
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:PCMTD2-MACROD2 (FusionGDB2 ID:63392) |
Fusion Gene Summary for PCMTD2-MACROD2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: PCMTD2-MACROD2 | Fusion gene ID: 63392 | Hgene | Tgene | Gene symbol | PCMTD2 | MACROD2 | Gene ID | 55251 | 140733 |
| Gene name | protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 | mono-ADP ribosylhydrolase 2 | |
| Synonyms | C20orf36 | C20orf133|C2orf133 | |
| Cytomap | 20q13.33 | 20p12.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein-L-isoaspartate O-methyltransferase domain-containing protein 2 | ADP-ribose glycohydrolase MACROD2MACRO domain containing 2MACRO domain-containing protein 2O-acetyl-ADP-ribose deacetylase MACROD2[Protein ADP-ribosylaspartate] hydrolase MACROD2[Protein ADP-ribosylglutamate] hydrolase[Protein ADP-ribosylglutamate] | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | A1Z1Q3 | |
| Ensembl transtripts involved in fusion gene | ENST00000299468, ENST00000308824, ENST00000369758, ENST00000266078, ENST00000609372, | ENST00000402914, ENST00000378058, ENST00000407045, ENST00000464883, ENST00000217246, ENST00000310348, | |
| Fusion gene scores | * DoF score | 9 X 5 X 7=315 | 37 X 34 X 11=13838 |
| # samples | 9 | 41 | |
| ** MAII score | log2(9/315*10)=-1.8073549220576 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(41/13838*10)=-5.07686772601114 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: PCMTD2 [Title/Abstract] AND MACROD2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | PCMTD2(62891625)-MACROD2(15210586), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | MACROD2 | GO:0042278 | purine nucleoside metabolic process | 21257746 |
| Tgene | MACROD2 | GO:0051725 | protein de-ADP-ribosylation | 23474712 |
 Fusion gene breakpoints across PCMTD2 (5'-gene) Fusion gene breakpoints across PCMTD2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
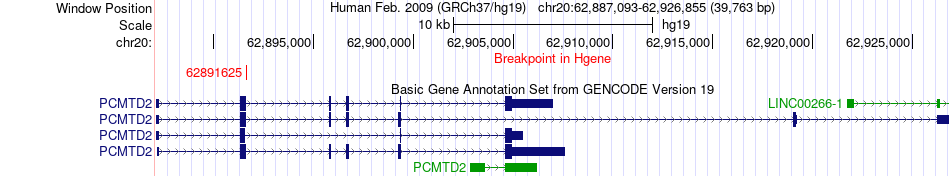 |
 Fusion gene breakpoints across MACROD2 (3'-gene) Fusion gene breakpoints across MACROD2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
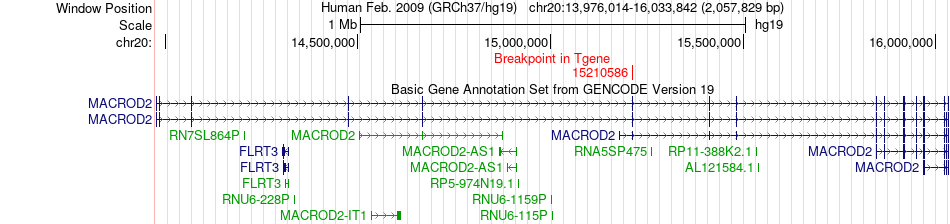 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-HC-7080-01A | PCMTD2 | chr20 | 62891625 | - | MACROD2 | chr20 | 15210586 | + |
| ChimerDB4 | PRAD | TCGA-HC-7080-01A | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| ChimerDB4 | PRAD | TCGA-HC-7080 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
Top |
Fusion Gene ORF analysis for PCMTD2-MACROD2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000299468 | ENST00000402914 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| 5CDS-5UTR | ENST00000308824 | ENST00000402914 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| 5CDS-5UTR | ENST00000369758 | ENST00000402914 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| 5CDS-intron | ENST00000299468 | ENST00000378058 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| 5CDS-intron | ENST00000299468 | ENST00000407045 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| 5CDS-intron | ENST00000299468 | ENST00000464883 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| 5CDS-intron | ENST00000308824 | ENST00000378058 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| 5CDS-intron | ENST00000308824 | ENST00000407045 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| 5CDS-intron | ENST00000308824 | ENST00000464883 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| 5CDS-intron | ENST00000369758 | ENST00000378058 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| 5CDS-intron | ENST00000369758 | ENST00000407045 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| 5CDS-intron | ENST00000369758 | ENST00000464883 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| In-frame | ENST00000299468 | ENST00000217246 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| In-frame | ENST00000299468 | ENST00000310348 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| In-frame | ENST00000308824 | ENST00000217246 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| In-frame | ENST00000308824 | ENST00000310348 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| In-frame | ENST00000369758 | ENST00000217246 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| In-frame | ENST00000369758 | ENST00000310348 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| intron-3CDS | ENST00000266078 | ENST00000217246 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| intron-3CDS | ENST00000266078 | ENST00000310348 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| intron-3CDS | ENST00000609372 | ENST00000217246 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| intron-3CDS | ENST00000609372 | ENST00000310348 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| intron-5UTR | ENST00000266078 | ENST00000402914 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| intron-5UTR | ENST00000609372 | ENST00000402914 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| intron-intron | ENST00000266078 | ENST00000378058 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| intron-intron | ENST00000266078 | ENST00000407045 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| intron-intron | ENST00000266078 | ENST00000464883 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| intron-intron | ENST00000609372 | ENST00000378058 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| intron-intron | ENST00000609372 | ENST00000407045 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
| intron-intron | ENST00000609372 | ENST00000464883 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000299468 | PCMTD2 | chr20 | 62891625 | + | ENST00000217246 | MACROD2 | chr20 | 15210586 | + | 4639 | 458 | 25 | 1317 | 430 |
| ENST00000299468 | PCMTD2 | chr20 | 62891625 | + | ENST00000310348 | MACROD2 | chr20 | 15210586 | + | 1387 | 458 | 25 | 1386 | 454 |
| ENST00000369758 | PCMTD2 | chr20 | 62891625 | + | ENST00000217246 | MACROD2 | chr20 | 15210586 | + | 4642 | 461 | 28 | 1320 | 430 |
| ENST00000369758 | PCMTD2 | chr20 | 62891625 | + | ENST00000310348 | MACROD2 | chr20 | 15210586 | + | 1390 | 461 | 28 | 1389 | 454 |
| ENST00000308824 | PCMTD2 | chr20 | 62891625 | + | ENST00000217246 | MACROD2 | chr20 | 15210586 | + | 4615 | 434 | 1 | 1293 | 430 |
| ENST00000308824 | PCMTD2 | chr20 | 62891625 | + | ENST00000310348 | MACROD2 | chr20 | 15210586 | + | 1363 | 434 | 1 | 1362 | 454 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000299468 | ENST00000217246 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + | 8.47E-05 | 0.99991524 |
| ENST00000299468 | ENST00000310348 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + | 0.00106908 | 0.9989309 |
| ENST00000369758 | ENST00000217246 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + | 8.46E-05 | 0.99991536 |
| ENST00000369758 | ENST00000310348 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + | 0.001080251 | 0.9989197 |
| ENST00000308824 | ENST00000217246 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + | 8.75E-05 | 0.9999125 |
| ENST00000308824 | ENST00000310348 | PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210586 | + | 0.001157137 | 0.99884284 |
Top |
Fusion Genomic Features for PCMTD2-MACROD2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210585 | + | 3.16E-06 | 0.9999968 |
| PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210585 | + | 3.16E-06 | 0.9999968 |
| PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210585 | + | 3.16E-06 | 0.9999968 |
| PCMTD2 | chr20 | 62891625 | + | MACROD2 | chr20 | 15210585 | + | 3.16E-06 | 0.9999968 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
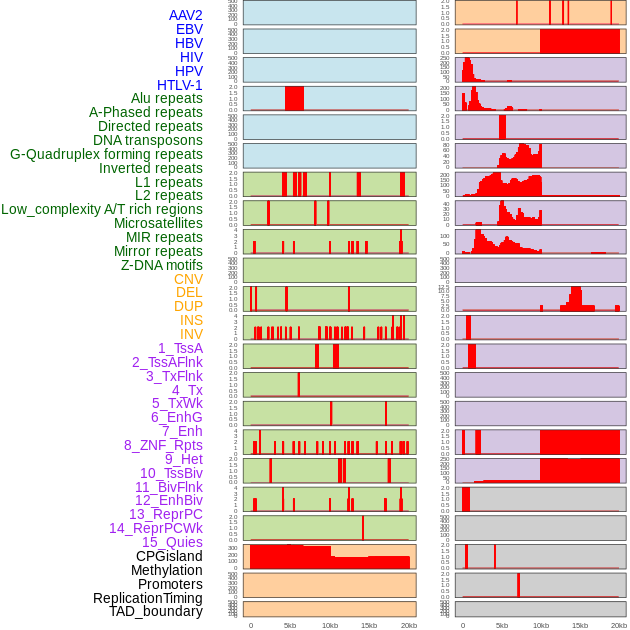 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
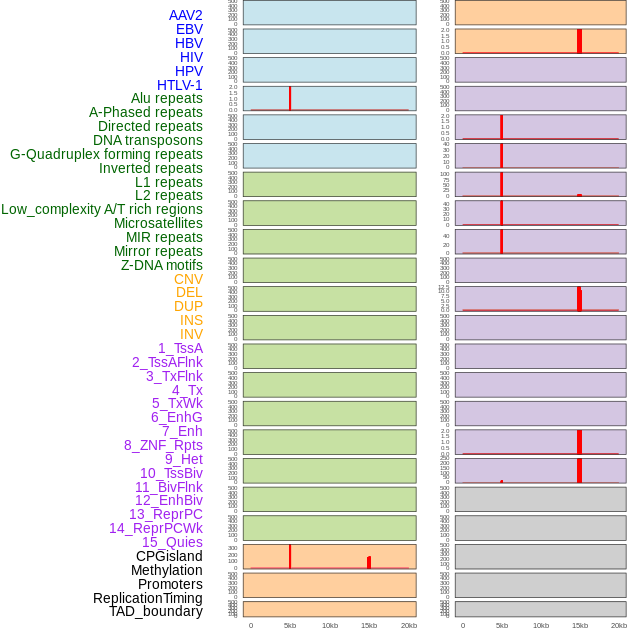 |
Top |
Fusion Protein Features for PCMTD2-MACROD2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr20:62891625/chr20:15210586) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | MACROD2 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Removes ADP-ribose from asparatate and glutamate residues in proteins bearing a single ADP-ribose moiety (PubMed:23474714, PubMed:23474712). Inactive towards proteins bearing poly-ADP-ribose (PubMed:23474714, PubMed:23474712). Deacetylates O-acetyl-ADP ribose, a signaling molecule generated by the deacetylation of acetylated lysine residues in histones and other proteins (PubMed:21257746). {ECO:0000269|PubMed:21257746, ECO:0000269|PubMed:23474712, ECO:0000269|PubMed:23474714}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000217246 | 4 | 17 | 247_388 | 139 | 426.0 | Compositional bias | Note=Glu-rich | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000310348 | 4 | 18 | 247_388 | 139 | 449.0 | Compositional bias | Note=Glu-rich | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000378058 | 0 | 10 | 247_388 | 0 | 214.0 | Compositional bias | Note=Glu-rich | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000378058 | 0 | 10 | 6_9 | 0 | 214.