
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ARHGEF7-CRYL1 (FusionGDB2 ID:6344) |
Fusion Gene Summary for ARHGEF7-CRYL1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ARHGEF7-CRYL1 | Fusion gene ID: 6344 | Hgene | Tgene | Gene symbol | ARHGEF7 | CRYL1 | Gene ID | 8874 | 51084 |
| Gene name | Rho guanine nucleotide exchange factor 7 | crystallin lambda 1 | |
| Synonyms | BETA-PIX|COOL-1|COOL1|Nbla10314|P50|P50BP|P85|P85COOL1|P85SPR|PAK3|PIXB | GDH|HEL30|gul3DH|lambda-CRY | |
| Cytomap | 13q34 | 13q12.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | rho guanine nucleotide exchange factor 7PAK-interacting exchange factor betaRho guanine nucleotide exchange factor (GEF) 7SH3 domain-containing proline-rich protein | lambda-crystallin homologL-gulonate 3-dehydrogenasecrystallin, lamda 1epididymis luminal protein 30testicular tissue protein Li 44 | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | Q14155 | Q9Y2S2 | |
| Ensembl transtripts involved in fusion gene | ENST00000317133, ENST00000370623, ENST00000375741, ENST00000218789, ENST00000375736, ENST00000544132, ENST00000375723, ENST00000375737, ENST00000375739, ENST00000426073, ENST00000478679, ENST00000483189, | ENST00000480748, ENST00000298248, ENST00000382812, | |
| Fusion gene scores | * DoF score | 25 X 17 X 12=5100 | 15 X 9 X 10=1350 |
| # samples | 26 | 17 | |
| ** MAII score | log2(26/5100*10)=-4.29391371871777 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(17/1350*10)=-2.98935275580049 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ARHGEF7 [Title/Abstract] AND CRYL1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ARHGEF7(111806338)-CRYL1(20987526), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ARHGEF7 | GO:0043547 | positive regulation of GTPase activity | 21048939 |
 Fusion gene breakpoints across ARHGEF7 (5'-gene) Fusion gene breakpoints across ARHGEF7 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
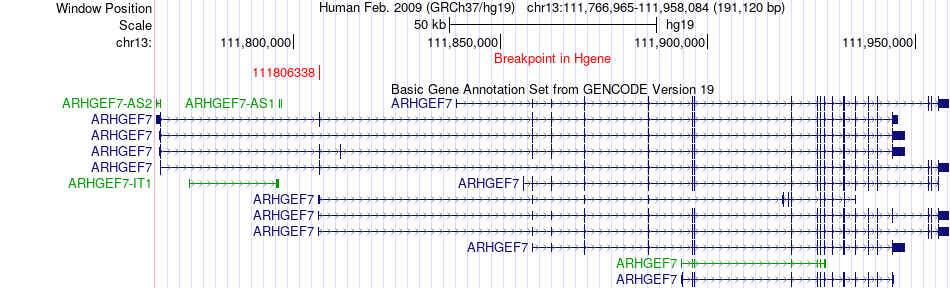 |
 Fusion gene breakpoints across CRYL1 (3'-gene) Fusion gene breakpoints across CRYL1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
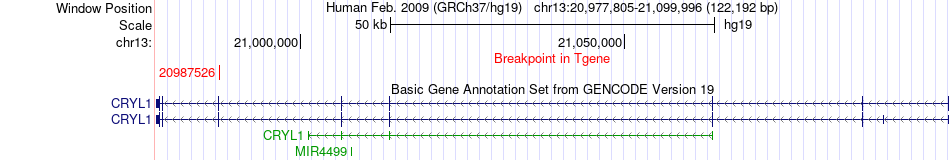 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | Non-Cancer | 235N | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
Top |
Fusion Gene ORF analysis for ARHGEF7-CRYL1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000317133 | ENST00000480748 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| 5CDS-intron | ENST00000370623 | ENST00000480748 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| 5CDS-intron | ENST00000375741 | ENST00000480748 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| 5UTR-3CDS | ENST00000218789 | ENST00000298248 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| 5UTR-3CDS | ENST00000218789 | ENST00000382812 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| 5UTR-3CDS | ENST00000375736 | ENST00000298248 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| 