
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:PDCD10-ARHGAP15 (FusionGDB2 ID:63577) |
Fusion Gene Summary for PDCD10-ARHGAP15 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: PDCD10-ARHGAP15 | Fusion gene ID: 63577 | Hgene | Tgene | Gene symbol | PDCD10 | ARHGAP15 | Gene ID | 11235 | 55843 |
| Gene name | programmed cell death 10 | Rho GTPase activating protein 15 | |
| Synonyms | CCM3|TFAR15 | BM046 | |
| Cytomap | 3q26.1 | 2q22.2-q22.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | programmed cell death protein 10TF-1 cell apoptosis-related protein 15apoptosis-related protein 15cerebral cavernous malformations 3 protein | rho GTPase-activating protein 15rho-type GTPase-activating protein 15 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q53QZ3 | |
| Ensembl transtripts involved in fusion gene | ENST00000392750, ENST00000461494, ENST00000470131, ENST00000471885, ENST00000473645, ENST00000487947, ENST00000497056, ENST00000487678, ENST00000492396, | ENST00000473426, ENST00000295095, ENST00000409869, | |
| Fusion gene scores | * DoF score | 4 X 4 X 3=48 | 13 X 10 X 9=1170 |
| # samples | 5 | 15 | |
| ** MAII score | log2(5/48*10)=0.0588936890535686 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(15/1170*10)=-2.96347412397489 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: PDCD10 [Title/Abstract] AND ARHGAP15 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | PDCD10(167437849)-ARHGAP15(143986148), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | PDCD10-ARHGAP15 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. PDCD10-ARHGAP15 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | PDCD10 | GO:0008284 | positive regulation of cell proliferation | 17360971|23541896 |
| Hgene | PDCD10 | GO:0030335 | positive regulation of cell migration | 23541896 |
| Hgene | PDCD10 | GO:0032874 | positive regulation of stress-activated MAPK cascade | 22652780 |
| Hgene | PDCD10 | GO:0042542 | response to hydrogen peroxide | 22291017 |
| Hgene | PDCD10 | GO:0043066 | negative regulation of apoptotic process | 17360971 |
| Hgene | PDCD10 | GO:0043406 | positive regulation of MAP kinase activity | 17360971 |
| Tgene | ARHGAP15 | GO:0008360 | regulation of cell shape | 12650940 |
 Fusion gene breakpoints across PDCD10 (5'-gene) Fusion gene breakpoints across PDCD10 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
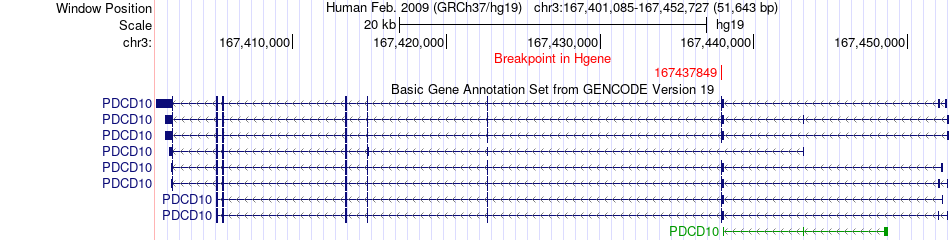 |
 Fusion gene breakpoints across ARHGAP15 (3'-gene) Fusion gene breakpoints across ARHGAP15 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
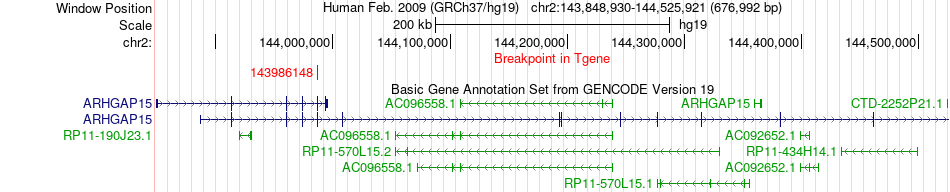 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChiTaRS5.0 | N/A | AV760860 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
Top |