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000402914 | 0 | 14 | 247_388 | 0 | 214.0 | Compositional bias | Note=Glu-rich | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000402914 | 0 | 14 | 6_9 | 0 | 214.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000407045 | 0 | 6 | 247_388 | 0 | 99.0 | Compositional bias | Note=Glu-rich | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000407045 | 0 | 6 | 6_9 | 0 | 99.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000378058 | 0 | 10 | 59_240 | 0 | 214.0 | Domain | Macro | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000402914 | 0 | 14 | 59_240 | 0 | 214.0 | Domain | Macro | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000407045 | 0 | 6 | 59_240 | 0 | 99.0 | Domain | Macro | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000217246 | 4 | 17 | 185_191 | 139 | 426.0 | Region | Substrate binding | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000310348 | 4 | 18 | 185_191 | 139 | 449.0 | Region | Substrate binding | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000378058 | 0 | 10 | 185_191 | 0 | 214.0 | Region | Substrate binding | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000378058 | 0 | 10 | 77_79 | 0 | 214.0 | Region | Substrate binding | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000378058 | 0 | 10 | 90_92 | 0 | 214.0 | Region | Substrate binding | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000378058 | 0 | 10 | 97_102 | 0 | 214.0 | Region | Substrate binding | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000402914 | 0 | 14 | 185_191 | 0 | 214.0 | Region | Substrate binding | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000402914 | 0 | 14 | 77_79 | 0 | 214.0 | Region | Substrate binding | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000402914 | 0 | 14 | 90_92 | 0 | 214.0 | Region | Substrate binding | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000402914 | 0 | 14 | 97_102 | 0 | 214.0 | Region | Substrate binding | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000407045 | 0 | 6 | 185_191 | 0 | 99.0 | Region | Substrate binding | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000407045 | 0 | 6 | 77_79 | 0 | 99.0 | Region | Substrate binding | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000407045 | 0 | 6 | 90_92 | 0 | 99.0 | Region | Substrate binding | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000407045 | 0 | 6 | 97_102 | 0 | 99.0 | Region | Substrate binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PCMTD2 | chr20:62891625 | chr20:15210586 | ENST00000308824 | + | 2 | 6 | 279_286 | 102 | 362.0 | Compositional bias | Note=Poly-Arg |
| Hgene | PCMTD2 | chr20:62891625 | chr20:15210586 | ENST00000369758 | + | 2 | 6 | 279_286 | 102 | 335.0 | Compositional bias | Note=Poly-Arg |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000217246 | 4 | 17 | 6_9 | 139 | 426.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000310348 | 4 | 18 | 6_9 | 139 | 449.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000217246 | 4 | 17 | 59_240 | 139 | 426.0 | Domain | Macro | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000310348 | 4 | 18 | 59_240 | 139 | 449.0 | Domain | Macro | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000217246 | 4 | 17 | 77_79 | 139 | 426.0 | Region | Substrate binding | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000217246 | 4 | 17 | 90_92 | 139 | 426.0 | Region | Substrate binding | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000217246 | 4 | 17 | 97_102 | 139 | 426.0 | Region | Substrate binding | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000310348 | 4 | 18 | 77_79 | 139 | 449.0 | Region | Substrate binding | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000310348 | 4 | 18 | 90_92 | 139 | 449.0 | Region | Substrate binding | |
| Tgene | MACROD2 | chr20:62891625 | chr20:15210586 | ENST00000310348 | 4 | 18 | 97_102 | 139 | 449.0 | Region | Substrate binding |
Top |