5UTR-3CDS | ENST00000375736 | ENST00000382812 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| 5UTR-3CDS | ENST00000544132 | ENST00000298248 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| 5UTR-3CDS | ENST00000544132 | ENST00000382812 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| 5UTR-intron | ENST00000218789 | ENST00000480748 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| 5UTR-intron | ENST00000375736 | ENST00000480748 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| 5UTR-intron | ENST00000544132 | ENST00000480748 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| In-frame | ENST00000317133 | ENST00000298248 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| In-frame | ENST00000317133 | ENST00000382812 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| In-frame | ENST00000370623 | ENST00000298248 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| In-frame | ENST00000370623 | ENST00000382812 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| In-frame | ENST00000375741 | ENST00000298248 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| In-frame | ENST00000375741 | ENST00000382812 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| intron-3CDS | ENST00000375723 | ENST00000298248 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| intron-3CDS | ENST00000375723 | ENST00000382812 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| intron-3CDS | ENST00000375737 | ENST00000298248 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| intron-3CDS | ENST00000375737 | ENST00000382812 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| intron-3CDS | ENST00000375739 | ENST00000298248 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| intron-3CDS | ENST00000375739 | ENST00000382812 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| intron-3CDS | ENST00000426073 | ENST00000298248 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| intron-3CDS | ENST00000426073 | ENST00000382812 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| intron-3CDS | ENST00000478679 | ENST00000298248 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| intron-3CDS | ENST00000478679 | ENST00000382812 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| intron-3CDS | ENST00000483189 | ENST00000298248 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| intron-3CDS | ENST00000483189 | ENST00000382812 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| intron-intron | ENST00000375723 | ENST00000480748 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| intron-intron | ENST00000375737 | ENST00000480748 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| intron-intron | ENST00000375739 | ENST00000480748 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| intron-intron | ENST00000426073 | ENST00000480748 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| intron-intron | ENST00000478679 | ENST00000480748 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