Fusion Gene ORF analysis for PDCD10-ARHGAP15 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000392750 | ENST00000473426 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| 5CDS-intron | ENST00000461494 | ENST00000473426 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| 5CDS-intron | ENST00000470131 | ENST00000473426 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| 5CDS-intron | ENST00000471885 | ENST00000473426 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| 5CDS-intron | ENST00000473645 | ENST00000473426 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| 5CDS-intron | ENST00000487947 | ENST00000473426 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| 5CDS-intron | ENST00000497056 | ENST00000473426 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| Frame-shift | ENST00000470131 | ENST00000295095 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| Frame-shift | ENST00000470131 | ENST00000409869 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| Frame-shift | ENST00000471885 | ENST00000295095 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| Frame-shift | ENST00000471885 | ENST00000409869 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| Frame-shift | ENST00000473645 | ENST00000295095 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| Frame-shift | ENST00000473645 | ENST00000409869 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| Frame-shift | ENST00000497056 | ENST00000295095 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| Frame-shift | ENST00000497056 | ENST00000409869 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| In-frame | ENST00000392750 | ENST00000295095 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| In-frame | ENST00000392750 | ENST00000409869 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| In-frame | ENST00000461494 | ENST00000295095 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| In-frame | ENST00000461494 | ENST00000409869 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| In-frame | ENST00000487947 | ENST00000295095 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| In-frame | ENST00000487947 | ENST00000409869 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| intron-3CDS | ENST00000487678 | ENST00000295095 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| intron-3CDS | ENST00000487678 | ENST00000409869 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| intron-3CDS | ENST00000492396 | ENST00000295095 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| intron-3CDS | ENST00000492396 | ENST00000409869 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| intron-intron | ENST00000487678 | ENST00000473426 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
| intron-intron | ENST00000492396 | ENST00000473426 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000392750 | PDCD10 | chr3 | 167437849 | - | ENST00000409869 | ARHGAP15 | chr2 | 143986148 | + | 3009 | 514 | 607 | 840 | 77 |
| ENST00000392750 | PDCD10 | chr3 | 167437849 | - | ENST00000295095 | ARHGAP15 | chr2 | 143986148 | + | 1826 | 514 | 446 | 1645 | 399 |
| ENST00000461494 | PDCD10 | chr3 | 167437849 | - | ENST00000409869 | ARHGAP15 | chr2 | 143986148 | + | 2820 | 325 | 418 | 651 | 77 |
| ENST00000461494 | PDCD10 | chr3 | 167437849 | - | ENST00000295095 | ARHGAP15 | chr2 | 143986148 | + | 1637 | 325 | 257 | 1456 | 399 |
