
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:PALM2-LPAR1 (FusionGDB2 ID:62494) |
Fusion Gene Summary for PALM2-LPAR1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: PALM2-LPAR1 | Fusion gene ID: 62494 | Hgene | Tgene | Gene symbol | PALM2 | LPAR1 | Gene ID | 445815 | 1902 |
| Gene name | PALM2 and AKAP2 fusion | lysophosphatidic acid receptor 1 | |
| Synonyms | AKAP-2|AKAP-KL|AKAP2|AKAPKL|MISP2|PALM2|PALM2-AKAP2|PRKA2 | EDG2|GPR26|Gpcr26|LPA1|Mrec1.3|VZG1|edg-2|rec.1.3|vzg-1 | |
| Cytomap | 9q31.3 | 9q31.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | A-kinase anchor protein 2PALM2-AKAP2 proteinparalemmin-2A kinase (PRKA) anchor protein 2A-kinase anchoring protein 2PALM2-AKAP2 fusionPALM2-AKAP2 readthroughparalemmin 2protein kinase A anchoring protein 2protein kinase A2 | lysophosphatidic acid receptor 1LPA receptor 1LPA-1endothelial differentiation, lysophosphatidic acid G-protein-coupled receptor, 2lysophosphatidic acid receptor Edg-2ventricular zone gene 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q92633 | |
| Ensembl transtripts involved in fusion gene | ENST00000314527, ENST00000374531, ENST00000448454, ENST00000483909, | ENST00000358883, ENST00000374430, ENST00000374431, ENST00000538760, ENST00000541779, | |
| Fusion gene scores | * DoF score | 4 X 4 X 4=64 | 10 X 6 X 6=360 |
| # samples | 5 | 11 | |
| ** MAII score | log2(5/64*10)=-0.356143810225275 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(11/360*10)=-1.71049338280502 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: PALM2 [Title/Abstract] AND LPAR1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | PALM2(112542813)-LPAR1(113638002), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | PALM2-LPAR1 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. PALM2-LPAR1 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | LPAR1 | GO:0007202 | activation of phospholipase C activity | 19306925 |
 Fusion gene breakpoints across PALM2 (5'-gene) Fusion gene breakpoints across PALM2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
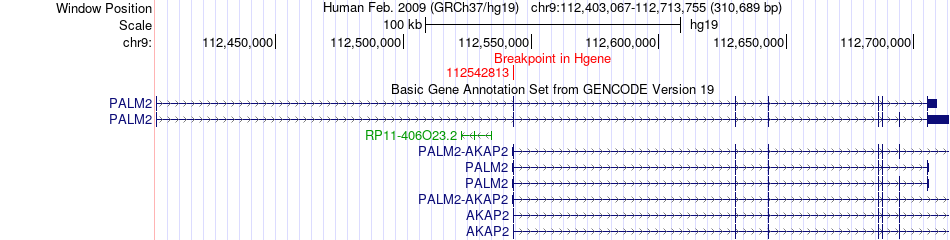 |
 Fusion gene breakpoints across LPAR1 (3'-gene) Fusion gene breakpoints across LPAR1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
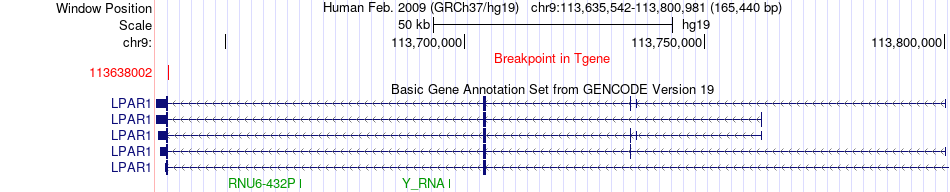 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | GBM | TCGA-06-5415-01A | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
Top |
Fusion Gene ORF analysis for PALM2-LPAR1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000314527 | ENST00000358883 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
| Frame-shift | ENST00000314527 | ENST00000374430 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
| Frame-shift | ENST00000314527 | ENST00000374431 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
| Frame-shift | ENST00000374531 | ENST00000358883 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
| Frame-shift | ENST00000374531 | ENST00000374430 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
| Frame-shift | ENST00000374531 | ENST00000374431 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
| Frame-shift | ENST00000448454 | ENST00000358883 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
| Frame-shift | ENST00000448454 | ENST00000374430 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
| Frame-shift | ENST00000448454 | ENST00000374431 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
| Frame-shift | ENST00000483909 | ENST00000358883 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
| Frame-shift | ENST00000483909 | ENST00000374430 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
| Frame-shift | ENST00000483909 | ENST00000374431 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
| In-frame | ENST00000314527 | ENST00000538760 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
| In-frame | ENST00000314527 | ENST00000541779 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
| In-frame | ENST00000374531 | ENST00000538760 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