| intron-intron | ENST00000483189 | ENST00000480748 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000317133 | ARHGEF7 | chr13 | 111806338 | + | ENST00000298248 | CRYL1 | chr13 | 20987526 | - | 1949 | 1160 | 710 | 1486 | 258 |
| ENST00000317133 | ARHGEF7 | chr13 | 111806338 | + | ENST00000382812 | CRYL1 | chr13 | 20987526 | - | 1945 | 1160 | 710 | 1486 | 258 |
| ENST00000375741 | ARHGEF7 | chr13 | 111806338 | + | ENST00000298248 | CRYL1 | chr13 | 20987526 | - | 1291 | 502 | 52 | 828 | 258 |
| ENST00000375741 | ARHGEF7 | chr13 | 111806338 | + | ENST00000382812 | CRYL1 | chr13 | 20987526 | - | 1287 | 502 | 52 | 828 | 258 |
| ENST00000370623 | ARHGEF7 | chr13 | 111806338 | + | ENST00000298248 | CRYL1 | chr13 | 20987526 | - | 1041 | 252 | 0 | 578 | 192 |
| ENST00000370623 | ARHGEF7 | chr13 | 111806338 | + | ENST00000382812 | CRYL1 | chr13 | 20987526 | - | 1037 | 252 | 0 | 578 | 192 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000317133 | ENST00000298248 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - | 0.011352372 | 0.98864764 |
| ENST00000317133 | ENST00000382812 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - | 0.011123719 | 0.9888763 |
| ENST00000375741 | ENST00000298248 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - | 0.007880698 | 0.9921193 |
| ENST00000375741 | ENST00000382812 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - | 0.007824055 | 0.992176 |
| ENST00000370623 | ENST00000298248 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - | 0.006880254 | 0.9931197 |
| ENST00000370623 | ENST00000382812 | ARHGEF7 | chr13 | 111806338 | + | CRYL1 | chr13 | 20987526 | - | 0.006909722 | 0.99309033 |
Top |
Fusion Genomic Features for ARHGEF7-CRYL1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
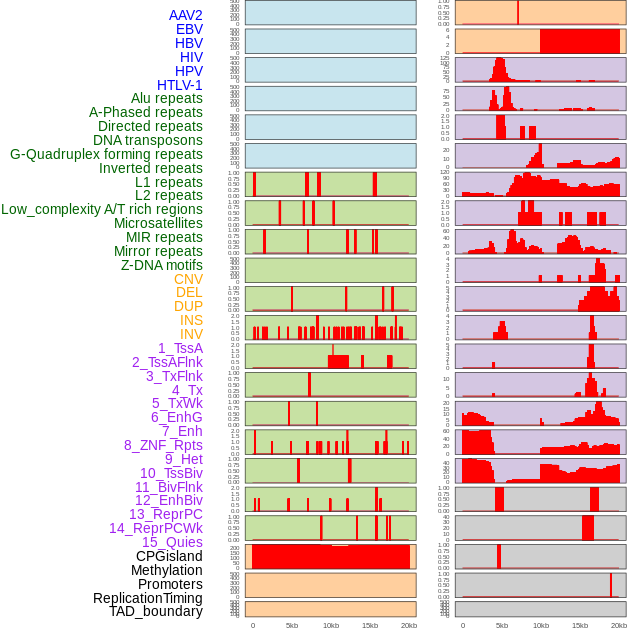 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for ARHGEF7-CRYL1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr13:111806338/chr13:20987526) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ARHGEF7 | CRYL1 |
| FUNCTION: Acts as a RAC1 guanine nucleotide exchange factor (GEF) and can induce membrane ruffling. Functions in cell migration, attachment and cell spreading. Promotes targeting of RAC1 to focal adhesions (By similarity). May function as a positive regulator of apoptosis. Downstream of NMDA receptors and CaMKK-CaMK1 signaling cascade, promotes the formation of spines and synapses in hippocampal neurons. {ECO:0000250, ECO:0000269|PubMed:18184567, ECO:0000269|PubMed:18716323, ECO:0000269|PubMed:19041750}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000317133 | + | 2 | 19 | 184_243 | 84 | 783.0 | Domain | SH3 |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000317133 | + | 2 | 19 | 1_133 | 84 | 783.0 | Domain | Calponin-homology (CH) |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000317133 | + | 2 | 19 | 271_451 | 84 | 783.0 | Domain | DH |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000317133 | + | 2 | 19 | 473_578 | 