPMASLYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQPADAERALDTMNFDVIKGKPVRIMWSQRD PSLRKSGVGNIFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGSKGYGFVHFETQEAAERAIEKMNGMLLNDRKVFVGRFKSRKERE AELGARAKEFTNVYIKNFGEDMDDERLKDLFGKFGPALSVKVMTDESGKSKGFGFVSFERHEDAQKAVDEMNGKELNGKQIYVGRAQKKV ERQTELKRKFEQMKQDRITRYQFNFVGKLLGPRGNSLKRLQEETLTKMSILGKGSMRDKAKEEELRKSGEAKYFHLNDDLHVLIEVFAPP AEAYARMGHALEEIKKFLIPDYNDEIRQAQLQELTYLNGGSENADVPVVRGKPTLRTRGVPAPAITRGRGGVTARPVGVVVPRGTPTPRG VLSTRGPVSRGRGLLTPRARGVPPTGYRPPPPPPTQETYGEYDYDDGYGTAYDEQSYDSYDNSYSTPAQSGADYYDYGHGLSEETYDSYG -------------------------------------------------------------- >62190_62190_2_PABPC1-KHDRBS3_PABPC1_chr8_101724880_ENST00000519004_KHDRBS3_chr8_136554897_ENST00000355849_length(transcript)=2464nt_BP=1104nt CCCTTCTCCCCGGCGGTTAGTGCTGAGAGTGCGGAGTGTGTGCTCCGGGCTCGGAACACACATTTATTATTAAAAAATCCAAAAAAAATC TAAAAAAATCTTTTAAAAAACCCCAAAAAAATTTACAAAAAATCCGCGTCTCCCCCGCCGGAGACTTTTATTTTTTTTCTTCCTCTTTTA TAAAATAACCCGGTGAAGCAGCCGAGACCGACCCGCCCGCCCGCGGCCCCGCAGCAGCTCCAAGAAGGAACCAAGAGACCGAGGCCTTCC CGCTGCCCGGACCCGACACCGCCACCCTCGCTCCCCGCCGGCAGCCGGCAGCCAGCGGCAGTGGATCGACCCCGTTCTGCGGCCGTTGAG GACATGATCACCCGCCGCTCCTTGGGCTACGCGTATGTGAACTTCCAGCAGCCGGCGGACGCGGAGCGTGCTTTGGACACCATGAATTTT GATGTTATAAAGGGCAAGCCAGTACGCATCATGTGGTCTCAGCGTGATCCATCACTTCGCAAAAGTGGAGTAGGCAACATATTCATTAAA AATCTGGACAAATCCATTGATAATAAAGCACTGTATGATACATTTTCTGCTTTTGGTAACATCCTTTCATGTAAGGTGGTTTGTGATGAA AATGGTTCCAAGGGCTATGGATTTGTACACTTTGAGACGCAGGAAGCAGCTGAAAGAGCTATTGAAAAAATGAATGGAATGCTCCTAAAT GATCGCAAAGTATTTGTTGGACGATTTAAGTCTCGTAAAGAACGAGAAGCTGAACTTGGAGCTAGGGCAAAAGAATTCACCAATGTTTAC ATCAAGAATTTTGGAGAAGACATGGATGATGAGCGCCTTAAGGATCTCTTTGGCAAGTTTGGGCCTGCCTTAAGTGTGAAAGTAATGACT GATGAAAGTGGAAAATCCAAAGGATTTGGATTTGTAAGCTTTGAAAGGCATGAAGATGCACAGAAAGCTGTGGATGAGATGAACGGAAAG GAGCTCAATGGAAAACAAATTTATGTTGGTCGAGCTCAGAAAAAGGTGGAACGGCAGACGGAACTTAAGCGCAAATTTGAACAGATGAAA CAAGATAGGATCACCAGATACCAGTTCAACTTTGTGGGGAAACTTTTGGGTCCACGTGGCAATTCTCTGAAGCGTTTACAAGAAGAAACC TTGACAAAAATGTCCATCCTTGGGAAAGGTTCCATGAGAGACAAGGCCAAGGAAGAAGAGTTGAGGAAAAGTGGAGAAGCGAAGTACTTC CATCTCAATGATGATCTCCATGTTCTCATTGAAGTGTTTGCCCCACCTGCAGAAGCTTATGCCAGGATGGGACATGCTTTGGAAGAAATC AAAAAGTTCCTCATCCCTGATTATAATGATGAGATCAGGCAAGCACAGCTCCAGGAGTTAACATATTTGAATGGTGGTTCAGAAAATGCA GATGTTCCAGTGGTTCGAGGGAAACCCACCTTGCGTACAAGAGGTGTACCAGCCCCAGCAATAACCAGGGGAAGGGGAGGAGTTACAGCC CGGCCAGTTGGAGTTGTAGTACCACGAGGGACGCCAACTCCCAGAGGAGTCCTGTCCACCCGAGGGCCAGTGAGTCGGGGAAGAGGACTT CTCACTCCCAGAGCAAGAGGAGTCCCCCCAACTGGGTACAGACCTCCACCGCCACCCCCGACACAAGAGACTTATGGAGAATATGACTAT