| In-frame | ENST00000374531 | ENST00000541779 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
| In-frame | ENST00000448454 | ENST00000538760 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
| In-frame | ENST00000448454 | ENST00000541779 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
| In-frame | ENST00000483909 | ENST00000538760 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
| In-frame | ENST00000483909 | ENST00000541779 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000374531 | PALM2 | chr9 | 112542813 | + | ENST00000541779 | LPAR1 | chr9 | 113638002 | - | 2584 | 125 | 2257 | 1922 | 111 |
| ENST00000374531 | PALM2 | chr9 | 112542813 | + | ENST00000538760 | LPAR1 | chr9 | 113638002 | - | 672 | 125 | 154 | 426 | 90 |
| ENST00000448454 | PALM2 | chr9 | 112542813 | + | ENST00000541779 | LPAR1 | chr9 | 113638002 | - | 2510 | 51 | 2183 | 1848 | 111 |
| ENST00000448454 | PALM2 | chr9 | 112542813 | + | ENST00000538760 | LPAR1 | chr9 | 113638002 | - | 598 | 51 | 80 | 352 | 90 |
| ENST00000483909 | PALM2 | chr9 | 112542813 | + | ENST00000541779 | LPAR1 | chr9 | 113638002 | - | 2590 | 131 | 67 | 432 | 121 |
| ENST00000483909 | PALM2 | chr9 | 112542813 | + | ENST00000538760 | LPAR1 | chr9 | 113638002 | - | 678 | 131 | 67 | 432 | 121 |
| ENST00000314527 | PALM2 | chr9 | 112542813 | + | ENST00000541779 | LPAR1 | chr9 | 113638002 | - | 2554 | 95 | 31 | 396 | 121 |
| ENST00000314527 | PALM2 | chr9 | 112542813 | + | ENST00000538760 | LPAR1 | chr9 | 113638002 | - | 642 | 95 | 31 | 396 | 121 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000374531 | ENST00000541779 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - | 0.689384 | 0.31061605 |
| ENST00000374531 | ENST00000538760 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - | 0.18232125 | 0.81767875 |
| ENST00000448454 | ENST00000541779 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - | 0.7970169 | 0.20298307 |
| ENST00000448454 | ENST00000538760 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - | 0.21420248 | 0.7857975 |
| ENST00000483909 | ENST00000541779 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - | 0.86210346 | 0.13789655 |
| ENST00000483909 | ENST00000538760 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - | 0.5555594 | 0.44444057 |
| ENST00000314527 | ENST00000541779 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - | 0.76421934 | 0.23578061 |
| ENST00000314527 | ENST00000538760 | PALM2 | chr9 | 112542813 | + | LPAR1 | chr9 | 113638002 | - | 0.5060285 | 0.4939715 |
Top |
Fusion Genomic Features for PALM2-LPAR1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
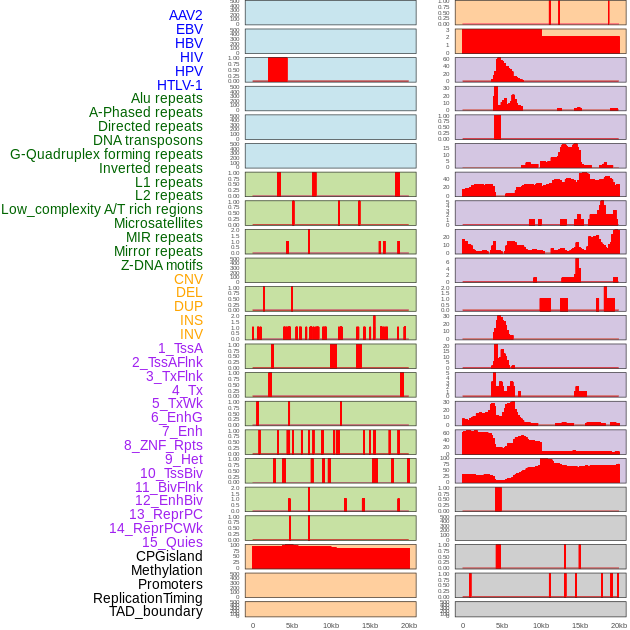 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for PALM2-LPAR1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:112542813/chr9:113638002) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | LPAR1 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Receptor for lysophosphatidic acid (LPA) (PubMed:9070858, PubMed:19306925, PubMed:25025571, PubMed:26091040). Plays a role in the reorganization of the actin cytoskeleton, cell migration, differentiation and proliferation, and thereby contributes to the responses to tissue damage and infectious agents. Activates downstream signaling cascades via the G(i)/G(o), G(12)/G(13), and G(q) families of heteromeric G proteins. Signaling inhibits adenylyl cyclase activity and decreases cellular cAMP levels (PubMed:26091040). Signaling triggers an increase of cytoplasmic Ca(2+) levels (PubMed:19656035, PubMed:19733258, PubMed:26091040). Activates RALA; this leads to the activation of phospholipase C (PLC) and the formation of inositol 1,4,5-trisphosphate (PubMed:19306925). Signaling mediates activation of down-stream MAP kinases (By similarity). Contributes to the regulation of cell shape. Promotes Rho-dependent reorganization of the actin cytoskeleton in neuronal cells and neurite retraction (PubMed:26091040). Promotes the activation of Rho and the formation of actin stress fibers (PubMed:26091040). Promotes formation of lamellipodia at the leading edge of migrating