84 | 783.0 | Domain | PH |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000370623 | + | 2 | 19 | 184_243 | 84 | 732.0 | Domain | SH3 |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000370623 | + | 2 | 19 | 1_133 | 84 | 732.0 | Domain | Calponin-homology (CH) |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000370623 | + | 2 | 19 | 271_451 | 84 | 732.0 | Domain | DH |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000370623 | + | 2 | 19 | 473_578 | 84 | 732.0 | Domain | PH |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000375723 | + | 1 | 17 | 184_243 | 0 | 626.0 | Domain | SH3 |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000375723 | + | 1 | 17 | 1_133 | 0 | 626.0 | Domain | Calponin-homology (CH) |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000375723 | + | 1 | 17 | 271_451 | 0 | 626.0 | Domain | DH |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000375723 | + | 1 | 17 | 473_578 | 0 | 626.0 | Domain | PH |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000375736 | + | 1 | 20 | 184_243 | 0 | 647.0 | Domain | SH3 |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000375736 | + | 1 | 20 | 1_133 | 0 | 647.0 | Domain | Calponin-homology (CH) |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000375736 | + | 1 | 20 | 271_451 | 0 | 647.0 | Domain | DH |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000375736 | + | 1 | 20 | 473_578 | 0 | 647.0 | Domain | PH |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000375739 | + | 1 | 18 | 184_243 | 0 | 754.0 | Domain | SH3 |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000375739 | + | 1 | 18 | 1_133 | 0 | 754.0 | Domain | Calponin-homology (CH) |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000375739 | + | 1 | 18 | 271_451 | 0 | 754.0 | Domain | DH |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000375739 | + | 1 | 18 | 473_578 | 0 | 754.0 | Domain | PH |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000375741 | + | 2 | 20 | 184_243 | 84 | 804.0 | Domain | SH3 |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000375741 | + | 2 | 20 | 1_133 | 84 | 804.0 | Domain | Calponin-homology (CH) |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000375741 | + | 2 | 20 | 271_451 | 84 | 804.0 | Domain | DH |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000375741 | + | 2 | 20 | 473_578 | 84 | 804.0 | Domain | PH |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000426073 | + | 1 | 20 | 184_243 | 0 | 647.0 | Domain | SH3 |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000426073 | + | 1 | 20 | 1_133 | 0 | 647.0 | Domain | Calponin-homology (CH) |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000426073 | + | 1 | 20 | 271_451 | 0 | 647.0 | Domain | DH |
| Hgene | ARHGEF7 | chr13:111806338 | chr13:20987526 | ENST00000426073 | + | 1 | 20 | 473_578 | 0 | 647.0 | Domain | PH |
| Tgene | CRYL1 | chr13:111806338 | chr13:20987526 | ENST00000298248 | 4 | 8 | 16_17 | 211 | 320.0 | Nucleotide binding | NAD | |