Fusion Gene Sequence for PCMTD2-MACROD2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >63392_63392_1_PCMTD2-MACROD2_PCMTD2_chr20_62891625_ENST00000299468_MACROD2_chr20_15210586_ENST00000217246_length(transcript)=4639nt_BP=458nt CGGCGGATGTTTACGGCGGCCGAGGTTGGAGCGGCGCTGCTCGGCCGCGGACACACGAGGGACGCGCCCGAGGAGCTGCAGGTGGCAGCC CAGGCGGTCCGAACCCGTCGGCCGGCCGAGCCTGGAGTATTGCCTAAGTGTAATCTTGAACATGGGCGGTGCTGTGAGTGCTGGTGAAGA CAATGATGAGCTGATAGATAATTTGAAAGAAGCACAGTATATCCGGACTGAGCTGGTAGAGCAGGCTTTCAGAGCTATCGATCGTGCAGA CTATTATCTTGAAGAATTTAAAGAAAATGCTTATAAAGACTTGGCATGGAAGCATGGAAACATTCACCTCTCAGCCCCGTGCATCTACTC GGAGGTGATGGAAGCCCTAGATCTGCAGCCTGGACTCTCGTTTCTGAACCTGGGCAGTGGCACTGGGTATCTCAGCTCCATGGTGGGCCT CATTCTAGATGTCATCCATACTGTAGGGCCAATAGCCAGGGGCCATATTAATGGTTCCCACAAGGAAGACCTTGCAAATTGCTATAAATC ATCTCTGAAGCTCGTGAAAGAAAATAACATCCGATCAGTTGCATTTCCCTGCATCTCAACAGGCATTTATGGCTTTCCCAACGAGCCTGC TGCAGTCATTGCCCTCAACACCATTAAGGAATGGCTTGCCAAGAATCACCATGAGGTGGATCGGATCATTTTCTGTGTCTTCTTAGAAGT TGACTTCAAAATCTACAAAAAGAAAATGAATGAGTTTTTCTCCGTAGACGATAATAATGAAGAAGAAGAGGATGTTGAAATGAAAGAAGA TTCAGATGAGAACGGTCCAGAGGAGAAGCAAAGTGTGGAAGAAATGGAAGAGCAGAGCCAAGATGCAGATGGTGTCAACACTGTCACTGT GCCCGGCCCTGCTTCAGAAGAGGCAGTTGAAGACTGTAAAGATGAAGATTTTGCAAAGGATGAAAATATTACAAAAGGCGGTGAAGTGAC AGATCATTCTGTGCGTGACCAAGATCATCCCGATGGACAAGAGAATGATTCAACGAAGAATGAAATAAAAATTGAAACAGAATCGCAGAG CTCATATATGGAAACAGAAGAACTTTCATCAAACCAAGAAGATGCCGTGATTGTGGAGCAACCAGAAGTGATTCCATTAACAGAGGACCA AGAAGAAAAAGAAGGTGAAAAAGCTCCAGGCGAGGACACACCTAGGATGCCTGGGAAAAGTGAAGGCTCCAGTGACCTAGAAAATACTCC AGGTCCTGATGCAGGGGCACAAGATGAAGCGAAGGAACAAAGAAATGGAACTAAATGACAATCCTCAGCATCGCAAGGCCTCTCCTGGCT CTGGGGGAGCTCGGGAAGATAGCAGCACACGCTGTGGAGGAGGGTGGGGGTGGGGGGAAGGCAAGTCCCATGGAAGGACGGGGAATCCTT TACTCTAATTTCTCCAGCTGCATTTTGTTCCGTTTATCTGCAGAAAAAGAAAGAAAAAAAAGAAAAAAAAAGTTTCCTTTAATTTGGTGG AGGGACCCATGTTGACGCATCTTTCAGGCATTATCCTTGTAATTTCTGTCTTTTCTCTTACAACTTTGCCCCAGGGTCACAGTGGCTTGA TTGAACACTCACATGTGTATCCTGGCCCCTGTCTGCTTTCTTGGTTATTTCACAAAGCTGGTCACACAGTGGTTTATTCAAAGGAAGGGG AGGAAGACAGTGGTTTGATAAGCTGCAGGATAAATTTTAGGAATCAATGAGCCCAGCAGCAGTATAATCCCCAGACAGAGGAGGCAGGAT AGAAAATGGGCAAAAGCCTCGGAAACCACTTGGAAAAGGTCTGGACAATGAGGTGAAAATATTTTCTTCAGGGTTCCCAAGGCACAATTT GTTCCAAGTGGCTAATGAGAAATATGGAAGCTGAATTTTTTCCAGAGCAGAGTGCAGAGGCATAACAGAAGGGTGGGCCCTGGCAGCCAT CTGGGTCTCTTCCTTCCTAACCATGGTGGCAGGTGCATCCTTCTTTGACACTGACTTTCAGCAGAGCTTACTTGGTTCATGAGGTCTTCA CATGGAGACTACCAGCAAGAGGTGACTCTCTGCTGCATAACTGTAAAGGATGGCCCTTTGCTAGGTGTTACAGTTAAAAGCTAAGAAAAG GGGCAGTGCATTTAGGACCCAAACATATGCCTATGAATATCAAAAGCTCCTCCTGAAATTGCTGTGAGTTTTCCATAAAAGAATATCCTG TCTTCACCCAAGGCTTGACAGCCCACAGAGTGGTCTCATTTGAAATTACAGGAAATTAGAGCTTTTGCTTGCAGTTCTGCCTTCCTGGCC TGTGTTTAAATGCTGTCACTTGTTTATGCCAAGTTCAAGGCTGATTCAATGGTTGGTCCCCTCACCCAGAAAACCCTGAAGGGGAGGATA CAGCTCTGAAGGGGGGCAGCAGTACTAAAAACCCAAGATGCCAGTGGTATAGTGGGCACAAGGGATGGCGACCATGAGGATGCCAGGCAT CATCACCAATATCTATCCTAGAGCCAGTATAAGGCCAGATGCCTACTTCCCACAGCCTCCCCGGGTTCCAAAGTCATGTCATTGTTTTCA GTGGAAACATATCGTTTGTTGCATATCTTCTTAAATCCATCTTCCTTGTAAGGGCTTTAGAACTAAAACTTACTTATATTGTTTTTCCTT AACAGAGGGAGAAAAATAGTGGATTATTATTTCTAAAATAAAAGGATGTTCTGCTTTCTAAATATCCCATCAAAATCTTCAGTTTTGCAC TTTTTTGATGGAAAATTCATCTTATCTTCCTATGACTTTGGTTTTAGCCTTTCTGAATTTGTTACCCCTTCTGGATGGCTTATTTGATAT ACTGGAATAGTTAACAAGCTATACTTCAGCATATGCACTATATTCTAACAAATTTTTTTTAATAAAATCAAGACATCAGCAAGAATGACA TTTACGTGACCTCATAATGTGGGATTATGGCCTTCTGTTGCTATTCCAGTTTGATATGGAAGCATCTATATCCTCTATTGCCGTTAGATG TTGTTGCTTTTCAGAAAAGTAACGAAAAGGCTCATTTTAAAGAATCCAAGAAACGATGTCATCCAAATATTGACAGTTTCTACATTTCAT GCCATCTTTATAACTCAATTGAAAGTTGCCGTCATTCTTGTGAAGTATTTGACAAGTGCAATCTGCTAGAAGCTCGTTTTTCTTGTGACT CCCAAATGTTAGTGCTACTTAGCCTCAGTAATGAGTTACAGTTGAAAAAAACATGAGGGAAACAGAGGGACAGAGATTTTCTAATGAACA ATGATGGAAGAGACCTAATGTCCTTGCTAGAAACAGCCAGGATGGAAATTATCCAGCCCTGGCATTCTCCTTATCATCAATGACAGTCAT TTTATTCATTTATTTCAAATGTGGGTGGGCTAGAAGTGGAAGGAGGGAATTCTCTCTGCCTAAAAATTCTAGAAGAATGAAAGTAATCTT TGTATCCAGGAAACTAAGAGAATGAGGAATAAATATCTTCAGCCCGACTCCTGAATTTGTTTATTCTTCCATCTATAATTAGATTGTGTT TTCATTTTTGCTTTGTCATGCTTTTTGGTTGTTATTTGGCTATACAGTTTTATGCTTTAAAACAAATGATAAAGTTAATTTCCAATTCAA TAGTGAAATATTAACAATCTAACTATAGCCAGATCAAAGACACCTGAACACAGAAAACCTTTATTTGCTGGTGCTGCCATTGCACAGGCT GTACAATGAAATAGATTTGAAAAGCTGATTGATTTTCCCGCACATAAATTCTGGATGTCAATTTCCAACCAAACTCCAATACAGCTATGT GGCATGAAGAGTTACAGGAGGGAGGGAGGAAAATAGCCCTATGTTAGTCATGTTTGCATACAGAGGATCAAAGTAGGCCTTCACCATAAT AGTTCTAATTAAAATGGTCCTCGCTGTAGGAGAGACAAAGGGGCTTTTCCTCTAGCTGGTAACTATTCAGATGATGGACAAGTCTTCTTT CATAAAAGATTACAAAGAAGGCATCCGAATCACTGTCTGTGATACTGGGTCACATATTAATCACTGCAGCTAATTGTAAATCTTTCTATG AAACACTGAAAAGCCTCTTTGTGAATTAATACAGTTCTGCTTGATGCACTTGATTTGAAAAGACATTTCTCTGTATGTGGCGCATGTCGG CTTTGCTTTGAAAAATAACAAAGTTAGCAGAATATGTTCAATATATTTTCTTGGGGAATAGGGTTTTTATTACATGATTCATTAAGGATT TGCCTTACCCTGACATTTGTGATATAAAGGAAAATCAGAAAAAAAGTAATTTTCTTGATCAAGATATGTTTTTACTTAATGCAAATAAAT GTAGTCTGTTGCTTGCAAGGAAAAAAAAATGGCTTCTGATATCTGGTATAAACTGCTAAATAGGATAATACGTGCCTCTTTTGTTAAACC GGCATTTAAATGCTGGACTGCTTCTAAATCTGTTTGTTTCTTTTCATCTGTGCCATACACTAAAAAACAACTGTTGCCTTCATACTATAT >63392_63392_1_PCMTD2-MACROD2_PCMTD2_chr20_62891625_ENST00000299468_MACROD2_chr20_15210586_ENST00000217246_length(amino