cells via activation of RAC1 (By similarity). Through its function as lysophosphatidic acid receptor, plays a role in chemotaxis and cell migration, including responses to injury and wounding (PubMed:18066075, PubMed:19656035, PubMed:19733258). Plays a role in triggering inflammation in response to bacterial lipopolysaccharide (LPS) via its interaction with CD14. Promotes cell proliferation in response to lysophosphatidic acid. Required for normal skeleton development. May play a role in osteoblast differentiation. Required for normal brain development. Required for normal proliferation, survival and maturation of newly formed neurons in the adult dentate gyrus. Plays a role in pain perception and in the initiation of neuropathic pain (By similarity). {ECO:0000250|UniProtKB:P61793, ECO:0000269|PubMed:18066075, ECO:0000269|PubMed:19306925, ECO:0000269|PubMed:19656035, ECO:0000269|PubMed:19733258, ECO:0000269|PubMed:25025571, ECO:0000269|PubMed:26091040, ECO:0000269|PubMed:9070858, ECO:0000305|PubMed:11093753, ECO:0000305|PubMed:9069262}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000358883 | 2 | 4 | 281_294 | 264 | 365.0 | Topological domain | Note=Extracellular | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000358883 | 2 | 4 | 316_364 | 264 | 365.0 | Topological domain | Note=Cytoplasmic | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374430 | 3 | 5 | 281_294 | 264 | 365.0 | Topological domain | Note=Extracellular | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374430 | 3 | 5 | 316_364 | 264 | 365.0 | Topological domain | Note=Cytoplasmic | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374431 | 3 | 5 | 281_294 | 264 | 365.0 | Topological domain | Note=Extracellular | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374431 | 3 | 5 | 316_364 | 264 | 365.0 | Topological domain | Note=Cytoplasmic | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000358883 | 2 | 4 | 295_315 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D7 | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374430 | 3 | 5 | 295_315 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D7 | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374431 | 3 | 5 | 295_315 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D7 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PALM2 | chr9:112542813 | chr9:113638002 | ENST00000314527 | + | 1 | 7 | 2_37 | 15 | 412.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | PALM2 | chr9:112542813 | chr9:113638002 | ENST00000314527 | + | 1 | 7 | 69_110 | 15 | 412.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | PALM2 | chr9:112542813 | chr9:113638002 | ENST00000374531 | + | 2 | 7 | 2_37 | 17 | 380.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | PALM2 | chr9:112542813 | chr9:113638002 | ENST00000374531 | + | 2 | 7 | 69_110 | 17 | 380.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000358883 | 2 | 4 | 124_129 | 264 | 365.0 | Region | Lysophosphatidic acid binding | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374430 | 3 | 5 | 124_129 | 264 | 365.0 | Region | Lysophosphatidic acid binding | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374431 | 3 | 5 | 124_129 | 264 | 365.0 | Region | Lysophosphatidic acid binding | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000358883 | 2 | 4 | 108_121 | 264 | 365.0 | Topological domain | Note=Extracellular | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000358883 | 2 | 4 | 145_163 | 264 | 365.0 | Topological domain | Note=Cytoplasmic | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000358883 | 2 | 4 | 185_204 | 264 | 365.0 | Topological domain | Note=Extracellular | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000358883 | 2 | 4 | 1_50 | 264 | 365.0 | Topological domain | Note=Extracellular | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000358883 | 2 | 4 | 226_255 | 264 | 365.0 | Topological domain | Note=Cytoplasmic | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000358883 | 2 | 4 | 76_83 | 264 | 365.0 | Topological domain | Note=Cytoplasmic | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374430 | 3 | 5 | 108_121 | 264 | 365.0 | Topological domain | Note=Extracellular | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374430 | 3 | 5 | 145_163 | 264 | 365.0 | Topological domain | Note=Cytoplasmic | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374430 | 3 | 5 | 185_204 | 264 | 365.0 | Topological domain | Note=Extracellular | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374430 | 3 | 5 | 1_50 | 264 | 365.0 | Topological domain | Note=Extracellular | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374430 | 3 | 5 | 226_255 | 264 | 365.0 | Topological domain | Note=Cytoplasmic | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374430 | 3 | 5 | 76_83 | 264 | 365.0 | Topological domain | Note=Cytoplasmic | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374431 | 3 | 5 | 108_121 | 264 | 365.0 | Topological domain | Note=Extracellular | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374431 | 3 | 5 | 145_163 | 264 | 365.0 | Topological domain | Note=Cytoplasmic | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374431 | 3 | 5 | 185_204 | 264 | 365.0 | Topological domain | Note=Extracellular | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374431 | 3 | 5 | 1_50 | 264 | 365.0 | Topological domain | Note=Extracellular | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374431 | 3 | 5 | 226_255 | 264 | 365.0 | Topological domain | Note=Cytoplasmic | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374431 | 3 | 5 | 76_83 | 264 | 365.0 | Topological domain | Note=Cytoplasmic | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000358883 | 2 | 4 | 122_144 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D3 | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000358883 | 2 | 4 | 164_184 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D4 | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000358883 | 2 | 4 | 205_225 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D5 | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000358883 | 2 | 4 | 256_280 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D6 | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000358883 | 2 | 4 | 51_75 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D1 | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000358883 | 2 | 4 | 84_107 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D2 | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374430 | 3 | 5 | 122_144 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D3 | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374430 | 3 | 5 | 164_184 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D4 | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374430 | 3 | 5 | 205_225 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D5 | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374430 | 3 | 5 | 256_280 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D6 | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374430 | 3 | 5 | 51_75 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D1 | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374430 | 3 | 5 | 84_107 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D2 | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374431 | 3 | 5 | 122_144 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D3 | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374431 | 3 | 5 | 164_184 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D4 | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374431 | 3 | 5 | 205_225 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D5 | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374431 | 3 | 5 | 256_280 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D6 | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374431 | 3 | 5 | 51_75 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D1 | |
| Tgene | LPAR1 | chr9:112542813 | chr9:113638002 | ENST00000374431 | 3 | 5 | 84_107 | 264 | 365.0 | Transmembrane | Note=Helical%3B Name%3D2 |
Top |