| Tgene | CRYL1 | chr13:111806338 | chr13:20987526 | ENST00000382812 | 5 | 9 | 16_17 | 189 | 298.0 | Nucleotide binding | NAD |
Top |
Fusion Gene Sequence for ARHGEF7-CRYL1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >6344_6344_1_ARHGEF7-CRYL1_ARHGEF7_chr13_111806338_ENST00000317133_CRYL1_chr13_20987526_ENST00000298248_length(transcript)=1949nt_BP=1160nt CCCCGAGACGCAGCCAGGCGCCCGGCCCGGCCCGGCCCGGCCCTCTGCACAGCCCCACTTCAGGCCACGCCGTTTCCTTGACCTTCCCCC GGCAGGGTCTGAAGCGTCACCGTCCGTCTAGAGCAGAGTCGCGCAGGCCCCTGCCCTGCCGGTGAGGTCGGCGCCGCGCTCCGGAGAGTC GGTTCCTCCCGATTCAGACCCATATGGCTTCAGATGGAACCATCATTTCTGTTCCGTCATGACCGGCTTGCAAGGTAGCAAAGCGAAACA AACGTCTCGTTTTCAGCCTGTCCCCGCCACCTTGAAATAAAACCGGCAAAAATAAAAAGAGCATTCTGGGAAGTACTTCATCATTTCAAC GCTCACGAATTCAAAACATAAACAAAGGCTTCCGAGTGCCCCGGCCAGGGGCGCGGGGCGCACGGCGGGCCCGGGGCAGGTAAGCGCAGG TGCGCGCCCGCCCCCACCCCCGGCTCCCTCCCCATCCGCTCCCCGCTCCCCTTCCCCTTTCCCTTCCCCGCCCGCTCCCAGCCGCCGCCG CCGCCCCGCGCACGGCCTGGAGCGGAGGCTGCGCAGGGCGCGGGGCGGCGCGGGCCGGGCGCGGGCCGGGCGGACGGCCGCGTCTTTCTT CTCCTGGCGGTGATGTCATTGGGCGACGGCGGCCGAGGCCGGGGGGCGGCGGCGGGCGCCCGCAGGTTCCCGAGCCGCTCCTGAGAAGGC GCCTGACAGCGGGCCGGGGCGCACGGAGAAGCGGGCCGGGCCGGACCTGCTGGGCCGCGCCGAGCCAATCGCCGGCGCCGGCCGCTCGAT GGGCGAGGCGGCGGCGGCGGCGGCGGGGGCCGCGGGCCGGGCCGCCGCTCCGAGGTGAAGGCGCGCGCCCCTCCCCGCCTGCCTCCCGGG CCGCAGCGATGAATTCCGCCGAGCAAACCGTTACGTGGCTCATCACTCTGGGGGTGCTGGAGTCGCCCAAAAAAACCATCTCGGACCCGG AGGGCTTTCTGCAGGCGTCGCTGAAGGATGGGGTGGTCCTCTGCAGGCTGCTGGAGCGCCTGCTCCCCGGGACCATCGAGAAAGTCTACC CCGAGCCCCGGAGCGAGAGCGAGTGCCTGAGCAACATCCGCGAGTTCCTGCGCGGCTGCGGGGCTTCCCTGCGGCTGGAGGAAGGAATCG TGTCTCCTAGTGACCTGGACCTTGTCATGTCAGAAGGGTTGGGCATGCGGTATGCATTCATTGGACCCCTGGAAACCATGCATCTCAATG CAGAAGGTATGTTAAGCTACTGCGACAGATACAGCGAAGGCATAAAACATGTCCTACAGACTTTTGGACCCATTCCAGAGTTTTCCAGGG CCACTGCTGAGAAGGTTAACCAGGACATGTGCATGAAGGTCCCTGATGACCCGGAGCACTTAGCTGCCAGGAGGCAGTGGAGGGACGAGT GCCTCATGAGACTCGCCAAGTTGAAGAGTCAAGTGCAGCCCCAGTGAATTTCTTGTAATGCAGCTTCCACTCCTCTCATTGGAGGCCCTA TTTGGGAACACTGCAAGCCCTTAATCAGCCCTCTGTGACATAGGTAGCAGCCCACGGAGATCCTAAGCTGGCTGTCTTGTGTGCAGCCTG AGTGGGGTGGTGCAGGCCGGTAGTCTGCCCGTCACTTTGGATCATAGCCCTGGGCCTGGCGGCACAGCAGCACTTGCGTTCTCGGGGCTG TCGATTTCCTGCCACCTGGGCAGATAACCTGGAGATTTTCACCTTTTCTTTTCAGCTTGATTGCATTTGACTATATTTTACAGCCAGTGA TTGTAGTTTCATGTTAATATGTGGCAAAATATTTTTGTAATTATTTTCTAATCCCTTTCTGAGTACTCTGGGGCCCTGCATTTATGAGGC >6344_6344_1_ARHGEF7-CRYL1_ARHGEF7_chr13_111806338_ENST00000317133_CRYL1_chr13_20987526_ENST00000298248_length(amino acids)=258AA_BP=145 MRRRLTAGRGARRSGPGRTCWAAPSQSPAPAARWARRRRRRRGPRAGPPLRGEGARPSPPASRAAAMNSAEQTVTWLITLGVLESPKKTI SDPEGFLQASLKDGVVLCRLLERLLPGTIEKVYPEPRSESECLSNIREFLRGCGASLRLEEGIVSPSDLDLVMSEGLGMRYAFIGPLETM -------------------------------------------------------------- >6344_6344_2_ARHGEF7-CRYL1_ARHGEF7_chr13_111806338_ENST00000317133_CRYL1_chr13_20987526_ENST00000382812_length(transcript)=1945nt_BP=1160nt CCCCGAGACGCAGCCAGGCGCCCGGCCCGGCCCGGCCCGGCCCTCTGCACAGCCCCACTTCAGGCCACGCCGTTTCCTTGACCTTCCCCC GGCAGGGTCTGAAGCGTCACCGTCCGTCTAGAGCAGAGTCGCGCAGGCCCCTGCCCTGCCGGTGAGGTCGGCGCCGCGCTCCGGAGAGTC