acids)=430AA_BP=144 MERRCSAADTRGTRPRSCRWQPRRSEPVGRPSLEYCLSVILNMGGAVSAGEDNDELIDNLKEAQYIRTELVEQAFRAIDRADYYLEEFKE NAYKDLAWKHGNIHLSAPCIYSEVMEALDLQPGLSFLNLGSGTGYLSSMVGLILDVIHTVGPIARGHINGSHKEDLANCYKSSLKLVKEN NIRSVAFPCISTGIYGFPNEPAAVIALNTIKEWLAKNHHEVDRIIFCVFLEVDFKIYKKKMNEFFSVDDNNEEEEDVEMKEDSDENGPEE KQSVEEMEEQSQDADGVNTVTVPGPASEEAVEDCKDEDFAKDENITKGGEVTDHSVRDQDHPDGQENDSTKNEIKIETESQSSYMETEEL -------------------------------------------------------------- >63392_63392_2_PCMTD2-MACROD2_PCMTD2_chr20_62891625_ENST00000299468_MACROD2_chr20_15210586_ENST00000310348_length(transcript)=1387nt_BP=458nt CGGCGGATGTTTACGGCGGCCGAGGTTGGAGCGGCGCTGCTCGGCCGCGGACACACGAGGGACGCGCCCGAGGAGCTGCAGGTGGCAGCC CAGGCGGTCCGAACCCGTCGGCCGGCCGAGCCTGGAGTATTGCCTAAGTGTAATCTTGAACATGGGCGGTGCTGTGAGTGCTGGTGAAGA CAATGATGAGCTGATAGATAATTTGAAAGAAGCACAGTATATCCGGACTGAGCTGGTAGAGCAGGCTTTCAGAGCTATCGATCGTGCAGA CTATTATCTTGAAGAATTTAAAGAAAATGCTTATAAAGACTTGGCATGGAAGCATGGAAACATTCACCTCTCAGCCCCGTGCATCTACTC GGAGGTGATGGAAGCCCTAGATCTGCAGCCTGGACTCTCGTTTCTGAACCTGGGCAGTGGCACTGGGTATCTCAGCTCCATGGTGGGCCT CATTCTAGATGTCATCCATACTGTAGGGCCAATAGCCAGGGGCCATATTAATGGTTCCCACAAGGAAGACCTTGCAAATTGCTATAAATC ATCTCTGAAGCTCGTGAAAGAAAATAACATCCGATCAGTTGCATTTCCCTGCATCTCAACAGGCATTTATGGCTTTCCCAACGAGCCTGC TGCAGTCATTGCCCTCAACACCATTAAGGAATGGCTTGCCAAGAATCACCATGAGGTGGATCGGATCATTTTCTGTGTCTTCTTAGAAGT TGACTTCAAAATCTACAAAAAGAAAATGAATGAGTTTTTCTCCGTAGACGATAATAATGAAGAAGAAGAGGATGTTGAAATGAAAGAAGA TTCAGATGAGAACGGTCCAGAGGAGAAGCAAAGTGTGGAAGAAATGGAAGAGCAGAGCCAAGATGCAGATGGTGTCAACACTGTCACTGT GCCCGGCCCTGCTTCAGAAGAGGCAGTTGAAGACTGTAAAGATGAAGATTTTGCAAAGGATGAAAATATTACAAAAGGCGGTGAAGTGAC AGATCATTCTGTGCGTGACCAAGATCATCCCGATGGACAAGAGAATGATTCAACGAAGAATGAAATAAAAATTGAAACAGAATCGCAGAG CTCATATATGGAAACAGAAGAACTTTCATCAAACCAAGAAGATGCCGTGATTGTGGAGCAACCAGAAGTGATTCCATTAACAGAGGACCA AGAAGAAAAAGAAGGTGAAAAAGCTCCAGGCGAGGACACACCTAGGATGCCTGGGAAAAGTGAAGGCTCCAGTGACCTAGAAAATACTCC AGGTCCTGATGTTGAAATGAATAGTCAGGTTGACAAGGTAAATGACCCAACAGAGAGTCAACAAGAAGATCAACTAATAGCAGGGGCACA >63392_63392_2_PCMTD2-MACROD2_PCMTD2_chr20_62891625_ENST00000299468_MACROD2_chr20_15210586_ENST00000310348_length(amino acids)=454AA_BP=144 MERRCSAADTRGTRPRSCRWQPRRSEPVGRPSLEYCLSVILNMGGAVSAGEDNDELIDNLKEAQYIRTELVEQAFRAIDRADYYLEEFKE NAYKDLAWKHGNIHLSAPCIYSEVMEALDLQPGLSFLNLGSGTGYLSSMVGLILDVIHTVGPIARGHINGSHKEDLANCYKSSLKLVKEN NIRSVAFPCISTGIYGFPNEPAAVIALNTIKEWLAKNHHEVDRIIFCVFLEVDFKIYKKKMNEFFSVDDNNEEEEDVEMKEDSDENGPEE KQSVEEMEEQSQDADGVNTVTVPGPASEEAVEDCKDEDFAKDENITKGGEVTDHSVRDQDHPDGQENDSTKNEIKIETESQSSYMETEEL SSNQEDAVIVEQPEVIPLTEDQEEKEGEKAPGEDTPRMPGKSEGSSDLENTPGPDVEMNSQVDKVNDPTESQQEDQLIAGAQDEAKEQRN -------------------------------------------------------------- >63392_63392_3_PCMTD2-MACROD2_PCMTD2_chr20_62891625_ENST00000308824_MACROD2_chr20_15210586_ENST00000217246_length(transcript)=4615nt_BP=434nt GTTGGAGCGGCGCTGCTCGGCCGCGGACACACGAGGGACGCGCCCGAGGAGCTGCAGGTGGCAGCCCAGGCGGTCCGAACCCGTCGGCCG GCCGAGCCTGGAGTATTGCCTAAGTGTAATCTTGAACATGGGCGGTGCTGTGAGTGCTGGTGAAGACAATGATGAGCTGATAGATAATTT GAAAGAAGCACAGTATATCCGGACTGAGCTGGTAGAGCAGGCTTTCAGAGCTATCGATCGTGCAGACTATTATCTTGAAGAATTTAAAGA AAATGCTTATAAAGACTTGGCATGGAAGCATGGAAACATTCACCTCTCAGCCCCGTGCATCTACTCGGAGGTGATGGAAGCCCTAGATCT GCAGCCTGGACTCTCGTTTCTGAACCTGGGCAGTGGCACTGGGTATCTCAGCTCCATGGTGGGCCTCATTCTAGATGTCATCCATACTGT AGGGCCAATAGCCAGGGGCCATATTAATGGTTCCCACAAGGAAGACCTTGCAAATTGCTATAAATCATCTCTGAAGCTCGTGAAAGAAAA TAACATCCGATCAGTTGCATTTCCCTGCATCTCAACAGGCATTTATGGCTTTCCCAACGAGCCTGCTGCAGTCATTGCCCTCAACACCAT TAAGGAATGGCTTGCCAAGAATCACCATGAGGTGGATCGGATCATTTTCTGTGTCTTCTTAGAAGTTGACTTCAAAATCTACAAAAAGAA AATGAATGAGTTTTTCTCCGTAGACGATAATAATGAAGAAGAAGAGGATGTTGAAATGAAAGAAGATTCAGATGAGAACGGTCCAGAGGA GAAGCAAAGTGTGGAAGAAATGGAAGAGCAGAGCCAAGATGCAGATGGTGTCAACACTGTCACTGTGCCCGGCCCTGCTTCAGAAGAGGC AGTTGAAGACTGTAAAGATGAAGATTTTGCAAAGGATGAAAATATTACAAAAGGCGGTGAAGTGACAGATCATTCTGTGCGTGACCAAGA TCATCCCGATGGACAAGAGAATGATTCAACGAAGAATGAAATAAAAATTGAAACAGAATCGCAGAGCTCATATATGGAAACAGAAGAACT TTCATCAAACCAAGAAGATGCCGTGATTGTGGAGCAACCAGAAGTGATTCCATTAACAGAGGACCAAGAAGAAAAAGAAGGTGAAAAAGC TCCAGGCGAGGACACACCTAGGATGCCTGGGAAAAGTGAAGGCTCCAGTGACCTAGAAAATACTCCAGGTCCTGATGCAGGGGCACAAGA TGAAGCGAAGGAACAAAGAAATGGAACTAAATGACAATCCTCAGCATCGCAAGGCCTCTCCTGGCTCTGGGGGAGCTCGGGAAGATAGCA GCACACGCTGTGGAGGAGGGTGGGGGTGGGGGGAAGGCAAGTCCCATGGAAGGACGGGGAATCCTTTACTCTAATTTCTCCAGCTGCATT TTGTTCCGTTTATCTGCAGAAAAAGAAAGAAAAAAAAGAAAAAAAAAGTTTCCTTTAATTTGGTGGAGGGACCCATGTTGACGCATCTTT CAGGCATTATCCTTGTAATTTCTGTCTTTTCTCTTACAACTTTGCCCCAGGGTCACAGTGGCTTGATTGAACACTCACATGTGTATCCTG