Fusion Gene Sequence for PALM2-LPAR1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >62494_62494_1_PALM2-LPAR1_PALM2_chr9_112542813_ENST00000314527_LPAR1_chr9_113638002_ENST00000538760_length(transcript)=642nt_BP=95nt GCGTGTGTTTTTTCTTTCCTCTTTCCCCTGCCTGTGTGCCCTTCTCCAGGATGGCAGAGGCGGAATTGCACAAGGAAAGGCTGCAAGCCA TAGCAGGGCCTTTATCATCTGCTGGACTCCTGGATTGGTTTTGTTACTTCTAGACGTGTGCTGTCCACAGTGCGACGTGCTGGCCTATGA GAAATTCTTCCTTCTCCTTGCTGAATTCAACTCTGCCATGAACCCCATCATTTACTCCTACCGCGACAAAGAAATGAGCGCCACCTTTAG GCAGATCCTCTGCTGCCAGCGCAGTGAGAACCCCACCGGCCCCACAGAAGGCTCAGACCGCTCGGCTTCCTCCCTCAACCACACCATCTT GGCTGGAGTTCACAGCAATGACCACTCTGTGGTTTAGAACGGAAACTGAGATGAGGAACCAGCCGTCCTCTCTTGGAGGATAAACAGCCT CCCCCTACCCAATTGCCAGGGCAAGGTGGGGTGTGAGAGAGGAGAAAAGTCAACTCATGTACTTAAACACTAACCAATGACAGTATTTGT TCCTGGACCCCACAAGACTTGATATATATTGAAAATTAGCTTATGTGACAACCCTCATCTTGATCCCCATCCCTTCTGAAAGTAGGAAGT >62494_62494_1_PALM2-LPAR1_PALM2_chr9_112542813_ENST00000314527_LPAR1_chr9_113638002_ENST00000538760_length(amino acids)=121AA_BP=22 MCALLQDGRGGIAQGKAASHSRAFIICWTPGLVLLLLDVCCPQCDVLAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILCCQRSEN -------------------------------------------------------------- >62494_62494_2_PALM2-LPAR1_PALM2_chr9_112542813_ENST00000314527_LPAR1_chr9_113638002_ENST00000541779_length(transcript)=2554nt_BP=95nt GCGTGTGTTTTTTCTTTCCTCTTTCCCCTGCCTGTGTGCCCTTCTCCAGGATGGCAGAGGCGGAATTGCACAAGGAAAGGCTGCAAGCCA TAGCAGGGCCTTTATCATCTGCTGGACTCCTGGATTGGTTTTGTTACTTCTAGACGTGTGCTGTCCACAGTGCGACGTGCTGGCCTATGA GAAATTCTTCCTTCTCCTTGCTGAATTCAACTCTGCCATGAACCCCATCATTTACTCCTACCGCGACAAAGAAATGAGCGCCACCTTTAG GCAGATCCTCTGCTGCCAGCGCAGTGAGAACCCCACCGGCCCCACAGAAGGCTCAGACCGCTCGGCTTCCTCCCTCAACCACACCATCTT GGCTGGAGTTCACAGCAATGACCACTCTGTGGTTTAGAACGGAAACTGAGATGAGGAACCAGCCGTCCTCTCTTGGAGGATAAACAGCCT CCCCCTACCCAATTGCCAGGGCAAGGTGGGGTGTGAGAGAGGAGAAAAGTCAACTCATGTACTTAAACACTAACCAATGACAGTATTTGT TCCTGGACCCCACAAGACTTGATATATATTGAAAATTAGCTTATGTGACAACCCTCATCTTGATCCCCATCCCTTCTGAAAGTAGGAAGT TGGAGCTCTTGCAATGGAATTCAAGAACAGACTCTGGAGTGTCCATTTAGACTACACTAACTAGACTTTTAAAAGATTTTGTGTGGTTTG GTGCAAGTCAGAATAAATTCTGGCTAGTTGAATCCACAACTTCATTTATATACAGGCTTCCCTTTTTTATTTTTAAAGGATACGTTTCAC TTAATAAACACGTTTATGCCTATCAGCATGTTTGTGATGGATGAGACTATGGACTGCTTTTAAACTACCATAATTCCATTTTTTCCCTTA CATAGGAAAACTGTAAGTTGGAATTATCTTTTGTTTAGAAAGCATGCATGTAATGTATGTATGCAGTATGCCTTACTTAAAAAGATTAAA AGGATACTAATGTTAAATCTTCTAGGAAATAGAACCTAGACTTCAAAGCCAGTATTTGTTTAGGTCATGAAGCAAACAATGCTCTAATCA CAATATTAACTGTTTAATTAAAATGTTGTAACAAGTATAAAACAGGGAATGTAAGTTTATTACCAAAGTGATATGTATTCCAAAAAAGTC ATAGAAGATGAAGCACTATAATATTGTTCCCATATATTTAAAATACCCAAGTACATTCTAATTACCAGTATATCAGAGGAAAATTTTCGT AGTCTTTGTAAAATAATATACTCATCATAGAAAACTTGAAAAATGCAGAAATGTATAAAAAAGCAAAAATGATTACTGATAATATCACAA CCCAGAAGTAACCACCTTTAAAAAGCAACCCCCATGTATGCCTATATGTGTATTGTATACTTTTTTTACATAATTGGAGTCATACTGTAA ACAGTTTTATAAGTAGATCTTTTTCATTGCAAAATTGCCACATTTTCTTATGGCATTAAAAATTTTACAAAAACATAATTTTAATGGCTA TATTATATTCCATTTAATGGATGCAACTCAGTTTATTTAACCATTCCCATGTTGTTAACTATTTAGGTTGTTTCTAATTTTCATTATTAT AAAGTTGCAGAAATTTGGTGTACATAAAACTGTCTCCATATAATTGATTATTAGGATATATTCCCATGAAGGATTCTTTTTTTAAAAAAA TGTGAAATGTCATCTTGTACTTACACCTTTCATGAAAAGGGATTTCCTGCTTTTGTACTGCATGGGTGGCAGTTGTGAGGAAAAGCCAGT CAAATGACCTTTTTACAAAAGAAATGCAGTGGTCACTTCAGTTGAGAGTGACTTTTTAATACAACAAGATCAACTAGAAGAATTCAACTG TCTCAAGAATCAAGGTACCCCAATATATCTCGCAATTCCAAACTTTGTTTGAGGGACTCGTTATCCAGCTCTTGGTAGCCACACCTGCAA TGTAAAATGGAAGAAAATGCAAAGAAACCAAATGTGCCGAGTGAATAAAGGATTGTCATATCAATTAGTGTGGGTCCATCTGTAGCAAAT GGGTTGAGTGTGTCAGTATGTGGTTGGTTACTGTGTATTCGCCAGGAATCACCCCGATAGGCTGCCACCCTATTAGGTGATACCTGTTTA ATATGTTGTCCAGGTAGACTAGTAGTTGCATCAGTTTGCTGTAACAAGTAACCAGTGAGGTAACACAGTGGTGAAGCAGGTCAGGGGAGG TCAGGAGGATGTCTGAGAGAAAGAAGTCCGGGAGATGAATGGCTGTCTAGGAAGGAGGATGTCAGTGCACGGTTAGTGTTTGAGCAGAGG GCAGACTTGTAAAGTACCTGTAGTGAAAAGAATGTGGGGACCCGATTAGCAGAAAGGTGTTTGCACGTACTTTATACAAAATACAGAATA CTTTATATTGGAAGTGAAAGAAATGAACGTGGACTTTTACACATGTGCATATTTTTCTGGAGGCTATATGGATTATTTGAAATGAAAAGC >62494_62494_2_PALM2-LPAR1_PALM2_chr9_112542813_ENST00000314527_LPAR1_chr9_113638002_ENST00000541779_length(amino acids)=121AA_BP=22 MCALLQDGRGGIAQGKAASHSRAFIICWTPGLVLLLLDVCCPQCDVLAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILCCQRSEN -------------------------------------------------------------- >62494_62494_3_PALM2-LPAR1_PALM2_chr9_112542813_ENST00000374531_LPAR1_chr9_113638002_ENST00000538760_length(transcript)=672nt_BP=125nt GTGCACTCGGCTCCGGGAGAGGCGAGCAGCGCCGGTGAGCCCCGCAGCAGCGCACCCGGCCGCGGAGCCCCGCGATGGAGATGGCAGAGG CGGAATTGCACAAGGAAAGGCTGCAAGCCATAGCAGGGCCTTTATCATCTGCTGGACTCCTGGATTGGTTTTGTTACTTCTAGACGTGTG CTGTCCACAGTGCGACGTGCTGGCCTATGAGAAATTCTTCCTTCTCCTTGCTGAATTCAACTCTGCCATGAACCCCATCATTTACTCCTA CCGCGACAAAGAAATGAGCGCCACCTTTAGGCAGATCCTCTGCTGCCAGCGCAGTGAGAACCCCACCGGCCCCACAGAAGGCTCAGACCG CTCGGCTTCCTCCCTCAACCACACCATCTTGGCTGGAGTTCACAGCAATGACCACTCTGTGGTTTAGAACGGAAACTGAGATGAGGAACC AGCCGTCCTCTCTTGGAGGATAAACAGCCTCCCCCTACCCAATTGCCAGGGCAAGGTGGGGTGTGAGAGAGGAGAAAAGTCAACTCATGT ACTTAAACACTAACCAATGACAGTATTTGTTCCTGGACCCCACAAGACTTGATATATATTGAAAATTAGCTTATGTGACAACCCTCATCT >62494_62494_3_PALM2-LPAR1_PALM2_chr9_112542813_ENST00000374531_LPAR1_chr9_113638002_ENST00000538760_length(amino acids)=90AA_BP= MVLLLLDVCCPQCDVLAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILCCQRSENPTGPTEGSDRSASSLNHTILAGVHSNDHSVV -------------------------------------------------------------- >62494_62494_4_PALM2-LPAR1_PALM2_chr9_112542813_ENST00000374531_LPAR1_chr9_113638002_ENST00000541779_length(transcript)=2584nt_BP=125nt GTGCACTCGGCTCCGGGAGAGGCGAGCAGCGCCGGTGAGCCCCGCAGCAGCGCACCCGGCCGCGGAGCCCCGCGATGGAGATGGCAGAGG CGGAATTGCACAAGGAAAGGCTGCAAGCCATAGCAGGGCCTTTATCATCTGCTGGACTCCTGGATTGGTTTTGTTACTTCTAGACGTGTG CTGTCCACAGTGCGACGTGCTGGCCTATGAGAAATTCTTCCTTCTCCTTGCTGAATTCAACTCTGCCATGAACCCCATCATTTACTCCTA CCGCGACAAAGAAATGAGCGCCACCTTTAGGCAGATCCTCTGCTGCCAGCGCAGTGAGAACCCCACCGGCCCCACAGAAGGCTCAGACCG CTCGGCTTCCTCCCTCAACCACACCATCTTGGCTGGAGTTCACAGCAATGACCACTCTGTGGTTTAGAACGGAAACTGAGATGAGGAACC AGCCGTCCTCTCTTGGAGGATAAACAGCCTCCCCCTACCCAATTGCCAGGGCAAGGTGGGGTGTGAGAGAGGAGAAAAGTCAACTCATGT ACTTAAACACTAACCAATGACAGTATTTGTTCCTGGACCCCACAAGACTTGATATATATTGAAAATTAGCTTATGTGACAACCCTCATCT TGATCCCCATCCCTTCTGAAAGTAGGAAGTTGGAGCTCTTGCAATGGAATTCAAGAACAGACTCTGGAGTGTCCATTTAGACTACACTAA CTAGACTTTTAAAAGATTTTGTGTGGTTTGGTGCAAGTCAGAATAAATTCTGGCTAGTTGAATCCACAACTTCATTTATATACAGGCTTC CCTTTTTTATTTTTAAAGGATACGTTTCACTTAATAAACACGTTTATGCCTATCAGCATGTTTGTGATGGATGAGACTATGGACTGCTTT TAAACTACCATAATTCCATTTTTTCCCTTACATAGGAAAACTGTAAGTTGGAATTATCTTTTGTTTAGAAAGCATGCATGTAATGTATGT ATGCAGTATGCCTTACTTAAAAAGATTAAAAGGATACTAATGTTAAATCTTCTAGGAAATAGAACCTAGACTTCAAAGCCAGTATTTGTT TAGGTCATGAAGCAAACAATGCTCTAATCACAATATTAACTGTTTAATTAAAATGTTGTAACAAGTATAAAACAGGGAATGTAAGTTTAT TACCAAAGTGATATGTATTCCAAAAAAGTCATAGAAGATGAAGCACTATAATATTGTTCCCATATATTTAAAATACCCAAGTACATTCTA ATTACCAGTATATCAGAGGAAAATTTTCGTAGTCTTTGTAAAATAATATACTCATCATAGAAAACTTGAAAAATGCAGAAATGTATAAAA AAGCAAAAATGATTACTGATAATATCACAACCCAGAAGTAACCACCTTTAAAAAGCAACCCCCATGTATGCCTATATGTGTATTGTATAC TTTTTTTACATAATTGGAGTCATACTGTAAACAGTTTTATAAGTAGATCTTTTTCATTGCAAAATTGCCACATTTTCTTATGGCATTAAA AATTTTACAAAAACATAATTTTAATGGCTATATTATATTCCATTTAATGGATGCAACTCAGTTTATTTAACCATTCCCATGTTGTTAACT ATTTAGGTTGTTTCTAATTTTCATTATTATAAAGTTGCAGAAATTTGGTGTACATAAAACTGTCTCCATATAATTGATTATTAGGATATA TTCCCATGAAGGATTCTTTTTTTAAAAAAATGTGAAATGTCATCTTGTACTTACACCTTTCATGAAAAGGGATTTCCTGCTTTTGTACTG CATGGGTGGCAGTTGTGAGGAAAAGCCAGTCAAATGACCTTTTTACAAAAGAAATGCAGTGGTCACTTCAGTTGAGAGTGACTTTTTAAT ACAACAAGATCAACTAGAAGAATTCAACTGTCTCAAGAATCAAGGTACCCCAATATATCTCGCAATTCCAAACTTTGTTTGAGGGACTCG TTATCCAGCTCTTGGTAGCCACACCTGCAATGTAAAATGGAAGAAAATGCAAAGAAACCAAATGTGCCGAGTGAATAAAGGATTGTCATA TCAATTAGTGTGGGTCCATCTGTAGCAAATGGGTTGAGTGTGTCAGTATGTGGTTGGTTACTGTGTATTCGCCAGGAATCACCCCGATAG GCTGCCACCCTATTAGGTGATACCTGTTTAATATGTTGTCCAGGTAGACTAGTAGTTGCATCAGTTTGCTGTAACAAGTAACCAGTGAGG TAACACAGTGGTGAAGCAGGTCAGGGGAGGTCAGGAGGATGTCTGAGAGAAAGAAGTCCGGGAGATGAATGGCTGTCTAGGAAGGAGGAT GTCAGTGCACGGTTAGTGTTTGAGCAGAGGGCAGACTTGTAAAGTACCTGTAGTGAAAAGAATGTGGGGACCCGATTAGCAGAAAGGTGT TTGCACGTACTTTATACAAAATACAGAATACTTTATATTGGAAGTGAAAGAAATGAACGTGGACTTTTACACATGTGCATATTTTTCTGG >62494_62494_4_PALM2-LPAR1_PALM2_chr9_112542813_ENST00000374531_LPAR1_chr9_113638002_ENST00000541779_length(amino acids)=111AA_BP= MCYLTGYLLQQTDATTSLPGQHIKQVSPNRVAAYRGDSWRIHSNQPHTDTLNPFATDGPTLIDMTILYSLGTFGFFAFSSILHCRCGYQE -------------------------------------------------------------- >62494_62494_5_PALM2-LPAR1_PALM2_chr9_112542813_ENST00000448454_LPAR1_chr9_113638002_ENST00000538760_length(transcript)=598nt_BP=51nt ATGGAGATGGCAGAGGCGGAATTGCACAAGGAAAGGCTGCAAGCCATAGCAGGGCCTTTATCATCTGCTGGACTCCTGGATTGGTTTTGT TACTTCTAGACGTGTGCTGTCCACAGTGCGACGTGCTGGCCTATGAGAAATTCTTCCTTCTCCTTGCTGAATTCAACTCTGCCATGAACC CCATCATTTACTCCTACCGCGACAAAGAAATGAGCGCCACCTTTAGGCAGATCCTCTGCTGCCAGCGCAGTGAGAACCCCACCGGCCCCA CAGAAGGCTCAGACCGCTCGGCTTCCTCCCTCAACCACACCATCTTGGCTGGAGTTCACAGCAATGACCACTCTGTGGTTTAGAACGGAA ACTGAGATGAGGAACCAGCCGTCCTCTCTTGGAGGATAAACAGCCTCCCCCTACCCAATTGCCAGGGCAAGGTGGGGTGTGAGAGAGGAG AAAAGTCAACTCATGTACTTAAACACTAACCAATGACAGTATTTGTTCCTGGACCCCACAAGACTTGATATATATTGAAAATTAGCTTAT >62494_62494_5_PALM2-LPAR1_PALM2_chr9_112542813_ENST00000448454_LPAR1_chr9_113638002_ENST00000538760_length(amino acids)=90AA_BP= MVLLLLDVCCPQCDVLAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILCCQRSENPTGPTEGSDRSASSLNHTILAGVHSNDHSVV -------------------------------------------------------------- >62494_62494_6_PALM2-LPAR1_PALM2_chr9_112542813_ENST00000448454_LPAR1_chr9_113638002_ENST00000541779_length(transcript)=2510nt_BP=51nt ATGGAGATGGCAGAGGCGGAATTGCACAAGGAAAGGCTGCAAGCCATAGCAGGGCCTTTATCATCTGCTGGACTCCTGGATTGGTTTTGT TACTTCTAGACGTGTGCTGTCCACAGTGCGACGTGCTGGCCTATGAGAAATTCTTCCTTCTCCTTGCTGAATTCAACTCTGCCATGAACC CCATCATTTACTCCTACCGCGACAAAGAAATGAGCGCCACCTTTAGGCAGATCCTCTGCTGCCAGCGCAGTGAGAACCCCACCGGCCCCA CAGAAGGCTCAGACCGCTCGGCTTCCTCCCTCAACCACACCATCTTGGCTGGAGTTCACAGCAATGACCACTCTGTGGTTTAGAACGGAA ACTGAGATGAGGAACCAGCCGTCCTCTCTTGGAGGATAAACAGCCTCCCCCTACCCAATTGCCAGGGCAAGGTGGGGTGTGAGAGAGGAG AAAAGTCAACTCATGTACTTAAACACTAACCAATGACAGTATTTGTTCCTGGACCCCACAAGACTTGATATATATTGAAAATTAGCTTAT GTGACAACCCTCATCTTGATCCCCATCCCTTCTGAAAGTAGGAAGTTGGAGCTCTTGCAATGGAATTCAAGAACAGACTCTGGAGTGTCC ATTTAGACTACACTAACTAGACTTTTAAAAGATTTTGTGTGGTTTGGTGCAAGTCAGAATAAATTCTGGCTAGTTGAATCCACAACTTCA TTTATATACAGGCTTCCCTTTTTTATTTTTAAAGGATACGTTTCACTTAATAAACACGTTTATGCCTATCAGCATGTTTGTGATGGATGA GACTATGGACTGCTTTTAAACTACCATAATTCCATTTTTTCCCTTACATAGGAAAACTGTAAGTTGGAATTATCTTTTGTTTAGAAAGCA TGCATGTAATGTATGTATGCAGTATGCCTTACTTAAAAAGATTAAAAGGATACTAATGTTAAATCTTCTAGGAAATAGAACCTAGACTTC AAAGCCAGTATTTGTTTAGGTCATGAAGCAAACAATGCTCTAATCACAATATTAACTGTTTAATTAAAATGTTGTAACAAGTATAAAACA GGGAATGTAAGTTTATTACCAAAGTGATATGTATTCCAAAAAAGTCATAGAAGATGAAGCACTATAATATTGTTCCCATATATTTAAAAT ACCCAAGTACATTCTAATTACCAGTATATCAGAGGAAAATTTTCGTAGTCTTTGTAAAATAATATACTCATCATAGAAAACTTGAAAAAT GCAGAAATGTATAAAAAAGCAAAAATGATTACTGATAATATCACAACCCAGAAGTAACCACCTTTAAAAAGCAACCCCCATGTATGCCTA TATGTGTATTGTATACTTTTTTTACATAATTGGAGTCATACTGTAAACAGTTTTATAAGTAGATCTTTTTCATTGCAAAATTGCCACATT TTCTTATGGCATTAAAAATTTTACAAAAACATAATTTTAATGGCTATATTATATTCCATTTAATGGATGCAACTCAGTTTATTTAACCAT TCCCATGTTGTTAACTATTTAGGTTGTTTCTAATTTTCATTATTATAAAGTTGCAGAAATTTGGTGTACATAAAACTGTCTCCATATAAT TGATTATTAGGATATATTCCCATGAAGGATTCTTTTTTTAAAAAAATGTGAAATGTCATCTTGTACTTACACCTTTCATGAAAAGGGATT TCCTGCTTTTGTACTGCATGGGTGGCAGTTGTGAGGAAAAGCCAGTCAAATGACCTTTTTACAAAAGAAATGCAGTGGTCACTTCAGTTG AGAGTGACTTTTTAATACAACAAGATCAACTAGAAGAATTCAACTGTCTCAAGAATCAAGGTACCCCAATATATCTCGCAATTCCAAACT TTGTTTGAGGGACTCGTTATCCAGCTCTTGGTAGCCACACCTGCAATGTAAAATGGAAGAAAATGCAAAGAAACCAAATGTGCCGAGTGA ATAAAGGATTGTCATATCAATTAGTGTGGGTCCATCTGTAGCAAATGGGTTGAGTGTGTCAGTATGTGGTTGGTTACTGTGTATTCGCCA GGAATCACCCCGATAGGCTGCCACCCTATTAGGTGATACCTGTTTAATATGTTGTCCAGGTAGACTAGTAGTTGCATCAGTTTGCTGTAA CAAGTAACCAGTGAGGTAACACAGTGGTGAAGCAGGTCAGGGGAGGTCAGGAGGATGTCTGAGAGAAAGAAGTCCGGGAGATGAATGGCT GTCTAGGAAGGAGGATGTCAGTGCACGGTTAGTGTTTGAGCAGAGGGCAGACTTGTAAAGTACCTGTAGTGAAAAGAATGTGGGGACCCG ATTAGCAGAAAGGTGTTTGCACGTACTTTATACAAAATACAGAATACTTTATATTGGAAGTGAAAGAAATGAACGTGGACTTTTACACAT >62494_62494_6_PALM2-LPAR1_PALM2_chr9_112542813_ENST00000448454_LPAR1_chr9_113638002_ENST00000541779_length(amino acids)=111AA_BP= MCYLTGYLLQQTDATTSLPGQHIKQVSPNRVAAYRGDSWRIHSNQPHTDTLNPFATDGPTLIDMTILYSLGTFGFFAFSSILHCRCGYQE -------------------------------------------------------------- >62494_62494_7_PALM2-LPAR1_PALM2_chr9_112542813_ENST00000483909_LPAR1_chr9_113638002_ENST00000538760_length(transcript)=678nt_BP=131nt CCCCGGGGTGCCCATCAGTTTCCTTGGGTGACTACAGCGTGTGTTTTTTCTTTCCTCTTTCCCCTGCCTGTGTGCCCTTCTCCAGGATGG