GGTTCCTCCCGATTCAGACCCATATGGCTTCAGATGGAACCATCATTTCTGTTCCGTCATGACCGGCTTGCAAGGTAGCAAAGCGAAACA AACGTCTCGTTTTCAGCCTGTCCCCGCCACCTTGAAATAAAACCGGCAAAAATAAAAAGAGCATTCTGGGAAGTACTTCATCATTTCAAC GCTCACGAATTCAAAACATAAACAAAGGCTTCCGAGTGCCCCGGCCAGGGGCGCGGGGCGCACGGCGGGCCCGGGGCAGGTAAGCGCAGG TGCGCGCCCGCCCCCACCCCCGGCTCCCTCCCCATCCGCTCCCCGCTCCCCTTCCCCTTTCCCTTCCCCGCCCGCTCCCAGCCGCCGCCG CCGCCCCGCGCACGGCCTGGAGCGGAGGCTGCGCAGGGCGCGGGGCGGCGCGGGCCGGGCGCGGGCCGGGCGGACGGCCGCGTCTTTCTT CTCCTGGCGGTGATGTCATTGGGCGACGGCGGCCGAGGCCGGGGGGCGGCGGCGGGCGCCCGCAGGTTCCCGAGCCGCTCCTGAGAAGGC GCCTGACAGCGGGCCGGGGCGCACGGAGAAGCGGGCCGGGCCGGACCTGCTGGGCCGCGCCGAGCCAATCGCCGGCGCCGGCCGCTCGAT GGGCGAGGCGGCGGCGGCGGCGGCGGGGGCCGCGGGCCGGGCCGCCGCTCCGAGGTGAAGGCGCGCGCCCCTCCCCGCCTGCCTCCCGGG CCGCAGCGATGAATTCCGCCGAGCAAACCGTTACGTGGCTCATCACTCTGGGGGTGCTGGAGTCGCCCAAAAAAACCATCTCGGACCCGG AGGGCTTTCTGCAGGCGTCGCTGAAGGATGGGGTGGTCCTCTGCAGGCTGCTGGAGCGCCTGCTCCCCGGGACCATCGAGAAAGTCTACC CCGAGCCCCGGAGCGAGAGCGAGTGCCTGAGCAACATCCGCGAGTTCCTGCGCGGCTGCGGGGCTTCCCTGCGGCTGGAGGAAGGAATCG TGTCTCCTAGTGACCTGGACCTTGTCATGTCAGAAGGGTTGGGCATGCGGTATGCATTCATTGGACCCCTGGAAACCATGCATCTCAATG CAGAAGGTATGTTAAGCTACTGCGACAGATACAGCGAAGGCATAAAACATGTCCTACAGACTTTTGGACCCATTCCAGAGTTTTCCAGGG CCACTGCTGAGAAGGTTAACCAGGACATGTGCATGAAGGTCCCTGATGACCCGGAGCACTTAGCTGCCAGGAGGCAGTGGAGGGACGAGT GCCTCATGAGACTCGCCAAGTTGAAGAGTCAAGTGCAGCCCCAGTGAATTTCTTGTAATGCAGCTTCCACTCCTCTCATTGGAGGCCCTA TTTGGGAACACTGCAAGCCCTTAATCAGCCCTCTGTGACATAGGTAGCAGCCCACGGAGATCCTAAGCTGGCTGTCTTGTGTGCAGCCTG AGTGGGGTGGTGCAGGCCGGTAGTCTGCCCGTCACTTTGGATCATAGCCCTGGGCCTGGCGGCACAGCAGCACTTGCGTTCTCGGGGCTG TCGATTTCCTGCCACCTGGGCAGATAACCTGGAGATTTTCACCTTTTCTTTTCAGCTTGATTGCATTTGACTATATTTTACAGCCAGTGA TTGTAGTTTCATGTTAATATGTGGCAAAATATTTTTGTAATTATTTTCTAATCCCTTTCTGAGTACTCTGGGGCCCTGCATTTATGAGGC >6344_6344_2_ARHGEF7-CRYL1_ARHGEF7_chr13_111806338_ENST00000317133_CRYL1_chr13_20987526_ENST00000382812_length(amino acids)=258AA_BP=145 MRRRLTAGRGARRSGPGRTCWAAPSQSPAPAARWARRRRRRRGPRAGPPLRGEGARPSPPASRAAAMNSAEQTVTWLITLGVLESPKKTI SDPEGFLQASLKDGVVLCRLLERLLPGTIEKVYPEPRSESECLSNIREFLRGCGASLRLEEGIVSPSDLDLVMSEGLGMRYAFIGPLETM -------------------------------------------------------------- >6344_6344_3_ARHGEF7-CRYL1_ARHGEF7_chr13_111806338_ENST00000370623_CRYL1_chr13_20987526_ENST00000298248_length(transcript)=1041nt_BP=252nt ATGAATTCCGCCGAGCAAACCGTTACGTGGCTCATCACTCTGGGGGTGCTGGAGTCGCCCAAAAAAACCATCTCGGACCCGGAGGGCTTT CTGCAGGCGTCGCTGAAGGATGGGGTGGTCCTCTGCAGGCTGCTGGAGCGCCTGCTCCCCGGGACCATCGAGAAAGTCTACCCCGAGCCC CGGAGCGAGAGCGAGTGCCTGAGCAACATCCGCGAGTTCCTGCGCGGCTGCGGGGCTTCCCTGCGGCTGGAGGAAGGAATCGTGTCTCCT AGTGACCTGGACCTTGTCATGTCAGAAGGGTTGGGCATGCGGTATGCATTCATTGGACCCCTGGAAACCATGCATCTCAATGCAGAAGGT ATGTTAAGCTACTGCGACAGATACAGCGAAGGCATAAAACATGTCCTACAGACTTTTGGACCCATTCCAGAGTTTTCCAGGGCCACTGCT GAGAAGGTTAACCAGGACATGTGCATGAAGGTCCCTGATGACCCGGAGCACTTAGCTGCCAGGAGGCAGTGGAGGGACGAGTGCCTCATG AGACTCGCCAAGTTGAAGAGTCAAGTGCAGCCCCAGTGAATTTCTTGTAATGCAGCTTCCACTCCTCTCATTGGAGGCCCTATTTGGGAA CACTGCAAGCCCTTAATCAGCCCTCTGTGACATAGGTAGCAGCCCACGGAGATCCTAAGCTGGCTGTCTTGTGTGCAGCCTGAGTGGGGT