GCCCCTGTCTGCTTTCTTGGTTATTTCACAAAGCTGGTCACACAGTGGTTTATTCAAAGGAAGGGGAGGAAGACAGTGGTTTGATAAGCT GCAGGATAAATTTTAGGAATCAATGAGCCCAGCAGCAGTATAATCCCCAGACAGAGGAGGCAGGATAGAAAATGGGCAAAAGCCTCGGAA ACCACTTGGAAAAGGTCTGGACAATGAGGTGAAAATATTTTCTTCAGGGTTCCCAAGGCACAATTTGTTCCAAGTGGCTAATGAGAAATA TGGAAGCTGAATTTTTTCCAGAGCAGAGTGCAGAGGCATAACAGAAGGGTGGGCCCTGGCAGCCATCTGGGTCTCTTCCTTCCTAACCAT GGTGGCAGGTGCATCCTTCTTTGACACTGACTTTCAGCAGAGCTTACTTGGTTCATGAGGTCTTCACATGGAGACTACCAGCAAGAGGTG ACTCTCTGCTGCATAACTGTAAAGGATGGCCCTTTGCTAGGTGTTACAGTTAAAAGCTAAGAAAAGGGGCAGTGCATTTAGGACCCAAAC ATATGCCTATGAATATCAAAAGCTCCTCCTGAAATTGCTGTGAGTTTTCCATAAAAGAATATCCTGTCTTCACCCAAGGCTTGACAGCCC ACAGAGTGGTCTCATTTGAAATTACAGGAAATTAGAGCTTTTGCTTGCAGTTCTGCCTTCCTGGCCTGTGTTTAAATGCTGTCACTTGTT TATGCCAAGTTCAAGGCTGATTCAATGGTTGGTCCCCTCACCCAGAAAACCCTGAAGGGGAGGATACAGCTCTGAAGGGGGGCAGCAGTA CTAAAAACCCAAGATGCCAGTGGTATAGTGGGCACAAGGGATGGCGACCATGAGGATGCCAGGCATCATCACCAATATCTATCCTAGAGC CAGTATAAGGCCAGATGCCTACTTCCCACAGCCTCCCCGGGTTCCAAAGTCATGTCATTGTTTTCAGTGGAAACATATCGTTTGTTGCAT ATCTTCTTAAATCCATCTTCCTTGTAAGGGCTTTAGAACTAAAACTTACTTATATTGTTTTTCCTTAACAGAGGGAGAAAAATAGTGGAT TATTATTTCTAAAATAAAAGGATGTTCTGCTTTCTAAATATCCCATCAAAATCTTCAGTTTTGCACTTTTTTGATGGAAAATTCATCTTA TCTTCCTATGACTTTGGTTTTAGCCTTTCTGAATTTGTTACCCCTTCTGGATGGCTTATTTGATATACTGGAATAGTTAACAAGCTATAC TTCAGCATATGCACTATATTCTAACAAATTTTTTTTAATAAAATCAAGACATCAGCAAGAATGACATTTACGTGACCTCATAATGTGGGA TTATGGCCTTCTGTTGCTATTCCAGTTTGATATGGAAGCATCTATATCCTCTATTGCCGTTAGATGTTGTTGCTTTTCAGAAAAGTAACG AAAAGGCTCATTTTAAAGAATCCAAGAAACGATGTCATCCAAATATTGACAGTTTCTACATTTCATGCCATCTTTATAACTCAATTGAAA GTTGCCGTCATTCTTGTGAAGTATTTGACAAGTGCAATCTGCTAGAAGCTCGTTTTTCTTGTGACTCCCAAATGTTAGTGCTACTTAGCC TCAGTAATGAGTTACAGTTGAAAAAAACATGAGGGAAACAGAGGGACAGAGATTTTCTAATGAACAATGATGGAAGAGACCTAATGTCCT TGCTAGAAACAGCCAGGATGGAAATTATCCAGCCCTGGCATTCTCCTTATCATCAATGACAGTCATTTTATTCATTTATTTCAAATGTGG GTGGGCTAGAAGTGGAAGGAGGGAATTCTCTCTGCCTAAAAATTCTAGAAGAATGAAAGTAATCTTTGTATCCAGGAAACTAAGAGAATG AGGAATAAATATCTTCAGCCCGACTCCTGAATTTGTTTATTCTTCCATCTATAATTAGATTGTGTTTTCATTTTTGCTTTGTCATGCTTT TTGGTTGTTATTTGGCTATACAGTTTTATGCTTTAAAACAAATGATAAAGTTAATTTCCAATTCAATAGTGAAATATTAACAATCTAACT ATAGCCAGATCAAAGACACCTGAACACAGAAAACCTTTATTTGCTGGTGCTGCCATTGCACAGGCTGTACAATGAAATAGATTTGAAAAG CTGATTGATTTTCCCGCACATAAATTCTGGATGTCAATTTCCAACCAAACTCCAATACAGCTATGTGGCATGAAGAGTTACAGGAGGGAG GGAGGAAAATAGCCCTATGTTAGTCATGTTTGCATACAGAGGATCAAAGTAGGCCTTCACCATAATAGTTCTAATTAAAATGGTCCTCGC TGTAGGAGAGACAAAGGGGCTTTTCCTCTAGCTGGTAACTATTCAGATGATGGACAAGTCTTCTTTCATAAAAGATTACAAAGAAGGCAT CCGAATCACTGTCTGTGATACTGGGTCACATATTAATCACTGCAGCTAATTGTAAATCTTTCTATGAAACACTGAAAAGCCTCTTTGTGA ATTAATACAGTTCTGCTTGATGCACTTGATTTGAAAAGACATTTCTCTGTATGTGGCGCATGTCGGCTTTGCTTTGAAAAATAACAAAGT TAGCAGAATATGTTCAATATATTTTCTTGGGGAATAGGGTTTTTATTACATGATTCATTAAGGATTTGCCTTACCCTGACATTTGTGATA TAAAGGAAAATCAGAAAAAAAGTAATTTTCTTGATCAAGATATGTTTTTACTTAATGCAAATAAATGTAGTCTGTTGCTTGCAAGGAAAA AAAAATGGCTTCTGATATCTGGTATAAACTGCTAAATAGGATAATACGTGCCTCTTTTGTTAAACCGGCATTTAAATGCTGGACTGCTTC TAAATCTGTTTGTTTCTTTTCATCTGTGCCATACACTAAAAAACAACTGTTGCCTTCATACTATATTTGTTAGAGCAGAATACAAATAAA >63392_63392_3_PCMTD2-MACROD2_PCMTD2_chr20_62891625_ENST00000308824_MACROD2_chr20_15210586_ENST00000217246_length(amino acids)=430AA_BP=144 LERRCSAADTRGTRPRSCRWQPRRSEPVGRPSLEYCLSVILNMGGAVSAGEDNDELIDNLKEAQYIRTELVEQAFRAIDRADYYLEEFKE NAYKDLAWKHGNIHLSAPCIYSEVMEALDLQPGLSFLNLGSGTGYLSSMVGLILDVIHTVGPIARGHINGSHKEDLANCYKSSLKLVKEN NIRSVAFPCISTGIYGFPNEPAAVIALNTIKEWLAKNHHEVDRIIFCVFLEVDFKIYKKKMNEFFSVDDNNEEEEDVEMKEDSDENGPEE KQSVEEMEEQSQDADGVNTVTVPGPASEEAVEDCKDEDFAKDENITKGGEVTDHSVRDQDHPDGQENDSTKNEIKIETESQSSYMETEEL -------------------------------------------------------------- >63392_63392_4_PCMTD2-MACROD2_PCMTD2_chr20_62891625_ENST00000308824_MACROD2_chr20_15210586_ENST00000310348_length(transcript)=1363nt_BP=434nt GTTGGAGCGGCGCTGCTCGGCCGCGGACACACGAGGGACGCGCCCGAGGAGCTGCAGGTGGCAGCCCAGGCGGTCCGAACCCGTCGGCCG GCCGAGCCTGGAGTATTGCCTAAGTGTAATCTTGAACATGGGCGGTGCTGTGAGTGCTGGTGAAGACAATGATGAGCTGATAGATAATTT GAAAGAAGCACAGTATATCCGGACTGAGCTGGTAGAGCAGGCTTTCAGAGCTATCGATCGTGCAGACTATTATCTTGAAGAATTTAAAGA AAATGCTTATAAAGACTTGGCATGGAAGCATGGAAACATTCACCTCTCAGCCCCGTGCATCTACTCGGAGGTGATGGAAGCCCTAGATCT GCAGCCTGGACTCTCGTTTCTGAACCTGGGCAGTGGCACTGGGTATCTCAGCTCCATGGTGGGCCTCATTCTAGATGTCATCCATACTGT AGGGCCAATAGCCAGGGGCCATATTAATGGTTCCCACAAGGAAGACCTTGCAAATTGCTATAAATCATCTCTGAAGCTCGTGAAAGAAAA TAACATCCGATCAGTTGCATTTCCCTGCATCTCAACAGGCATTTATGGCTTTCCCAACGAGCCTGCTGCAGTCATTGCCCTCAACACCAT TAAGGAATGGCTTGCCAAGAATCACCATGAGGTGGATCGGATCATTTTCTGTGTCTTCTTAGAAGTTGACTTCAAAATCTACAAAAAGAA AATGAATGAGTTTTTCTCCGTAGACGATAATAATGAAGAAGAAGAGGATGTTGAAATGAAAGAAGATTCAGATGAGAACGGTCCAGAGGA