CAGAGGCGGAATTGCACAAGGAAAGGCTGCAAGCCATAGCAGGGCCTTTATCATCTGCTGGACTCCTGGATTGGTTTTGTTACTTCTAGA CGTGTGCTGTCCACAGTGCGACGTGCTGGCCTATGAGAAATTCTTCCTTCTCCTTGCTGAATTCAACTCTGCCATGAACCCCATCATTTA CTCCTACCGCGACAAAGAAATGAGCGCCACCTTTAGGCAGATCCTCTGCTGCCAGCGCAGTGAGAACCCCACCGGCCCCACAGAAGGCTC AGACCGCTCGGCTTCCTCCCTCAACCACACCATCTTGGCTGGAGTTCACAGCAATGACCACTCTGTGGTTTAGAACGGAAACTGAGATGA GGAACCAGCCGTCCTCTCTTGGAGGATAAACAGCCTCCCCCTACCCAATTGCCAGGGCAAGGTGGGGTGTGAGAGAGGAGAAAAGTCAAC TCATGTACTTAAACACTAACCAATGACAGTATTTGTTCCTGGACCCCACAAGACTTGATATATATTGAAAATTAGCTTATGTGACAACCC >62494_62494_7_PALM2-LPAR1_PALM2_chr9_112542813_ENST00000483909_LPAR1_chr9_113638002_ENST00000538760_length(amino acids)=121AA_BP=22 MCALLQDGRGGIAQGKAASHSRAFIICWTPGLVLLLLDVCCPQCDVLAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILCCQRSEN -------------------------------------------------------------- >62494_62494_8_PALM2-LPAR1_PALM2_chr9_112542813_ENST00000483909_LPAR1_chr9_113638002_ENST00000541779_length(transcript)=2590nt_BP=131nt CCCCGGGGTGCCCATCAGTTTCCTTGGGTGACTACAGCGTGTGTTTTTTCTTTCCTCTTTCCCCTGCCTGTGTGCCCTTCTCCAGGATGG CAGAGGCGGAATTGCACAAGGAAAGGCTGCAAGCCATAGCAGGGCCTTTATCATCTGCTGGACTCCTGGATTGGTTTTGTTACTTCTAGA CGTGTGCTGTCCACAGTGCGACGTGCTGGCCTATGAGAAATTCTTCCTTCTCCTTGCTGAATTCAACTCTGCCATGAACCCCATCATTTA CTCCTACCGCGACAAAGAAATGAGCGCCACCTTTAGGCAGATCCTCTGCTGCCAGCGCAGTGAGAACCCCACCGGCCCCACAGAAGGCTC AGACCGCTCGGCTTCCTCCCTCAACCACACCATCTTGGCTGGAGTTCACAGCAATGACCACTCTGTGGTTTAGAACGGAAACTGAGATGA GGAACCAGCCGTCCTCTCTTGGAGGATAAACAGCCTCCCCCTACCCAATTGCCAGGGCAAGGTGGGGTGTGAGAGAGGAGAAAAGTCAAC TCATGTACTTAAACACTAACCAATGACAGTATTTGTTCCTGGACCCCACAAGACTTGATATATATTGAAAATTAGCTTATGTGACAACCC TCATCTTGATCCCCATCCCTTCTGAAAGTAGGAAGTTGGAGCTCTTGCAATGGAATTCAAGAACAGACTCTGGAGTGTCCATTTAGACTA CACTAACTAGACTTTTAAAAGATTTTGTGTGGTTTGGTGCAAGTCAGAATAAATTCTGGCTAGTTGAATCCACAACTTCATTTATATACA GGCTTCCCTTTTTTATTTTTAAAGGATACGTTTCACTTAATAAACACGTTTATGCCTATCAGCATGTTTGTGATGGATGAGACTATGGAC TGCTTTTAAACTACCATAATTCCATTTTTTCCCTTACATAGGAAAACTGTAAGTTGGAATTATCTTTTGTTTAGAAAGCATGCATGTAAT GTATGTATGCAGTATGCCTTACTTAAAAAGATTAAAAGGATACTAATGTTAAATCTTCTAGGAAATAGAACCTAGACTTCAAAGCCAGTA TTTGTTTAGGTCATGAAGCAAACAATGCTCTAATCACAATATTAACTGTTTAATTAAAATGTTGTAACAAGTATAAAACAGGGAATGTAA GTTTATTACCAAAGTGATATGTATTCCAAAAAAGTCATAGAAGATGAAGCACTATAATATTGTTCCCATATATTTAAAATACCCAAGTAC ATTCTAATTACCAGTATATCAGAGGAAAATTTTCGTAGTCTTTGTAAAATAATATACTCATCATAGAAAACTTGAAAAATGCAGAAATGT ATAAAAAAGCAAAAATGATTACTGATAATATCACAACCCAGAAGTAACCACCTTTAAAAAGCAACCCCCATGTATGCCTATATGTGTATT GTATACTTTTTTTACATAATTGGAGTCATACTGTAAACAGTTTTATAAGTAGATCTTTTTCATTGCAAAATTGCCACATTTTCTTATGGC ATTAAAAATTTTACAAAAACATAATTTTAATGGCTATATTATATTCCATTTAATGGATGCAACTCAGTTTATTTAACCATTCCCATGTTG TTAACTATTTAGGTTGTTTCTAATTTTCATTATTATAAAGTTGCAGAAATTTGGTGTACATAAAACTGTCTCCATATAATTGATTATTAG GATATATTCCCATGAAGGATTCTTTTTTTAAAAAAATGTGAAATGTCATCTTGTACTTACACCTTTCATGAAAAGGGATTTCCTGCTTTT GTACTGCATGGGTGGCAGTTGTGAGGAAAAGCCAGTCAAATGACCTTTTTACAAAAGAAATGCAGTGGTCACTTCAGTTGAGAGTGACTT TTTAATACAACAAGATCAACTAGAAGAATTCAACTGTCTCAAGAATCAAGGTACCCCAATATATCTCGCAATTCCAAACTTTGTTTGAGG GACTCGTTATCCAGCTCTTGGTAGCCACACCTGCAATGTAAAATGGAAGAAAATGCAAAGAAACCAAATGTGCCGAGTGAATAAAGGATT GTCATATCAATTAGTGTGGGTCCATCTGTAGCAAATGGGTTGAGTGTGTCAGTATGTGGTTGGTTACTGTGTATTCGCCAGGAATCACCC CGATAGGCTGCCACCCTATTAGGTGATACCTGTTTAATATGTTGTCCAGGTAGACTAGTAGTTGCATCAGTTTGCTGTAACAAGTAACCA GTGAGGTAACACAGTGGTGAAGCAGGTCAGGGGAGGTCAGGAGGATGTCTGAGAGAAAGAAGTCCGGGAGATGAATGGCTGTCTAGGAAG GAGGATGTCAGTGCACGGTTAGTGTTTGAGCAGAGGGCAGACTTGTAAAGTACCTGTAGTGAAAAGAATGTGGGGACCCGATTAGCAGAA AGGTGTTTGCACGTACTTTATACAAAATACAGAATACTTTATATTGGAAGTGAAAGAAATGAACGTGGACTTTTACACATGTGCATATTT >62494_62494_8_PALM2-LPAR1_PALM2_chr9_112542813_ENST00000483909_LPAR1_chr9_113638002_ENST00000541779_length(amino acids)=121AA_BP=22 MCALLQDGRGGIAQGKAASHSRAFIICWTPGLVLLLLDVCCPQCDVLAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILCCQRSEN -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for PALM2-LPAR1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for PALM2-LPAR1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for PALM2-LPAR1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
