
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:PAK1-ARHGEF18 (FusionGDB2 ID:62397) |
Fusion Gene Summary for PAK1-ARHGEF18 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: PAK1-ARHGEF18 | Fusion gene ID: 62397 | Hgene | Tgene | Gene symbol | PAK1 | ARHGEF18 | Gene ID | 5585 | 23370 |
| Gene name | protein kinase N1 | Rho/Rac guanine nucleotide exchange factor 18 | |
| Synonyms | DBK|PAK-1|PAK1|PKN|PKN-ALPHA|PRK1|PRKCL1 | P114-RhoGEF|RP78|SA-RhoGEF|p114RhoGEF | |
| Cytomap | 19p13.12 | 19p13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | serine/threonine-protein kinase N1protease-activated kinase 1protein kinase C-like 1protein kinase C-like PKNprotein kinase C-related kinase 1protein kinase PKN-alphaserine-threonine kinase Nserine/threonine protein kinase N | rho guanine nucleotide exchange factor 18114 kDa Rho-specific guanine nucleotide exchange factorRho-specific guanine nucleotide exchange factor p114Rho/Rac guanine nucleotide exchange factor (GEF) 18septin-associated RhoGEF | |
| Modification date | 20200329 | 20200320 | |
| UniProtAcc | . | Q6ZSZ5 | |
| Ensembl transtripts involved in fusion gene | ENST00000525542, ENST00000278568, ENST00000356341, ENST00000528203, ENST00000530617, | ENST00000319670, ENST00000359920, | |
| Fusion gene scores | * DoF score | 31 X 18 X 9=5022 | 8 X 8 X 2=128 |
| # samples | 36 | 8 | |
| ** MAII score | log2(36/5022*10)=-3.80219321694183 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(8/128*10)=-0.678071905112638 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: PAK1 [Title/Abstract] AND ARHGEF18 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | PAK1(77043775)-ARHGEF18(7521213), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | PAK1-ARHGEF18 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. PAK1-ARHGEF18 seems lost the major protein functional domain in Hgene partner, which is a kinase due to the frame-shifted ORF. PAK1-ARHGEF18 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | PAK1 | GO:0006357 | regulation of transcription by RNA polymerase II | 12514133 |
| Hgene | PAK1 | GO:0006468 | protein phosphorylation | 17332740 |
| Hgene | PAK1 | GO:0035407 | histone H3-T11 phosphorylation | 18066052 |
| Tgene | ARHGEF18 | GO:0007264 | small GTPase mediated signal transduction | 14512443 |
| Tgene | ARHGEF18 | GO:0008360 | regulation of cell shape | 14512443 |
| Tgene | ARHGEF18 | GO:0030036 | actin cytoskeleton organization | 14512443 |
 Fusion gene breakpoints across PAK1 (5'-gene) Fusion gene breakpoints across PAK1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
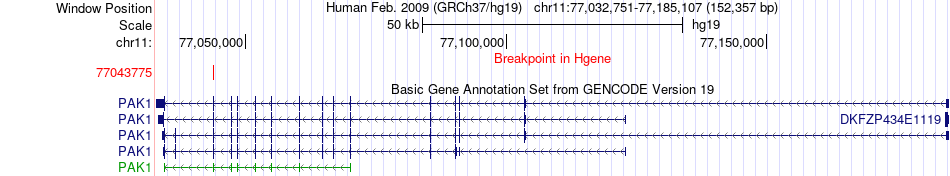 |
 Fusion gene breakpoints across ARHGEF18 (3'-gene) Fusion gene breakpoints across ARHGEF18 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
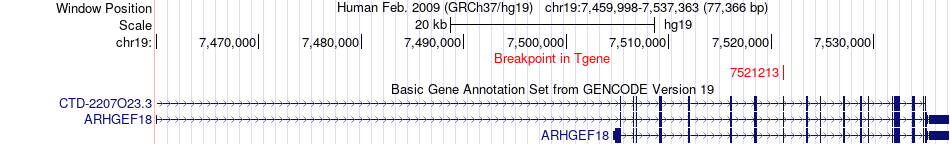 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-AQ-A04L-01B | PAK1 | chr11 | 77043775 | - | ARHGEF18 | chr19 | 7521213 | + |
Top |