GATGATGGATATGGCACTGCTTATGATGAACAGAGTTATGATTCCTATGATAACAGCTATAGCACCCCAGCCCAAAGTGGTGCTGATTAC TATGATTACGGACATGGACTCAGTGAGGAGACTTATGATTCCTACGGGCAAGAAGAGTGGACTAACTCAAGACACAAGGCACCTTCAGCG AGGACAGCAAAGGGCGTCTACAGAGACCAGCCATATGGCAGATACTGATTGTACTGTCTGATGTTGTGAAATAGCCAATCTCCACCAGTC CTGTATACTGTTCAAAGTAATTTTTTTCTATGAACAATCCCTTTTTAAATAAATCAGAATGCTTAAAATCTGAATGGATGGAACTTAAAA CTACTTTGTTGAAACATCAACCTGGGCAGAAAAAAAAAAAAAAAGACATGTAAAATTTTGTTATTTCCAGTCTGTATATGAAAAAATAGG TCATCAAAAGGAAAAAAAATAACTTTGATTAACTAGTGTTAAACAAAAAATAGGTTTACTAAATATGTTAATCTATTCTTTTAACATAAG CCTCACCTTTCATTTTAAAGGTTTCCATAGAATTTAGTTATTTTATCTTTCAGCCATATGCTAGTTTTTTTTTCTCTTGCCAACATGCGT AAAAAGGGAAGCCAATTACAAGTGCAAATAATGTGGTATTCTTTTGTAACTCAAGTCTTGAAATGTTCTGTAGTGTTAAGCAAAGTCTCC >62190_62190_2_PABPC1-KHDRBS3_PABPC1_chr8_101724880_ENST00000519004_KHDRBS3_chr8_136554897_ENST00000355849_length(amino acids)=524AA_BP=247 MITRRSLGYAYVNFQQPADAERALDTMNFDVIKGKPVRIMWSQRDPSLRKSGVGNIFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDEN GSKGYGFVHFETQEAAERAIEKMNGMLLNDRKVFVGRFKSRKEREAELGARAKEFTNVYIKNFGEDMDDERLKDLFGKFGPALSVKVMTD ESGKSKGFGFVSFERHEDAQKAVDEMNGKELNGKQIYVGRAQKKVERQTELKRKFEQMKQDRITRYQFNFVGKLLGPRGNSLKRLQEETL TKMSILGKGSMRDKAKEEELRKSGEAKYFHLNDDLHVLIEVFAPPAEAYARMGHALEEIKKFLIPDYNDEIRQAQLQELTYLNGGSENAD VPVVRGKPTLRTRGVPAPAITRGRGGVTARPVGVVVPRGTPTPRGVLSTRGPVSRGRGLLTPRARGVPPTGYRPPPPPPTQETYGEYDYD -------------------------------------------------------------- >62190_62190_3_PABPC1-KHDRBS3_PABPC1_chr8_101724880_ENST00000522387_KHDRBS3_chr8_136554897_ENST00000355849_length(transcript)=2456nt_BP=1096nt AAATCTTTTAAAAAACCCCAAAAAAATTTACAAAAAATCCGCGTCTCCCCCGCCGGAGACTTTTATTTTTTTTCTTCCTCTTTTATAAAA TAACCCGGTGAAGCAGCCGAGACCGACCCGCCCGCCCGCGGCCCCGCAGCAGCTCCAAGAAGGAACCAAGAGACCGAGGCCTTCCCGCTG CCCGGACCCGACACCGCCACCCTCGCTCCCCGCCGGCAGCCGGCAGCCAGCGGCAGTGGATCGACCCCGTTCTGCGGCCGTTGAGTAGTT TTCAATTCCGGTCGCGGCCTGTGGCCCTGCGGGCAGCCGTGCCGAGATGAACCCCAGTGCCCCCAGCTACCCCATGGCCTCGCTCTACGT GGGGGACCTCCACCCCGACGTGACCGAGGCGATGCTCTACGAGAAGTTCAGCCCGGCCGGGCCCATCCTCTCCATCCGGGTCTGCAGGGA CATGATCACCCGCCGCTCCTTGGGCTACGCGTATGTGAACTTCCAGCAGCCGGCGGACGCGGAGCGTGCTTTGGACACCATGAATTTTGA TGTTATAAAGGGCAAGCCAGTACGCATCATGTGGTCTCAGCGTGATCCATCACTTCGCAAAAGTGGAGTGGTTTGTGATGAAAATGGTTC CAAGGGCTATGGATTTGTACACTTTGAGACGCAGGAAGCAGCTGAAAGAGCTATTGAAAAAATGAATGGAATGCTCCTAAATGATCGCAA AGTATTTGTTGGACGATTTAAGTCTCGTAAAGAACGAGAAGCTGAACTTGGAGCTAGGGCAAAAGAATTCACCAATGTTTACATCAAGAA TTTTGGAGAAGACATGGATGATGAGCGCCTTAAGGATCTCTTTGGCAAGTTTGGGCCTGCCTTAAGTGTGAAAGTAATGACTGATGAAAG