GAAGCAAAGTGTGGAAGAAATGGAAGAGCAGAGCCAAGATGCAGATGGTGTCAACACTGTCACTGTGCCCGGCCCTGCTTCAGAAGAGGC AGTTGAAGACTGTAAAGATGAAGATTTTGCAAAGGATGAAAATATTACAAAAGGCGGTGAAGTGACAGATCATTCTGTGCGTGACCAAGA TCATCCCGATGGACAAGAGAATGATTCAACGAAGAATGAAATAAAAATTGAAACAGAATCGCAGAGCTCATATATGGAAACAGAAGAACT TTCATCAAACCAAGAAGATGCCGTGATTGTGGAGCAACCAGAAGTGATTCCATTAACAGAGGACCAAGAAGAAAAAGAAGGTGAAAAAGC TCCAGGCGAGGACACACCTAGGATGCCTGGGAAAAGTGAAGGCTCCAGTGACCTAGAAAATACTCCAGGTCCTGATGTTGAAATGAATAG TCAGGTTGACAAGGTAAATGACCCAACAGAGAGTCAACAAGAAGATCAACTAATAGCAGGGGCACAAGATGAAGCGAAGGAACAAAGAAA >63392_63392_4_PCMTD2-MACROD2_PCMTD2_chr20_62891625_ENST00000308824_MACROD2_chr20_15210586_ENST00000310348_length(amino acids)=454AA_BP=144 LERRCSAADTRGTRPRSCRWQPRRSEPVGRPSLEYCLSVILNMGGAVSAGEDNDELIDNLKEAQYIRTELVEQAFRAIDRADYYLEEFKE NAYKDLAWKHGNIHLSAPCIYSEVMEALDLQPGLSFLNLGSGTGYLSSMVGLILDVIHTVGPIARGHINGSHKEDLANCYKSSLKLVKEN NIRSVAFPCISTGIYGFPNEPAAVIALNTIKEWLAKNHHEVDRIIFCVFLEVDFKIYKKKMNEFFSVDDNNEEEEDVEMKEDSDENGPEE KQSVEEMEEQSQDADGVNTVTVPGPASEEAVEDCKDEDFAKDENITKGGEVTDHSVRDQDHPDGQENDSTKNEIKIETESQSSYMETEEL SSNQEDAVIVEQPEVIPLTEDQEEKEGEKAPGEDTPRMPGKSEGSSDLENTPGPDVEMNSQVDKVNDPTESQQEDQLIAGAQDEAKEQRN -------------------------------------------------------------- >63392_63392_5_PCMTD2-MACROD2_PCMTD2_chr20_62891625_ENST00000369758_MACROD2_chr20_15210586_ENST00000217246_length(transcript)=4642nt_BP=461nt CGGCGGCGGATGTTTACGGCGGCCGAGGTTGGAGCGGCGCTGCTCGGCCGCGGACACACGAGGGACGCGCCCGAGGAGCTGCAGGTGGCA GCCCAGGCGGTCCGAACCCGTCGGCCGGCCGAGCCTGGAGTATTGCCTAAGTGTAATCTTGAACATGGGCGGTGCTGTGAGTGCTGGTGA AGACAATGATGAGCTGATAGATAATTTGAAAGAAGCACAGTATATCCGGACTGAGCTGGTAGAGCAGGCTTTCAGAGCTATCGATCGTGC AGACTATTATCTTGAAGAATTTAAAGAAAATGCTTATAAAGACTTGGCATGGAAGCATGGAAACATTCACCTCTCAGCCCCGTGCATCTA CTCGGAGGTGATGGAAGCCCTAGATCTGCAGCCTGGACTCTCGTTTCTGAACCTGGGCAGTGGCACTGGGTATCTCAGCTCCATGGTGGG CCTCATTCTAGATGTCATCCATACTGTAGGGCCAATAGCCAGGGGCCATATTAATGGTTCCCACAAGGAAGACCTTGCAAATTGCTATAA ATCATCTCTGAAGCTCGTGAAAGAAAATAACATCCGATCAGTTGCATTTCCCTGCATCTCAACAGGCATTTATGGCTTTCCCAACGAGCC TGCTGCAGTCATTGCCCTCAACACCATTAAGGAATGGCTTGCCAAGAATCACCATGAGGTGGATCGGATCATTTTCTGTGTCTTCTTAGA AGTTGACTTCAAAATCTACAAAAAGAAAATGAATGAGTTTTTCTCCGTAGACGATAATAATGAAGAAGAAGAGGATGTTGAAATGAAAGA AGATTCAGATGAGAACGGTCCAGAGGAGAAGCAAAGTGTGGAAGAAATGGAAGAGCAGAGCCAAGATGCAGATGGTGTCAACACTGTCAC TGTGCCCGGCCCTGCTTCAGAAGAGGCAGTTGAAGACTGTAAAGATGAAGATTTTGCAAAGGATGAAAATATTACAAAAGGCGGTGAAGT GACAGATCATTCTGTGCGTGACCAAGATCATCCCGATGGACAAGAGAATGATTCAACGAAGAATGAAATAAAAATTGAAACAGAATCGCA GAGCTCATATATGGAAACAGAAGAACTTTCATCAAACCAAGAAGATGCCGTGATTGTGGAGCAACCAGAAGTGATTCCATTAACAGAGGA CCAAGAAGAAAAAGAAGGTGAAAAAGCTCCAGGCGAGGACACACCTAGGATGCCTGGGAAAAGTGAAGGCTCCAGTGACCTAGAAAATAC TCCAGGTCCTGATGCAGGGGCACAAGATGAAGCGAAGGAACAAAGAAATGGAACTAAATGACAATCCTCAGCATCGCAAGGCCTCTCCTG GCTCTGGGGGAGCTCGGGAAGATAGCAGCACACGCTGTGGAGGAGGGTGGGGGTGGGGGGAAGGCAAGTCCCATGGAAGGACGGGGAATC CTTTACTCTAATTTCTCCAGCTGCATTTTGTTCCGTTTATCTGCAGAAAAAGAAAGAAAAAAAAGAAAAAAAAAGTTTCCTTTAATTTGG TGGAGGGACCCATGTTGACGCATCTTTCAGGCATTATCCTTGTAATTTCTGTCTTTTCTCTTACAACTTTGCCCCAGGGTCACAGTGGCT TGATTGAACACTCACATGTGTATCCTGGCCCCTGTCTGCTTTCTTGGTTATTTCACAAAGCTGGTCACACAGTGGTTTATTCAAAGGAAG GGGAGGAAGACAGTGGTTTGATAAGCTGCAGGATAAATTTTAGGAATCAATGAGCCCAGCAGCAGTATAATCCCCAGACAGAGGAGGCAG GATAGAAAATGGGCAAAAGCCTCGGAAACCACTTGGAAAAGGTCTGGACAATGAGGTGAAAATATTTTCTTCAGGGTTCCCAAGGCACAA TTTGTTCCAAGTGGCTAATGAGAAATATGGAAGCTGAATTTTTTCCAGAGCAGAGTGCAGAGGCATAACAGAAGGGTGGGCCCTGGCAGC CATCTGGGTCTCTTCCTTCCTAACCATGGTGGCAGGTGCATCCTTCTTTGACACTGACTTTCAGCAGAGCTTACTTGGTTCATGAGGTCT TCACATGGAGACTACCAGCAAGAGGTGACTCTCTGCTGCATAACTGTAAAGGATGGCCCTTTGCTAGGTGTTACAGTTAAAAGCTAAGAA AAGGGGCAGTGCATTTAGGACCCAAACATATGCCTATGAATATCAAAAGCTCCTCCTGAAATTGCTGTGAGTTTTCCATAAAAGAATATC CTGTCTTCACCCAAGGCTTGACAGCCCACAGAGTGGTCTCATTTGAAATTACAGGAAATTAGAGCTTTTGCTTGCAGTTCTGCCTTCCTG GCCTGTGTTTAAATGCTGTCACTTGTTTATGCCAAGTTCAAGGCTGATTCAATGGTTGGTCCCCTCACCCAGAAAACCCTGAAGGGGAGG ATACAGCTCTGAAGGGGGGCAGCAGTACTAAAAACCCAAGATGCCAGTGGTATAGTGGGCACAAGGGATGGCGACCATGAGGATGCCAGG CATCATCACCAATATCTATCCTAGAGCCAGTATAAGGCCAGATGCCTACTTCCCACAGCCTCCCCGGGTTCCAAAGTCATGTCATTGTTT TCAGTGGAAACATATCGTTTGTTGCATATCTTCTTAAATCCATCTTCCTTGTAAGGGCTTTAGAACTAAAACTTACTTATATTGTTTTTC CTTAACAGAGGGAGAAAAATAGTGGATTATTATTTCTAAAATAAAAGGATGTTCTGCTTTCTAAATATCCCATCAAAATCTTCAGTTTTG CACTTTTTTGATGGAAAATTCATCTTATCTTCCTATGACTTTGGTTTTAGCCTTTCTGAATTTGTTACCCCTTCTGGATGGCTTATTTGA TATACTGGAATAGTTAACAAGCTATACTTCAGCATATGCACTATATTCTAACAAATTTTTTTTAATAAAATCAAGACATCAGCAAGAATG ACATTTACGTGACCTCATAATGTGGGATTATGGCCTTCTGTTGCTATTCCAGTTTGATATGGAAGCATCTATATCCTCTATTGCCGTTAG