Fusion Gene ORF analysis for PAK1-ARHGEF18 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5UTR-3CDS | ENST00000525542 | ENST00000319670 | PAK1 | chr11 | 77043775 | - | ARHGEF18 | chr19 | 7521213 | + |
| 5UTR-3CDS | ENST00000525542 | ENST00000359920 | PAK1 | chr11 | 77043775 | - | ARHGEF18 | chr19 | 7521213 | + |
| Frame-shift | ENST00000278568 | ENST00000319670 | PAK1 | chr11 | 77043775 | - | ARHGEF18 | chr19 | 7521213 | + |
| Frame-shift | ENST00000278568 | ENST00000359920 | PAK1 | chr11 | 77043775 | - | ARHGEF18 | chr19 | 7521213 | + |
| Frame-shift | ENST00000356341 | ENST00000319670 | PAK1 | chr11 | 77043775 | - | ARHGEF18 | chr19 | 7521213 | + |
| Frame-shift | ENST00000356341 | ENST00000359920 | PAK1 | chr11 | 77043775 | - | ARHGEF18 | chr19 | 7521213 | + |
| Frame-shift | ENST00000528203 | ENST00000319670 | PAK1 | chr11 | 77043775 | - | ARHGEF18 | chr19 | 7521213 | + |
| Frame-shift | ENST00000528203 | ENST00000359920 | PAK1 | chr11 | 77043775 | - | ARHGEF18 | chr19 | 7521213 | + |
| In-frame | ENST00000530617 | ENST00000319670 | PAK1 | chr11 | 77043775 | - | ARHGEF18 | chr19 | 7521213 | + |
| In-frame | ENST00000530617 | ENST00000359920 | PAK1 | chr11 | 77043775 | - | ARHGEF18 | chr19 | 7521213 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000530617 | PAK1 | chr11 | 77043775 | - | ENST00000319670 | ARHGEF18 | chr19 | 7521213 | + | 5803 | 1863 | 1868 | 3844 | 658 |
| ENST00000530617 | PAK1 | chr11 | 77043775 | - | ENST00000359920 | ARHGEF18 | chr19 | 7521213 | + | 5803 | 1863 | 1868 | 3844 | 658 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000530617 | ENST00000319670 | PAK1 | chr11 | 77043775 | - | ARHGEF18 | chr19 | 7521213 | + | 0.016259726 | 0.98374027 |
| ENST00000530617 | ENST00000359920 | PAK1 | chr11 | 77043775 | - | ARHGEF18 | chr19 | 7521213 | + | 0.016259726 | 0.98374027 |
Top |
Fusion Genomic Features for PAK1-ARHGEF18 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| PAK1 | chr11 | 77043774 | - | ARHGEF18 | chr19 | 7521212 | + | 2.14E-05 | 0.99997866 |
| PAK1 | chr11 | 77043774 | - | ARHGEF18 | chr19 | 7521212 | + | 2.14E-05 | 0.99997866 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
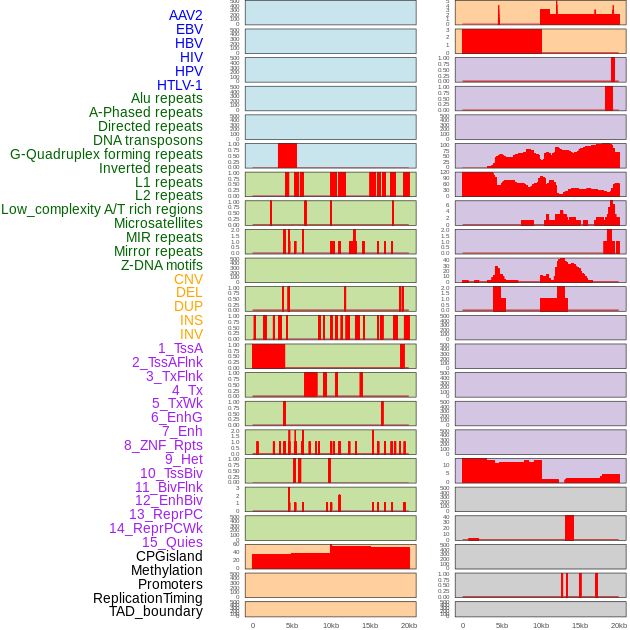 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
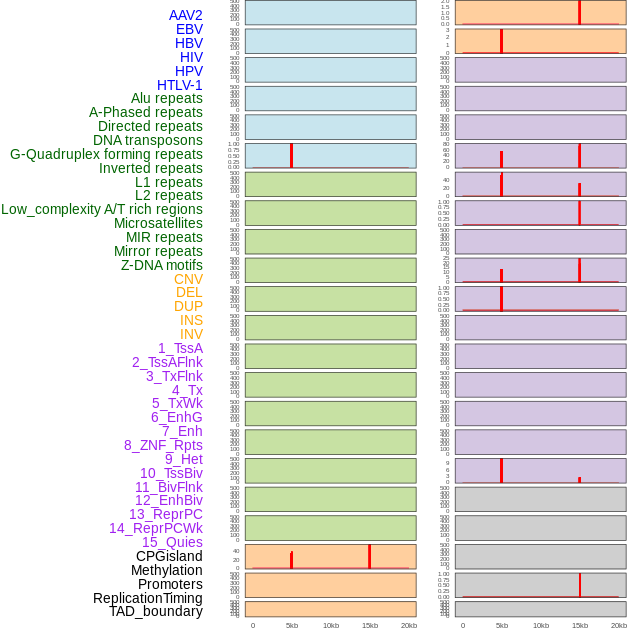 |
Top |
Fusion Protein Features for PAK1-ARHGEF18 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:77043775/chr19:7521213) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | ARHGEF18 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Acts as guanine nucleotide exchange factor (GEF) for RhoA GTPases. Its activation induces formation of actin stress fibers. Also acts as a GEF for RAC1, inducing production of reactive oxygen species (ROS). Does not act as a GEF for CDC42. The G protein beta-gamma (Gbetagamma) subunits of heterotrimeric G proteins act as activators, explaining the integrated effects of LPA and other G-protein coupled receptor agonists on actin