GGTGCAGGCCGGTAGTCTGCCCGTCACTTTGGATCATAGCCCTGGGCCTGGCGGCACAGCAGCACTTGCGTTCTCGGGGCTGTCGATTTC CTGCCACCTGGGCAGATAACCTGGAGATTTTCACCTTTTCTTTTCAGCTTGATTGCATTTGACTATATTTTACAGCCAGTGATTGTAGTT TCATGTTAATATGTGGCAAAATATTTTTGTAATTATTTTCTAATCCCTTTCTGAGTACTCTGGGGCCCTGCATTTATGAGGCACCTACCT >6344_6344_3_ARHGEF7-CRYL1_ARHGEF7_chr13_111806338_ENST00000370623_CRYL1_chr13_20987526_ENST00000298248_length(amino acids)=192AA_BP=79 MNSAEQTVTWLITLGVLESPKKTISDPEGFLQASLKDGVVLCRLLERLLPGTIEKVYPEPRSESECLSNIREFLRGCGASLRLEEGIVSP SDLDLVMSEGLGMRYAFIGPLETMHLNAEGMLSYCDRYSEGIKHVLQTFGPIPEFSRATAEKVNQDMCMKVPDDPEHLAARRQWRDECLM -------------------------------------------------------------- >6344_6344_4_ARHGEF7-CRYL1_ARHGEF7_chr13_111806338_ENST00000370623_CRYL1_chr13_20987526_ENST00000382812_length(transcript)=1037nt_BP=252nt ATGAATTCCGCCGAGCAAACCGTTACGTGGCTCATCACTCTGGGGGTGCTGGAGTCGCCCAAAAAAACCATCTCGGACCCGGAGGGCTTT CTGCAGGCGTCGCTGAAGGATGGGGTGGTCCTCTGCAGGCTGCTGGAGCGCCTGCTCCCCGGGACCATCGAGAAAGTCTACCCCGAGCCC CGGAGCGAGAGCGAGTGCCTGAGCAACATCCGCGAGTTCCTGCGCGGCTGCGGGGCTTCCCTGCGGCTGGAGGAAGGAATCGTGTCTCCT AGTGACCTGGACCTTGTCATGTCAGAAGGGTTGGGCATGCGGTATGCATTCATTGGACCCCTGGAAACCATGCATCTCAATGCAGAAGGT ATGTTAAGCTACTGCGACAGATACAGCGAAGGCATAAAACATGTCCTACAGACTTTTGGACCCATTCCAGAGTTTTCCAGGGCCACTGCT GAGAAGGTTAACCAGGACATGTGCATGAAGGTCCCTGATGACCCGGAGCACTTAGCTGCCAGGAGGCAGTGGAGGGACGAGTGCCTCATG AGACTCGCCAAGTTGAAGAGTCAAGTGCAGCCCCAGTGAATTTCTTGTAATGCAGCTTCCACTCCTCTCATTGGAGGCCCTATTTGGGAA CACTGCAAGCCCTTAATCAGCCCTCTGTGACATAGGTAGCAGCCCACGGAGATCCTAAGCTGGCTGTCTTGTGTGCAGCCTGAGTGGGGT GGTGCAGGCCGGTAGTCTGCCCGTCACTTTGGATCATAGCCCTGGGCCTGGCGGCACAGCAGCACTTGCGTTCTCGGGGCTGTCGATTTC CTGCCACCTGGGCAGATAACCTGGAGATTTTCACCTTTTCTTTTCAGCTTGATTGCATTTGACTATATTTTACAGCCAGTGATTGTAGTT TCATGTTAATATGTGGCAAAATATTTTTGTAATTATTTTCTAATCCCTTTCTGAGTACTCTGGGGCCCTGCATTTATGAGGCACCTACCT >6344_6344_4_ARHGEF7-CRYL1_ARHGEF7_chr13_111806338_ENST00000370623_CRYL1_chr13_20987526_ENST00000382812_length(amino acids)=192AA_BP=79 MNSAEQTVTWLITLGVLESPKKTISDPEGFLQASLKDGVVLCRLLERLLPGTIEKVYPEPRSESECLSNIREFLRGCGASLRLEEGIVSP SDLDLVMSEGLGMRYAFIGPLETMHLNAEGMLSYCDRYSEGIKHVLQTFGPIPEFSRATAEKVNQDMCMKVPDDPEHLAARRQWRDECLM -------------------------------------------------------------- >6344_6344_5_ARHGEF7-CRYL1_ARHGEF7_chr13_111806338_ENST00000375741_CRYL1_chr13_20987526_ENST00000298248_length(transcript)=1291nt_BP=502nt GCGGCCGAGGCCGGGGGGCGGCGGCGGGCGCCCGCAGGTTCCCGAGCCGCTCCTGAGAAGGCGCCTGACAGCGGGCCGGGGCGCACGGAG AAGCGGGCCGGGCCGGACCTGCTGGGCCGCGCCGAGCCAATCGCCGGCGCCGGCCGCTCGATGGGCGAGGCGGCGGCGGCGGCGGCGGGG GCCGCGGGCCGGGCCGCCGCTCCGAGGTGAAGGCGCGCGCCCCTCCCCGCCTGCCTCCCGGGCCGCAGCGATGAATTCCGCCGAGCAAAC CGTTACGTGGCTCATCACTCTGGGGGTGCTGGAGTCGCCCAAAAAAACCATCTCGGACCCGGAGGGCTTTCTGCAGGCGTCGCTGAAGGA TGGGGTGGTCCTCTGCAGGCTGCTGGAGCGCCTGCTCCCCGGGACCATCGAGAAAGTCTACCCCGAGCCCCGGAGCGAGAGCGAGTGCCT GAGCAACATCCGCGAGTTCCTGCGCGGCTGCGGGGCTTCCCTGCGGCTGGAGGAAGGAATCGTGTCTCCTAGTGACCTGGACCTTGTCAT GTCAGAAGGGTTGGGCATGCGGTATGCATTCATTGGACCCCTGGAAACCATGCATCTCAATGCAGAAGGTATGTTAAGCTACTGCGACAG