ATGTTGTTGCTTTTCAGAAAAGTAACGAAAAGGCTCATTTTAAAGAATCCAAGAAACGATGTCATCCAAATATTGACAGTTTCTACATTT CATGCCATCTTTATAACTCAATTGAAAGTTGCCGTCATTCTTGTGAAGTATTTGACAAGTGCAATCTGCTAGAAGCTCGTTTTTCTTGTG ACTCCCAAATGTTAGTGCTACTTAGCCTCAGTAATGAGTTACAGTTGAAAAAAACATGAGGGAAACAGAGGGACAGAGATTTTCTAATGA ACAATGATGGAAGAGACCTAATGTCCTTGCTAGAAACAGCCAGGATGGAAATTATCCAGCCCTGGCATTCTCCTTATCATCAATGACAGT CATTTTATTCATTTATTTCAAATGTGGGTGGGCTAGAAGTGGAAGGAGGGAATTCTCTCTGCCTAAAAATTCTAGAAGAATGAAAGTAAT CTTTGTATCCAGGAAACTAAGAGAATGAGGAATAAATATCTTCAGCCCGACTCCTGAATTTGTTTATTCTTCCATCTATAATTAGATTGT GTTTTCATTTTTGCTTTGTCATGCTTTTTGGTTGTTATTTGGCTATACAGTTTTATGCTTTAAAACAAATGATAAAGTTAATTTCCAATT CAATAGTGAAATATTAACAATCTAACTATAGCCAGATCAAAGACACCTGAACACAGAAAACCTTTATTTGCTGGTGCTGCCATTGCACAG GCTGTACAATGAAATAGATTTGAAAAGCTGATTGATTTTCCCGCACATAAATTCTGGATGTCAATTTCCAACCAAACTCCAATACAGCTA TGTGGCATGAAGAGTTACAGGAGGGAGGGAGGAAAATAGCCCTATGTTAGTCATGTTTGCATACAGAGGATCAAAGTAGGCCTTCACCAT AATAGTTCTAATTAAAATGGTCCTCGCTGTAGGAGAGACAAAGGGGCTTTTCCTCTAGCTGGTAACTATTCAGATGATGGACAAGTCTTC TTTCATAAAAGATTACAAAGAAGGCATCCGAATCACTGTCTGTGATACTGGGTCACATATTAATCACTGCAGCTAATTGTAAATCTTTCT ATGAAACACTGAAAAGCCTCTTTGTGAATTAATACAGTTCTGCTTGATGCACTTGATTTGAAAAGACATTTCTCTGTATGTGGCGCATGT CGGCTTTGCTTTGAAAAATAACAAAGTTAGCAGAATATGTTCAATATATTTTCTTGGGGAATAGGGTTTTTATTACATGATTCATTAAGG ATTTGCCTTACCCTGACATTTGTGATATAAAGGAAAATCAGAAAAAAAGTAATTTTCTTGATCAAGATATGTTTTTACTTAATGCAAATA AATGTAGTCTGTTGCTTGCAAGGAAAAAAAAATGGCTTCTGATATCTGGTATAAACTGCTAAATAGGATAATACGTGCCTCTTTTGTTAA ACCGGCATTTAAATGCTGGACTGCTTCTAAATCTGTTTGTTTCTTTTCATCTGTGCCATACACTAAAAAACAACTGTTGCCTTCATACTA >63392_63392_5_PCMTD2-MACROD2_PCMTD2_chr20_62891625_ENST00000369758_MACROD2_chr20_15210586_ENST00000217246_length(amino acids)=430AA_BP=144 MERRCSAADTRGTRPRSCRWQPRRSEPVGRPSLEYCLSVILNMGGAVSAGEDNDELIDNLKEAQYIRTELVEQAFRAIDRADYYLEEFKE NAYKDLAWKHGNIHLSAPCIYSEVMEALDLQPGLSFLNLGSGTGYLSSMVGLILDVIHTVGPIARGHINGSHKEDLANCYKSSLKLVKEN NIRSVAFPCISTGIYGFPNEPAAVIALNTIKEWLAKNHHEVDRIIFCVFLEVDFKIYKKKMNEFFSVDDNNEEEEDVEMKEDSDENGPEE KQSVEEMEEQSQDADGVNTVTVPGPASEEAVEDCKDEDFAKDENITKGGEVTDHSVRDQDHPDGQENDSTKNEIKIETESQSSYMETEEL -------------------------------------------------------------- >63392_63392_6_PCMTD2-MACROD2_PCMTD2_chr20_62891625_ENST00000369758_MACROD2_chr20_15210586_ENST00000310348_length(transcript)=1390nt_BP=461nt CGGCGGCGGATGTTTACGGCGGCCGAGGTTGGAGCGGCGCTGCTCGGCCGCGGACACACGAGGGACGCGCCCGAGGAGCTGCAGGTGGCA GCCCAGGCGGTCCGAACCCGTCGGCCGGCCGAGCCTGGAGTATTGCCTAAGTGTAATCTTGAACATGGGCGGTGCTGTGAGTGCTGGTGA AGACAATGATGAGCTGATAGATAATTTGAAAGAAGCACAGTATATCCGGACTGAGCTGGTAGAGCAGGCTTTCAGAGCTATCGATCGTGC AGACTATTATCTTGAAGAATTTAAAGAAAATGCTTATAAAGACTTGGCATGGAAGCATGGAAACATTCACCTCTCAGCCCCGTGCATCTA CTCGGAGGTGATGGAAGCCCTAGATCTGCAGCCTGGACTCTCGTTTCTGAACCTGGGCAGTGGCACTGGGTATCTCAGCTCCATGGTGGG CCTCATTCTAGATGTCATCCATACTGTAGGGCCAATAGCCAGGGGCCATATTAATGGTTCCCACAAGGAAGACCTTGCAAATTGCTATAA ATCATCTCTGAAGCTCGTGAAAGAAAATAACATCCGATCAGTTGCATTTCCCTGCATCTCAACAGGCATTTATGGCTTTCCCAACGAGCC TGCTGCAGTCATTGCCCTCAACACCATTAAGGAATGGCTTGCCAAGAATCACCATGAGGTGGATCGGATCATTTTCTGTGTCTTCTTAGA AGTTGACTTCAAAATCTACAAAAAGAAAATGAATGAGTTTTTCTCCGTAGACGATAATAATGAAGAAGAAGAGGATGTTGAAATGAAAGA AGATTCAGATGAGAACGGTCCAGAGGAGAAGCAAAGTGTGGAAGAAATGGAAGAGCAGAGCCAAGATGCAGATGGTGTCAACACTGTCAC TGTGCCCGGCCCTGCTTCAGAAGAGGCAGTTGAAGACTGTAAAGATGAAGATTTTGCAAAGGATGAAAATATTACAAAAGGCGGTGAAGT GACAGATCATTCTGTGCGTGACCAAGATCATCCCGATGGACAAGAGAATGATTCAACGAAGAATGAAATAAAAATTGAAACAGAATCGCA GAGCTCATATATGGAAACAGAAGAACTTTCATCAAACCAAGAAGATGCCGTGATTGTGGAGCAACCAGAAGTGATTCCATTAACAGAGGA CCAAGAAGAAAAAGAAGGTGAAAAAGCTCCAGGCGAGGACACACCTAGGATGCCTGGGAAAAGTGAAGGCTCCAGTGACCTAGAAAATAC TCCAGGTCCTGATGTTGAAATGAATAGTCAGGTTGACAAGGTAAATGACCCAACAGAGAGTCAACAAGAAGATCAACTAATAGCAGGGGC >63392_63392_6_PCMTD2-MACROD2_PCMTD2_chr20_62891625_ENST00000369758_MACROD2_chr20_15210586_ENST00000310348_length(amino acids)=454AA_BP=144 MERRCSAADTRGTRPRSCRWQPRRSEPVGRPSLEYCLSVILNMGGAVSAGEDNDELIDNLKEAQYIRTELVEQAFRAIDRADYYLEEFKE NAYKDLAWKHGNIHLSAPCIYSEVMEALDLQPGLSFLNLGSGTGYLSSMVGLILDVIHTVGPIARGHINGSHKEDLANCYKSSLKLVKEN NIRSVAFPCISTGIYGFPNEPAAVIALNTIKEWLAKNHHEVDRIIFCVFLEVDFKIYKKKMNEFFSVDDNNEEEEDVEMKEDSDENGPEE KQSVEEMEEQSQDADGVNTVTVPGPASEEAVEDCKDEDFAKDENITKGGEVTDHSVRDQDHPDGQENDSTKNEIKIETESQSSYMETEEL SSNQEDAVIVEQPEVIPLTEDQEEKEGEKAPGEDTPRMPGKSEGSSDLENTPGPDVEMNSQVDKVNDPTESQQEDQLIAGAQDEAKEQRN -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for PCMTD2-MACROD2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for PCMTD2-MACROD2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for PCMTD2-MACROD2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