stress fiber formation, cell shape change and ROS production. Required for EPB41L4B-mediated regulation of the circumferential actomyosin belt in epithelial cells (PubMed:22006950). {ECO:0000269|PubMed:11085924, ECO:0000269|PubMed:14512443, ECO:0000269|PubMed:15558029, ECO:0000269|PubMed:22006950, ECO:0000269|PubMed:28132693}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PAK1 | chr11:77043775 | chr19:7521213 | ENST00000278568 | - | 14 | 16 | 75_88 | 517 | 554.0 | Domain | CRIB |
| Hgene | PAK1 | chr11:77043775 | chr19:7521213 | ENST00000356341 | - | 14 | 15 | 75_88 | 517 | 546.0 | Domain | CRIB |
| Hgene | PAK1 | chr11:77043775 | chr19:7521213 | ENST00000278568 | - | 14 | 16 | 276_284 | 517 | 554.0 | Nucleotide binding | Note=ATP |
| Hgene | PAK1 | chr11:77043775 | chr19:7521213 | ENST00000278568 | - | 14 | 16 | 345_347 | 517 | 554.0 | Nucleotide binding | Note=ATP |
| Hgene | PAK1 | chr11:77043775 | chr19:7521213 | ENST00000356341 | - | 14 | 15 | 276_284 | 517 | 546.0 | Nucleotide binding | Note=ATP |
| Hgene | PAK1 | chr11:77043775 | chr19:7521213 | ENST00000356341 | - | 14 | 15 | 345_347 | 517 | 546.0 | Nucleotide binding | Note=ATP |
| Hgene | PAK1 | chr11:77043775 | chr19:7521213 | ENST00000278568 | - | 14 | 16 | 70_140 | 517 | 554.0 | Region | Note=Autoregulatory region |
| Hgene | PAK1 | chr11:77043775 | chr19:7521213 | ENST00000278568 | - | 14 | 16 | 75_105 | 517 | 554.0 | Region | Note=GTPase-binding |
| Hgene | PAK1 | chr11:77043775 | chr19:7521213 | ENST00000356341 | - | 14 | 15 | 70_140 | 517 | 546.0 | Region | Note=Autoregulatory region |
| Hgene | PAK1 | chr11:77043775 | chr19:7521213 | ENST00000356341 | - | 14 | 15 | 75_105 | 517 | 546.0 | Region | Note=GTPase-binding |
| Tgene | ARHGEF18 | chr11:77043775 | chr19:7521213 | ENST00000319670 | 7 | 21 | 1038_1148 | 355 | 1618.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | ARHGEF18 | chr11:77043775 | chr19:7521213 | ENST00000359920 | 6 | 20 | 1038_1148 | 513 | 1556.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | ARHGEF18 | chr11:77043775 | chr19:7521213 | ENST00000319670 | 7 | 21 | 1042_1142 | 355 | 1618.0 | Compositional bias | Arg-rich | |
| Tgene | ARHGEF18 | chr11:77043775 | chr19:7521213 | ENST00000319670 | 7 | 21 | 1292_1346 | 355 | 1618.0 | Compositional bias | Pro-rich | |
| Tgene | ARHGEF18 | chr11:77043775 | chr19:7521213 | ENST00000359920 | 6 | 20 | 1042_1142 | 513 | 1556.0 | Compositional bias | Arg-rich | |
| Tgene | ARHGEF18 | chr11:77043775 | chr19:7521213 | ENST00000359920 | 6 | 20 | 1292_1346 | 513 | 1556.0 | Compositional bias | Pro-rich | |
| Tgene | ARHGEF18 | chr11:77043775 | chr19:7521213 | ENST00000319670 | 7 | 21 | 447_644 | 355 | 1618.0 | Domain | DH | |
| Tgene | ARHGEF18 | chr11:77043775 | chr19:7521213 | ENST00000319670 | 7 | 21 | 684_786 | 355 | 1618.0 | Domain | PH | |
| Tgene | ARHGEF18 | chr11:77043775 | chr19:7521213 | ENST00000359920 | 6 | 20 | 684_786 | 513 | 1556.0 | Domain | PH |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PAK1 | chr11:77043775 | chr19:7521213 | ENST00000278568 | - | 14 | 16 | 270_521 | 517 | 554.0 | Domain | Protein kinase |