TGGAAAATCCAAAGGATTTGGATTTGTAAGCTTTGAAAGGCATGAAGATGCACAGAAAGCTGTGGATGAGATGAACGGAAAGGAGCTCAA TGGAAAACAAATTTATGTTGGTCGAGCTCAGAAAAAGGTGGAACGGCAGACGGAACTTAAGCGCAAATTTGAACAGATGAAACAAGATAG GATCACCAGATACCAGTTCAACTTTGTGGGGAAACTTTTGGGTCCACGTGGCAATTCTCTGAAGCGTTTACAAGAAGAAACCTTGACAAA AATGTCCATCCTTGGGAAAGGTTCCATGAGAGACAAGGCCAAGGAAGAAGAGTTGAGGAAAAGTGGAGAAGCGAAGTACTTCCATCTCAA TGATGATCTCCATGTTCTCATTGAAGTGTTTGCCCCACCTGCAGAAGCTTATGCCAGGATGGGACATGCTTTGGAAGAAATCAAAAAGTT CCTCATCCCTGATTATAATGATGAGATCAGGCAAGCACAGCTCCAGGAGTTAACATATTTGAATGGTGGTTCAGAAAATGCAGATGTTCC AGTGGTTCGAGGGAAACCCACCTTGCGTACAAGAGGTGTACCAGCCCCAGCAATAACCAGGGGAAGGGGAGGAGTTACAGCCCGGCCAGT TGGAGTTGTAGTACCACGAGGGACGCCAACTCCCAGAGGAGTCCTGTCCACCCGAGGGCCAGTGAGTCGGGGAAGAGGACTTCTCACTCC CAGAGCAAGAGGAGTCCCCCCAACTGGGTACAGACCTCCACCGCCACCCCCGACACAAGAGACTTATGGAGAATATGACTATGATGATGG ATATGGCACTGCTTATGATGAACAGAGTTATGATTCCTATGATAACAGCTATAGCACCCCAGCCCAAAGTGGTGCTGATTACTATGATTA CGGACATGGACTCAGTGAGGAGACTTATGATTCCTACGGGCAAGAAGAGTGGACTAACTCAAGACACAAGGCACCTTCAGCGAGGACAGC AAAGGGCGTCTACAGAGACCAGCCATATGGCAGATACTGATTGTACTGTCTGATGTTGTGAAATAGCCAATCTCCACCAGTCCTGTATAC TGTTCAAAGTAATTTTTTTCTATGAACAATCCCTTTTTAAATAAATCAGAATGCTTAAAATCTGAATGGATGGAACTTAAAACTACTTTG TTGAAACATCAACCTGGGCAGAAAAAAAAAAAAAAAGACATGTAAAATTTTGTTATTTCCAGTCTGTATATGAAAAAATAGGTCATCAAA AGGAAAAAAAATAACTTTGATTAACTAGTGTTAAACAAAAAATAGGTTTACTAAATATGTTAATCTATTCTTTTAACATAAGCCTCACCT TTCATTTTAAAGGTTTCCATAGAATTTAGTTATTTTATCTTTCAGCCATATGCTAGTTTTTTTTTCTCTTGCCAACATGCGTAAAAAGGG AAGCCAATTACAAGTGCAAATAATGTGGTATTCTTTTGTAACTCAAGTCTTGAAATGTTCTGTAGTGTTAAGCAAAGTCTCCTCTTGCTT >62190_62190_3_PABPC1-KHDRBS3_PABPC1_chr8_101724880_ENST00000522387_KHDRBS3_chr8_136554897_ENST00000355849_length(amino acids)=537AA_BP=260 MNPSAPSYPMASLYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQPADAERALDTMNFDVIKGKPVRIMWSQRD PSLRKSGVVCDENGSKGYGFVHFETQEAAERAIEKMNGMLLNDRKVFVGRFKSRKEREAELGARAKEFTNVYIKNFGEDMDDERLKDLFG KFGPALSVKVMTDESGKSKGFGFVSFERHEDAQKAVDEMNGKELNGKQIYVGRAQKKVERQTELKRKFEQMKQDRITRYQFNFVGKLLGP RGNSLKRLQEETLTKMSILGKGSMRDKAKEEELRKSGEAKYFHLNDDLHVLIEVFAPPAEAYARMGHALEEIKKFLIPDYNDEIRQAQLQ ELTYLNGGSENADVPVVRGKPTLRTRGVPAPAITRGRGGVTARPVGVVVPRGTPTPRGVLSTRGPVSRGRGLLTPRARGVPPTGYRPPPP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for PABPC1-KHDRBS3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for PABPC1-KHDRBS3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for PABPC1-KHDRBS3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