ATACAGCGAAGGCATAAAACATGTCCTACAGACTTTTGGACCCATTCCAGAGTTTTCCAGGGCCACTGCTGAGAAGGTTAACCAGGACAT GTGCATGAAGGTCCCTGATGACCCGGAGCACTTAGCTGCCAGGAGGCAGTGGAGGGACGAGTGCCTCATGAGACTCGCCAAGTTGAAGAG TCAAGTGCAGCCCCAGTGAATTTCTTGTAATGCAGCTTCCACTCCTCTCATTGGAGGCCCTATTTGGGAACACTGCAAGCCCTTAATCAG CCCTCTGTGACATAGGTAGCAGCCCACGGAGATCCTAAGCTGGCTGTCTTGTGTGCAGCCTGAGTGGGGTGGTGCAGGCCGGTAGTCTGC CCGTCACTTTGGATCATAGCCCTGGGCCTGGCGGCACAGCAGCACTTGCGTTCTCGGGGCTGTCGATTTCCTGCCACCTGGGCAGATAAC CTGGAGATTTTCACCTTTTCTTTTCAGCTTGATTGCATTTGACTATATTTTACAGCCAGTGATTGTAGTTTCATGTTAATATGTGGCAAA ATATTTTTGTAATTATTTTCTAATCCCTTTCTGAGTACTCTGGGGCCCTGCATTTATGAGGCACCTACCTTCATTTTGCTAACGCTTATT >6344_6344_5_ARHGEF7-CRYL1_ARHGEF7_chr13_111806338_ENST00000375741_CRYL1_chr13_20987526_ENST00000298248_length(amino acids)=258AA_BP=145 MRRRLTAGRGARRSGPGRTCWAAPSQSPAPAARWARRRRRRRGPRAGPPLRGEGARPSPPASRAAAMNSAEQTVTWLITLGVLESPKKTI SDPEGFLQASLKDGVVLCRLLERLLPGTIEKVYPEPRSESECLSNIREFLRGCGASLRLEEGIVSPSDLDLVMSEGLGMRYAFIGPLETM -------------------------------------------------------------- >6344_6344_6_ARHGEF7-CRYL1_ARHGEF7_chr13_111806338_ENST00000375741_CRYL1_chr13_20987526_ENST00000382812_length(transcript)=1287nt_BP=502nt GCGGCCGAGGCCGGGGGGCGGCGGCGGGCGCCCGCAGGTTCCCGAGCCGCTCCTGAGAAGGCGCCTGACAGCGGGCCGGGGCGCACGGAG AAGCGGGCCGGGCCGGACCTGCTGGGCCGCGCCGAGCCAATCGCCGGCGCCGGCCGCTCGATGGGCGAGGCGGCGGCGGCGGCGGCGGGG GCCGCGGGCCGGGCCGCCGCTCCGAGGTGAAGGCGCGCGCCCCTCCCCGCCTGCCTCCCGGGCCGCAGCGATGAATTCCGCCGAGCAAAC CGTTACGTGGCTCATCACTCTGGGGGTGCTGGAGTCGCCCAAAAAAACCATCTCGGACCCGGAGGGCTTTCTGCAGGCGTCGCTGAAGGA TGGGGTGGTCCTCTGCAGGCTGCTGGAGCGCCTGCTCCCCGGGACCATCGAGAAAGTCTACCCCGAGCCCCGGAGCGAGAGCGAGTGCCT GAGCAACATCCGCGAGTTCCTGCGCGGCTGCGGGGCTTCCCTGCGGCTGGAGGAAGGAATCGTGTCTCCTAGTGACCTGGACCTTGTCAT GTCAGAAGGGTTGGGCATGCGGTATGCATTCATTGGACCCCTGGAAACCATGCATCTCAATGCAGAAGGTATGTTAAGCTACTGCGACAG ATACAGCGAAGGCATAAAACATGTCCTACAGACTTTTGGACCCATTCCAGAGTTTTCCAGGGCCACTGCTGAGAAGGTTAACCAGGACAT GTGCATGAAGGTCCCTGATGACCCGGAGCACTTAGCTGCCAGGAGGCAGTGGAGGGACGAGTGCCTCATGAGACTCGCCAAGTTGAAGAG TCAAGTGCAGCCCCAGTGAATTTCTTGTAATGCAGCTTCCACTCCTCTCATTGGAGGCCCTATTTGGGAACACTGCAAGCCCTTAATCAG CCCTCTGTGACATAGGTAGCAGCCCACGGAGATCCTAAGCTGGCTGTCTTGTGTGCAGCCTGAGTGGGGTGGTGCAGGCCGGTAGTCTGC CCGTCACTTTGGATCATAGCCCTGGGCCTGGCGGCACAGCAGCACTTGCGTTCTCGGGGCTGTCGATTTCCTGCCACCTGGGCAGATAAC CTGGAGATTTTCACCTTTTCTTTTCAGCTTGATTGCATTTGACTATATTTTACAGCCAGTGATTGTAGTTTCATGTTAATATGTGGCAAA ATATTTTTGTAATTATTTTCTAATCCCTTTCTGAGTACTCTGGGGCCCTGCATTTATGAGGCACCTACCTTCATTTTGCTAACGCTTATT >6344_6344_6_ARHGEF7-CRYL1_ARHGEF7_chr13_111806338_ENST00000375741_CRYL1_chr13_20987526_ENST00000382812_length(amino acids)=258AA_BP=145 MRRRLTAGRGARRSGPGRTCWAAPSQSPAPAARWARRRRRRRGPRAGPPLRGEGARPSPPASRAAAMNSAEQTVTWLITLGVLESPKKTI SDPEGFLQASLKDGVVLCRLLERLLPGTIEKVYPEPRSESECLSNIREFLRGCGASLRLEEGIVSPSDLDLVMSEGLGMRYAFIGPLETM -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ARHGEF7-CRYL1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ARHGEF7-CRYL1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ARHGEF7-CRYL1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