| Hgene | PAK1 | chr11:77043775 | chr19:7521213 | ENST00000356341 | - | 14 | 15 | 270_521 | 517 | 546.0 | Domain | Protein kinase |
| Tgene | ARHGEF18 | chr11:77043775 | chr19:7521213 | ENST00000359920 | 6 | 20 | 447_644 | 513 | 1556.0 | Domain | DH | |
| Tgene | ARHGEF18 | chr11:77043775 | chr19:7521213 | ENST00000319670 | 7 | 21 | 310_334 | 355 | 1618.0 | Zinc finger | C2H2-type%3B degenerate | |
| Tgene | ARHGEF18 | chr11:77043775 | chr19:7521213 | ENST00000359920 | 6 | 20 | 310_334 | 513 | 1556.0 | Zinc finger | C2H2-type%3B degenerate |
Top |
Fusion Gene Sequence for PAK1-ARHGEF18 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >62397_62397_1_PAK1-ARHGEF18_PAK1_chr11_77043775_ENST00000530617_ARHGEF18_chr19_7521213_ENST00000319670_length(transcript)=5803nt_BP=1863nt TGCTTTGCAGAAAAGGAGAAGCGCGCGGCTTCCTCCCGCCTCCTTCCCTCCTTTCTTCCCCCGCCCCTTCTGTTTCGCCAACCCCGACGC GGCGCCTTCGCTGCCGCCCTGCGAGAGCGCGAGCGTGCTGCACCTCGGGAGCTGGCGGCTGAGCTGCCAGGAGCCCAGCCCAGCTGAATC GAGCGGAGCCGGTGGAGCAGGGGCCTTGTGGGTACCCGGTTGGGCAGGGAGAGGTGCGGCTCTGCGACGGAAACAATCGCCAGAGATGCC GGGGCTAGCCTTCCCCACCAGTAGCTGCTGCTGGTGGTGACAATGTCAAATAACGGCCTAGACATTCAAGACAAACCCCCAGCCCCTCCG ATGAGAAATACCAGCACTATGATTGGAGCCGGCAGCAAAGATGCTGGAACCCTAAACCATGGTTCTAAACCTCTGCCTCCAAACCCAGAG GAGAAGAAAAAGAAGGACCGATTTTACCGATCCATTTTACCTGGAGATAAAACAAATAAAAAGAAAGAGAAAGAGCGGCCAGAGATTTCT CTCCCTTCAGATTTTGAACACACAATTCATGTCGGTTTTGATGCTGTCACAGGGGAGTTTACGGGAATGCCAGAGCAGTGGGCCCGCTTG CTTCAGACATCAAATATCACTAAGTCGGAGCAGAAGAAAAACCCGCAGGCTGTTCTGGATGTGTTGGAGTTTTACAACTCGAAGAAGACA TCCAACAGCCAGAAATACATGAGCTTTACAGATAAGTCAGCTGAGGATTACAATTCTTCTAATGCCTTGAATGTGAAGGCTGTGTCTGAG ACTCCTGCAGTGCCACCAGTTTCAGAAGATGAGGATGATGATGATGATGATGCTACCCCACCACCAGTGATTGCTCCACGCCCAGAGCAC ACAAAATCTGTATACACACGGTCTGTGATTGAACCACTTCCTGTCACTCCAACTCGGGACGTGGCTACATCTCCCATTTCACCTACTGAA AATAACACCACTCCACCAGATGCTTTGACCCGGAATACTGAGAAGCAGAAGAAGAAGCCTAAAATGTCTGATGAGGAGATCTTGGAGAAA TTACGAAGCATAGTGAGTGTGGGCGATCCTAAGAAGAAATATACACGGTTTGAGAAGATTGGACAAGGTGCTTCAGGCACCGTGTACACA GCAATGGATGTGGCCACAGGACAGGAGGTGGCCATTAAGCAGATGAATCTTCAGCAGCAGCCCAAGAAAGAGCTGATTATTAATGAGATC CTGGTCATGAGGGAAAACAAGAACCCAAACATTGTGAATTACTTGGACAGTTACCTCGTGGGAGATGAGCTGTGGGTTGTTATGGAATAC TTGGCTGGAGGCTCCTTGACAGATGTGGTGACAGAAACTTGCATGGATGAAGGCCAAATTGCAGCTGTGTGCCGTGAGTGTCTGCAGGCT CTGGAGTTCTTGCATTCGAACCAGGTCATTCACAGAGACATCAAGAGTGACAATATTCTGTTGGGAATGGATGGCTCTGTCAAGCTAACT GACTTTGGATTCTGTGCACAGATAACCCCAGAGCAGAGCAAACGGAGCACCATGGTAGGAACCCCATACTGGATGGCACCAGAGGTTGTG ACACGAAAGGCCTATGGGCCCAAGGTTGACATCTGGTCCCTGGGCATCATGGCCATCGAAATGATTGAAGGGGAGCCTCCATACCTCAAT GAAAACCCTCTGAGAGCCTTGTACCTCATTGCCACCAATGGGACCCCAGAACTTCAGAACCCAGAGAAGCTGTCAGCTATCTTCCGGGAC TTTCTGAACCGCTGTCTCGAGATGGATGTGGAGAAGAGAGGTTCAGCTAAAGAGCTGCTACAGATATCCTGGCTATCCTGCTGACCGACG TACTTTTGCTGCTACAAGAAAAAGATCAGAAATACGTCTTTGCTTCTGTGGACTCAAAGCCACCCGTCATCTCGTTACAAAAGCTCATCG TGAGGGAAGTGGCCAACGAGGAGAAAGCGATGTTTCTGATCAGCGCCTCCTTGCAAGGGCCGGAGATGTATGAAATCTACACGAGCTCCA AAGAGGACAGGAACGCCTGGATGGCCCACATCCAAAGGGCTGTGGAGAGCTGCCCTGACGAGGAGGAGGGGCCCTTCAGCCTGCCCGAAG AGGAAAGGAAGGTGGTCGAGGCCCGCGCCACGAGACTCCGGGACTTTCAAGAGCGGTTGAGCATGAAAGACCAGCTGATCGCACAGAGCC TCCTAGAGAAACAGCAGATCTACCTGGAGATGGCCGAGATGGGCGGCCTCGAAGACCTGCCCCAGCCCCGAGGCCTATTCCGTGGAGGGG ACCCATCCGAGACCCTGCAGGGGGAGCTAATTCTCAAGTCGGCCATGAGCGAGATCGAGGGCATCCAGAGCCTGATCTGCAGGCAGCTGG GCAGCGCCAACGGCCAGGCGGAAGACGGAGGCAGCTCCACAGGCCCGCCCAGGAGGGCTGAGACCTTCGCGGGCTACGACTGCACAAACA GCCCCACCAAGAATGGCAGTTTCAAGAAGAAAGTCAGCAGCACTGACCCCAGGCCCCGAGACTGGCGAGGCCCCCCAAACAGCCCGGACT TGAAGCTCAGTGACAGTGACATTCCTGGGAGCTCTGAGGAATCGCCGCAGGTGGTGGAGGCGCCAGGCACGGAATCCGATCCCCGTCTGC CCACCGTCCTGGAGTCGGAGCTTGTCCAGCGGATCCAGACACTGTCCCAGCTGCTCCTGAACCTTCAGGCGGTAATCGCCCACCAGGACA GCTATGTGGAGACGCAGCGGGCTGCCATCCAGGAGCGGGAGAAGCAGTTCCGGCTGCAGTCGACGCGTGGGAACCTGCTGCTGGAGCAGG AGCGGCAACGCAACTTCGAGAAGCAGCGGGAGGAGCGCGCGGCCCTGGAGAAGCTGCAGAGCCAGCTGCGGCACGAGCAGCAGCGCTGGG AGCGCGAGCGCCAGTGGCAGCACCAGGAGCTGGAGCGTGCGGGCGCGCGGCTGCAGGAGCGCGAGGGCGAGGCGCGGCAGCTACGCGAGC GGCTGGAGCAGGAGCGGGCCGAGCTGGAGCGCCAGCGCCAGGCCTACCAGCACGACCTGGAGCGGCTGCGCGAGGCCCAGCGTGCCGTGG AGCGCGAGCGGGAGCGCCTGGAGCTGCTGCGCCGCCTCAAGAAGCAGAACACCGCGCCAGGCGCGCTGCCGCCCGACACACTGGCCGAGG CCCAGCCCCCAAGCCACCCTCCCAGCTTCAACGGGGAAGGGCTGGAGGGCCCTCGTGTGAGCATGCTGCCATCCGGCGTGGGGCCAGAGT ACGCAGAGCGCCCCGAGGTGGCTCGCCGGGACAGCGCCCCCACCGAGAACCGGCTGGCCAAGAGCGATGTGCCCATCCAGCTGCTCAGCG CCACCAACCAGTTCCAGAGGCAGGCGGCCGTGCAGCAGCAGATCCCCACCAAGCTGGCGGCCTCCACCAAGGGTGGCAAGGACAAGGGCG GCAAGAGCAGGGGCTCTCAGCGCTGGGAGAGCTCAGCGTCCTTCGACCTGAAGCAGCAGCTGCTGCTCAACAAGCTCATGGGGAAAGATG AGAGCACCTCACGGAACCGCCGCTCGCTGAGCCCTATCCTGCCCGGCAGACACAGTCCTGCGCCCCCACCAGACCCTGGCTTCCCCGCCC CGAGCCCACCGCCAGCTGACAGCCCCTCCGAGGGCTTCTCTCTCAAGGCCGGGGGCACAGCCCTCCTGCCCGGGCCCCCAGCTCCCTCGC CACTGCCGGCCACACCACTCAGCGCCAAGGAGGACGCCAGCAAAGAAGACGTCATCTTCTTCTAAAAGGGCCGTGACTCAAGGAAAGTTT TTAATGGAAAGTTGAGCCAGAACTAAACCAGGGAGCTGTCTGAAATCATAGCACCCCATCCGGGTGGCGGGGAGATCAACTCCGAGCTGT TTTTCCGAGGCAGTGAGGAACGGTGCCGGCTCTGCACGGAGCTGAGGACAGGACAGACCTTGCTTTGAGAAGGAGCTGCCGGCCGGGGCC ACGCTCCACAGCCGCCGCGCGACAGTGGAGCCAAGGGTTAGGGCACCAGGAGGGGCCAGGTGGCGTCGGCAGCATCTGTCCCCAGAATCA GGCAGAATCCACTTCCCAAACAGAGCCCCACGCAGGTTCACCATGAACCTCAGGGTCAGGGAATGAGCCAGGCACGGGGGCATGGGCAGA GAGGGCCACGGGGCAGGGCCCACTGAGGGAACATCAGTGGCCCTCCAGTCAGGTTCTGTGGGTTTGGAAGCCCATCGTGAAAGGGGCTGA CCTTTGCCCCTTTTTACTTGGCATTGGTTTTGAAACCAGCTGTTTCCCAAACTCTGCTTCCCAAGGGCAACCGTTGCTGTTCACACGCTC AGCCTGTCTGGGGGAGCGGGCCTCTAGCTTCAGCCAGGGCGGGTACACACCCTGGGCACAGGGTCCTCAGCCCCCGGGAAATGAGCTCCC AGGGCTGGCGTCCCACCTTCCAGGTGGGGGCTGGCACATCACAGACTGTCGAGAGCGCCATGTCCCAGGGCATGCAGAGGATGCACCTAG AGACGTTGCAGCAAGTGGACAAGTGGCCGCTGTGCGGGCCCCTCGCTTGTAGTGAGCTGTTGCAGCTTACGGTCCGTTCCCTGGAGGGGT GGAGGAAGGAGGTGTTGGGCAGCATCAAAGGTGCTGGGACATCCCAGGGTGGTGAGATCCATCCACGATCCAGCTCCGGTGGAGAAAGGG CCCATGTCAAGCCTTGTTCTGCACCCCAAGCATTGGTGGTAGGACTGGGTCCTGGCTGATCGTCCTTGTTCCCAGTGGGGTACATGTGAG CCCCTGCCAGGGCCAAGTCCTTCTCCCGAACCCAGGGTCCTGGGAACTGCAGATCCCGGGGGGATTCAGCCCTTCTCCCACTGTGCTGGC AGAGGCACTCCTGTGACGCTGAATACAGTGAACAGGGACATTCCCGCCACTCGGGGACAGATGGGCACAAGGGAGGGGAAACTCCATCAG GAAGTGCTCCCCTGGGCAGAGGCGCCCACTGGGTGCTGTGGGCTCAGGAGGGGGCGGGGCAGGAGCTGGTGCCAACCGGGAACCAGAGCC CCACAGCCATACAGCCCATTGGTGACAAGGTCCTGAGAACACAGTGGCCAGGTGTCCCCAGGCTCCTGGCCCCTCCGACGACCTCAACTC TGCCCAGCCCGGTCCCTGGCCATCAGCGACGCTGTCCGCCCCCCGTCAGATCCCATGTGTGCCATGTTTATCATCAGTGTTTTGTATTTT TGTACTGAGTATCGGAGCACTTTACAGAAGCTGACTGTACATTCCTGTTCTGTTGTGAAGAGAACATTCCCAGACCCTGGCACCCTCCTG AGCCGGCGTGTGCCGGTCCAGCCCTCCGAGATGCCACAATTCCTTGGATGGGGGAGAAGTTCAAGGAATTTCTGCTCGGCCACGCGGTGG GAACCCCGCGTCCCCGCCATGTGGCAGAGGGGTCTCAGTCGTGCTAGGCATCGGGCGGCAGCGCCGACAGCCCTTCCCTCGCCAGTGCCC CTCGGCCACTCCTGGGTTGGAGCCCGATTTTATTTGTAAAGTTGACAGTCGAGCAAATGTTCCTATTTTCGTGGGATCTGCACACGTCTT TGTCAGTTGTGGTCATGATCTTAGTCACCTGCTAATTATTTTTACAATGATTACAACATTTCCTCACTGCGGGATATTTCTGACCCGCTT >62397_62397_1_PAK1-ARHGEF18_PAK1_chr11_77043775_ENST00000530617_ARHGEF18_chr19_7521213_ENST00000319670_length(amino acids)=658AA_BP=1 MAILLTDVLLLLQEKDQKYVFASVDSKPPVISLQKLIVREVANEEKAMFLISASLQGPEMYEIYTSSKEDRNAWMAHIQRAVESCPDEEE GPFSLPEEERKVVEARATRLRDFQERLSMKDQLIAQSLLEKQQIYLEMAEMGGLEDLPQPRGLFRGGDPSETLQGELILKSAMSEIEGIQ SLICRQLGSANGQAEDGGSSTGPPRRAETFAGYDCTNSPTKNGSFKKKVSSTDPRPRDWRGPPNSPDLKLSDSDIPGSSEESPQVVEAPG TESDPRLPTVLESELVQRIQTLSQLLLNLQAVIAHQDSYVETQRAAIQEREKQFRLQSTRGNLLLEQERQRNFEKQREERAALEKLQSQL RHEQQRWERERQWQHQELERAGARLQEREGEARQLRERLEQERAELERQRQAYQHDLERLREAQRAVERERERLELLRRLKKQNTAPGAL PPDTLAEAQPPSHPPSFNGEGLEGPRVSMLPSGVGPEYAERPEVARRDSAPTENRLAKSDVPIQLLSATNQFQRQAAVQQQIPTKLAAST KGGKDKGGKSRGSQRWESSASFDLKQQLLLNKLMGKDESTSRNRRSLSPILPGRHSPAPPPDPGFPAPSPPPADSPSEGFSLKAGGTALL -------------------------------------------------------------- >62397_62397_2_PAK1-ARHGEF18_PAK1_chr11_77043775_ENST00000530617_ARHGEF18_chr19_7521213_ENST00000359920_length(transcript)=5803nt_BP=1863nt TGCTTTGCAGAAAAGGAGAAGCGCGCGGCTTCCTCCCGCCTCCTTCCCTCCTTTCTTCCCCCGCCCCTTCTGTTTCGCCAACCCCGACGC GGCGCCTTCGCTGCCGCCCTGCGAGAGCGCGAGCGTGCTGCACCTCGGGAGCTGGCGGCTGAGCTGCCAGGAGCCCAGCCCAGCTGAATC GAGCGGAGCCGGTGGAGCAGGGGCCTTGTGGGTACCCGGTTGGGCAGGGAGAGGTGCGGCTCTGCGACGGAAACAATCGCCAGAGATGCC GGGGCTAGCCTTCCCCACCAGTAGCTGCTGCTGGTGGTGACAATGTCAAATAACGGCCTAGACATTCAAGACAAACCCCCAGCCCCTCCG ATGAGAAATACCAGCACTATGATTGGAGCCGGCAGCAAAGATGCTGGAACCCTAAACCATGGTTCTAAACCTCTGCCTCCAAACCCAGAG GAGAAGAAAAAGAAGGACCGATTTTACCGATCCATTTTACCTGGAGATAAAACAAATAAAAAGAAAGAGAAAGAGCGGCCAGAGATTTCT CTCCCTTCAGATTTTGAACACACAATTCATGTCGGTTTTGATGCTGTCACAGGGGAGTTTACGGGAATGCCAGAGCAGTGGGCCCGCTTG CTTCAGACATCAAATATCACTAAGTCGGAGCAGAAGAAAAACCCGCAGGCTGTTCTGGATGTGTTGGAGTTTTACAACTCGAAGAAGACA TCCAACAGCCAGAAATACATGAGCTTTACAGATAAGTCAGCTGAGGATTACAATTCTTCTAATGCCTTGAATGTGAAGGCTGTGTCTGAG ACTCCTGCAGTGCCACCAGTTTCAGAAGATGAGGATGATGATGATGATGATGCTACCCCACCACCAGTGATTGCTCCACGCCCAGAGCAC ACAAAATCTGTATACACACGGTCTGTGATTGAACCACTTCCTGTCACTCCAACTCGGGACGTGGCTACATCTCCCATTTCACCTACTGAA AATAACACCACTCCACCAGATGCTTTGACCCGGAATACTGAGAAGCAGAAGAAGAAGCCTAAAATGTCTGATGAGGAGATCTTGGAGAAA TTACGAAGCATAGTGAGTGTGGGCGATCCTAAGAAGAAATATACACGGTTTGAGAAGATTGGACAAGGTGCTTCAGGCACCGTGTACACA GCAATGGATGTGGCCACAGGACAGGAGGTGGCCATTAAGCAGATGAATCTTCAGCAGCAGCCCAAGAAAGAGCTGATTATTAATGAGATC CTGGTCATGAGGGAAAACAAGAACCCAAACATTGTGAATTACTTGGACAGTTACCTCGTGGGAGATGAGCTGTGGGTTGTTATGGAATAC TTGGCTGGAGGCTCCTTGACAGATGTGGTGACAGAAACTTGCATGGATGAAGGCCAAATTGCAGCTGTGTGCCGTGAGTGTCTGCAGGCT CTGGAGTTCTTGCATTCGAACCAGGTCATTCACAGAGACATCAAGAGTGACAATATTCTGTTGGGAATGGATGGCTCTGTCAAGCTAACT GACTTTGGATTCTGTGCACAGATAACCCCAGAGCAGAGCAAACGGAGCACCATGGTAGGAACCCCATACTGGATGGCACCAGAGGTTGTG ACACGAAAGGCCTATGGGCCCAAGGTTGACATCTGGTCCCTGGGCATCATGGCCATCGAAATGATTGAAGGGGAGCCTCCATACCTCAAT GAAAACCCTCTGAGAGCCTTGTACCTCATTGCCACCAATGGGACCCCAGAACTTCAGAACCCAGAGAAGCTGTCAGCTATCTTCCGGGAC TTTCTGAACCGCTGTCTCGAGATGGATGTGGAGAAGAGAGGTTCAGCTAAAGAGCTGCTACAGATATCCTGGCTATCCTGCTGACCGACG TACTTTTGCTGCTACAAGAAAAAGATCAGAAATACGTCTTTGCTTCTGTGGACTCAAAGCCACCCGTCATCTCGTTACAAAAGCTCATCG TGAGGGAAGTGGCCAACGAGGAGAAAGCGATGTTTCTGATCAGCGCCTCCTTGCAAGGGCCGGAGATGTATGAAATCTACACGAGCTCCA AAGAGGACAGGAACGCCTGGATGGCCCACATCCAAAGGGCTGTGGAGAGCTGCCCTGACGAGGAGGAGGGGCCCTTCAGCCTGCCCGAAG AGGAAAGGAAGGTGGTCGAGGCCCGCGCCACGAGACTCCGGGACTTTCAAGAGCGGTTGAGCATGAAAGACCAGCTGATCGCACAGAGCC TCCTAGAGAAACAGCAGATCTACCTGGAGATGGCCGAGATGGGCGGCCTCGAAGACCTGCCCCAGCCCCGAGGCCTATTCCGTGGAGGGG ACCCATCCGAGACCCTGCAGGGGGAGCTAATTCTCAAGTCGGCCATGAGCGAGATCGAGGGCATCCAGAGCCTGATCTGCAGGCAGCTGG GCAGCGCCAACGGCCAGGCGGAAGACGGAGGCAGCTCCACAGGCCCGCCCAGGAGGGCTGAGACCTTCGCGGGCTACGACTGCACAAACA GCCCCACCAAGAATGGCAGTTTCAAGAAGAAAGTCAGCAGCACTGACCCCAGGCCCCGAGACTGGCGAGGCCCCCCAAACAGCCCGGACT TGAAGCTCAGTGACAGTGACATTCCTGGGAGCTCTGAGGAATCGCCGCAGGTGGTGGAGGCGCCAGGCACGGAATCCGATCCCCGTCTGC CCACCGTCCTGGAGTCGGAGCTTGTCCAGCGGATCCAGACACTGTCCCAGCTGCTCCTGAACCTTCAGGCGGTAATCGCCCACCAGGACA GCTATGTGGAGACGCAGCGGGCTGCCATCCAGGAGCGGGAGAAGCAGTTCCGGCTGCAGTCGACGCGTGGGAACCTGCTGCTGGAGCAGG AGCGGCAACGCAACTTCGAGAAGCAGCGGGAGGAGCGCGCGGCCCTGGAGAAGCTGCAGAGCCAGCTGCGGCACGAGCAGCAGCGCTGGG AGCGCGAGCGCCAGTGGCAGCACCAGGAGCTGGAGCGTGCGGGCGCGCGGCTGCAGGAGCGCGAGGGCGAGGCGCGGCAGCTACGCGAGC GGCTGGAGCAGGAGCGGGCCGAGCTGGAGCGCCAGCGCCAGGCCTACCAGCACGACCTGGAGCGGCTGCGCGAGGCCCAGCGTGCCGTGG AGCGCGAGCGGGAGCGCCTGGAGCTGCTGCGCCGCCTCAAGAAGCAGAACACCGCGCCAGGCGCGCTGCCGCCCGACACACTGGCCGAGG CCCAGCCCCCAAGCCACCCTCCCAGCTTCAACGGGGAAGGGCTGGAGGGCCCTCGTGTGAGCATGCTGCCATCCGGCGTGGGGCCAGAGT ACGCAGAGCGCCCCGAGGTGGCTCGCCGGGACAGCGCCCCCACCGAGAACCGGCTGGCCAAGAGCGATGTGCCCATCCAGCTGCTCAGCG CCACCAACCAGTTCCAGAGGCAGGCGGCCGTGCAGCAGCAGATCCCCACCAAGCTGGCGGCCTCCACCAAGGGTGGCAAGGACAAGGGCG GCAAGAGCAGGGGCTCTCAGCGCTGGGAGAGCTCAGCGTCCTTCGACCTGAAGCAGCAGCTGCTGCTCAACAAGCTCATGGGGAAAGATG AGAGCACCTCACGGAACCGCCGCTCGCTGAGCCCTATCCTGCCCGGCAGACACAGTCCTGCGCCCCCACCAGACCCTGGCTTCCCCGCCC CGAGCCCACCGCCAGCTGACAGCCCCTCCGAGGGCTTCTCTCTCAAGGCCGGGGGCACAGCCCTCCTGCCCGGGCCCCCAGCTCCCTCGC CACTGCCGGCCACACCACTCAGCGCCAAGGAGGACGCCAGCAAAGAAGACGTCATCTTCTTCTAAAAGGGCCGTGACTCAAGGAAAGTTT TTAATGGAAAGTTGAGCCAGAACTAAACCAGGGAGCTGTCTGAAATCATAGCACCCCATCCGGGTGGCGGGGAGATCAACTCCGAGCTGT TTTTCCGAGGCAGTGAGGAACGGTGCCGGCTCTGCACGGAGCTGAGGACAGGACAGACCTTGCTTTGAGAAGGAGCTGCCGGCCGGGGCC ACGCTCCACAGCCGCCGCGCGACAGTGGAGCCAAGGGTTAGGGCACCAGGAGGGGCCAGGTGGCGTCGGCAGCATCTGTCCCCAGAATCA GGCAGAATCCACTTCCCAAACAGAGCCCCACGCAGGTTCACCATGAACCTCAGGGTCAGGGAATGAGCCAGGCACGGGGGCATGGGCAGA GAGGGCCACGGGGCAGGGCCCACTGAGGGAACATCAGTGGCCCTCCAGTCAGGTTCTGTGGGTTTGGAAGCCCATCGTGAAAGGGGCTGA CCTTTGCCCCTTTTTACTTGGCATTGGTTTTGAAACCAGCTGTTTCCCAAACTCTGCTTCCCAAGGGCAACCGTTGCTGTTCACACGCTC AGCCTGTCTGGGGGAGCGGGCCTCTAGCTTCAGCCAGGGCGGGTACACACCCTGGGCACAGGGTCCTCAGCCCCCGGGAAATGAGCTCCC AGGGCTGGCGTCCCACCTTCCAGGTGGGGGCTGGCACATCACAGACTGTCGAGAGCGCCATGTCCCAGGGCATGCAGAGGATGCACCTAG AGACGTTGCAGCAAGTGGACAAGTGGCCGCTGTGCGGGCCCCTCGCTTGTAGTGAGCTGTTGCAGCTTACGGTCCGTTCCCTGGAGGGGT GGAGGAAGGAGGTGTTGGGCAGCATCAAAGGTGCTGGGACATCCCAGGGTGGTGAGATCCATCCACGATCCAGCTCCGGTGGAGAAAGGG CCCATGTCAAGCCTTGTTCTGCACCCCAAGCATTGGTGGTAGGACTGGGTCCTGGCTGATCGTCCTTGTTCCCAGTGGGGTACATGTGAG CCCCTGCCAGGGCCAAGTCCTTCTCCCGAACCCAGGGTCCTGGGAACTGCAGATCCCGGGGGGATTCAGCCCTTCTCCCACTGTGCTGGC AGAGGCACTCCTGTGACGCTGAATACAGTGAACAGGGACATTCCCGCCACTCGGGGACAGATGGGCACAAGGGAGGGGAAACTCCATCAG GAAGTGCTCCCCTGGGCAGAGGCGCCCACTGGGTGCTGTGGGCTCAGGAGGGGGCGGGGCAGGAGCTGGTGCCAACCGGGAACCAGAGCC CCACAGCCATACAGCCCATTGGTGACAAGGTCCTGAGAACACAGTGGCCAGGTGTCCCCAGGCTCCTGGCCCCTCCGACGACCTCAACTC TGCCCAGCCCGGTCCCTGGCCATCAGCGACGCTGTCCGCCCCCCGTCAGATCCCATGTGTGCCATGTTTATCATCAGTGTTTTGTATTTT TGTACTGAGTATCGGAGCACTTTACAGAAGCTGACTGTACATTCCTGTTCTGTTGTGAAGAGAACATTCCCAGACCCTGGCACCCTCCTG AGCCGGCGTGTGCCGGTCCAGCCCTCCGAGATGCCACAATTCCTTGGATGGGGGAGAAGTTCAAGGAATTTCTGCTCGGCCACGCGGTGG GAACCCCGCGTCCCCGCCATGTGGCAGAGGGGTCTCAGTCGTGCTAGGCATCGGGCGGCAGCGCCGACAGCCCTTCCCTCGCCAGTGCCC CTCGGCCACTCCTGGGTTGGAGCCCGATTTTATTTGTAAAGTTGACAGTCGAGCAAATGTTCCTATTTTCGTGGGATCTGCACACGTCTT TGTCAGTTGTGGTCATGATCTTAGTCACCTGCTAATTATTTTTACAATGATTACAACATTTCCTCACTGCGGGATATTTCTGACCCGCTT >62397_62397_2_PAK1-ARHGEF18_PAK1_chr11_77043775_ENST00000530617_ARHGEF18_chr19_7521213_ENST00000359920_length(amino acids)=658AA_BP=1 MAILLTDVLLLLQEKDQKYVFASVDSKPPVISLQKLIVREVANEEKAMFLISASLQGPEMYEIYTSSKEDRNAWMAHIQRAVESCPDEEE GPFSLPEEERKVVEARATRLRDFQERLSMKDQLIAQSLLEKQQIYLEMAEMGGLEDLPQPRGLFRGGDPSETLQGELILKSAMSEIEGIQ SLICRQLGSANGQAEDGGSSTGPPRRAETFAGYDCTNSPTKNGSFKKKVSSTDPRPRDWRGPPNSPDLKLSDSDIPGSSEESPQVVEAPG TESDPRLPTVLESELVQRIQTLSQLLLNLQAVIAHQDSYVETQRAAIQEREKQFRLQSTRGNLLLEQERQRNFEKQREERAALEKLQSQL RHEQQRWERERQWQHQELERAGARLQEREGEARQLRERLEQERAELERQRQAYQHDLERLREAQRAVERERERLELLRRLKKQNTAPGAL PPDTLAEAQPPSHPPSFNGEGLEGPRVSMLPSGVGPEYAERPEVARRDSAPTENRLAKSDVPIQLLSATNQFQRQAAVQQQIPTKLAAST KGGKDKGGKSRGSQRWESSASFDLKQQLLLNKLMGKDESTSRNRRSLSPILPGRHSPAPPPDPGFPAPSPPPADSPSEGFSLKAGGTALL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for PAK1-ARHGEF18 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Hgene | PAK1 | chr11:77043775 | chr19:7521213 | ENST00000278568 | - | 14 | 16 | 132_270 | 517.0 | 554.0 | CRIPAK |
| Hgene | PAK1 | chr11:77043775 | chr19:7521213 | ENST00000356341 | - | 14 | 15 | 132_270 | 517.0 | 546.0 | CRIPAK |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for PAK1-ARHGEF18 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for PAK1-ARHGEF18 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