| ENST00000487947 | PDCD10 | chr3 | 167437849 | - | ENST00000409869 | ARHGAP15 | chr2 | 143986148 | + | 2731 | 236 | 329 | 562 | 77 |
| ENST00000487947 | PDCD10 | chr3 | 167437849 | - | ENST00000295095 | ARHGAP15 | chr2 | 143986148 | + | 1548 | 236 | 168 | 1367 | 399 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000392750 | ENST00000409869 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + | 0.57373726 | 0.42626277 |
| ENST00000392750 | ENST00000295095 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + | 0.001243721 | 0.9987563 |
| ENST00000461494 | ENST00000409869 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + | 0.6896707 | 0.31032935 |
| ENST00000461494 | ENST00000295095 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + | 0.001427839 | 0.9985721 |
| ENST00000487947 | ENST00000409869 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + | 0.943834 | 0.05616599 |
| ENST00000487947 | ENST00000295095 | PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986148 | + | 0.001919461 | 0.9980805 |
Top |
Fusion Genomic Features for PDCD10-ARHGAP15 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986149 | + | 1.97E-07 | 0.99999976 |
| PDCD10 | chr3 | 167437849 | - | ARHGAP15 | chr2 | 143986149 | + | 1.97E-07 | 0.99999976 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
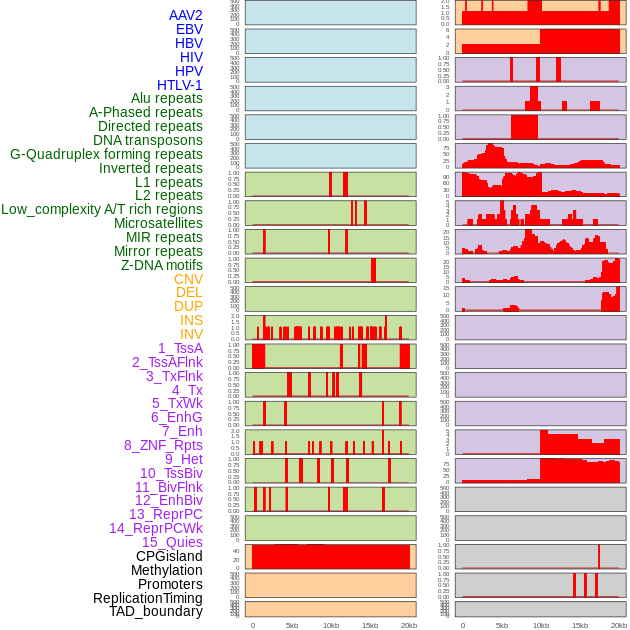 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
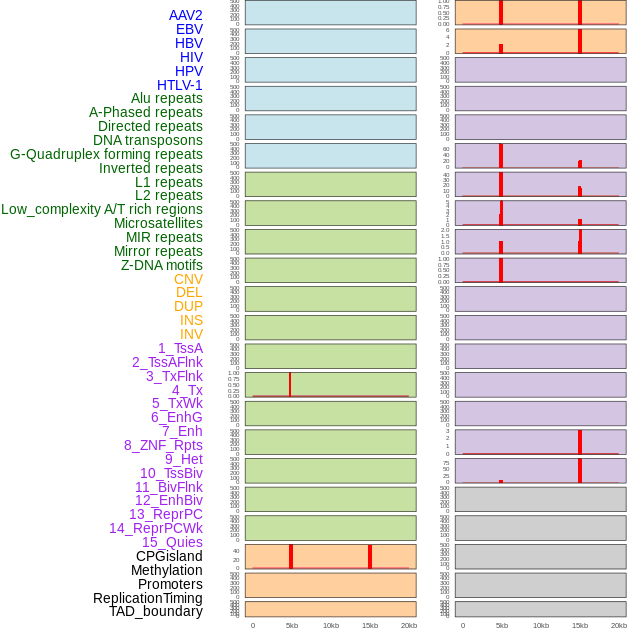 |
Top |
Fusion Protein Features for PDCD10-ARHGAP15 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:167437849/chr2:143986148) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | ARHGAP15 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: GTPase activator for the Rho-type GTPases by converting them to an inactive GDP-bound state. Has activity toward RAC1. Overexpression results in an increase in actin stress fibers and cell contraction. {ECO:0000269|PubMed:12650940}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ARHGAP15 | chr3:167437849 | chr2:143986148 | ENST00000295095 | 3 | 14 | 281_470 | 98 | 476.0 | Domain | Rho-GAP |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ARHGAP15 | chr3:167437849 | chr2:143986148 | ENST00000295095 | 3 | 14 | 79_189 | 98 | 476.0 | Domain | PH |
Top |
Fusion Gene Sequence for PDCD10-ARHGAP15 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >63577_63577_1_PDCD10-ARHGAP15_PDCD10_chr3_167437849_ENST00000392750_ARHGAP15_chr2_143986148_ENST00000295095_length(transcript)=1826nt_BP=514nt AAGTCAGTGCCTCAGTTGCTGGTAAGGTGTAGCTGTTCTGTCCCAGTCCCCGTAGCTCTGCACCGAGCAGAAGAGGTCTAGGGTCACGCA GGGGCCATGGCTGAGGCACTTGGGCTTGGTTCTGGCAGGCGGAGCGGACGGGGCTGAGAGCGGGGCCCTACGGGCGGAAGAGGAGCACTG GAAGAAGGAAGAGACAAATGTTGGGGTCTACGGATTCAGTTCCAGTTTAATGATGTGGCCCTCTAAGGAGATAATTTTGCCTTAAACAGA AAGACTTGACCCTCTTAGGTGTCATACCTAAGATCAGTGTGTTTTTTGTGTCCAATTCTTTTATCACCAAAAAAGAGAAGAAATATTGCA GTGAATGAAGATTCCTCTGCATTTTAGCACTGCTTTTTCAACTGTAGTTGGCTTTTGAATGAGGATGACAATGGAAGAGATGAAGAATGA AGCTGAGACCACATCCATGGTTTCTATGCCCCTCTATGCAGTCATGTATCCTGTGTTTAATGAGGAAAAACTGGTCTACTTCCTGGATTG TTCTTTCTAGTCGAAGAATTGAATTTTACAAAGAATCCAAGCAACAGGCTCTGTCCAATATGAAAACTGGGCACAAACCAGAAAGTGTGG ATTTGTGTGGAGCACACATTGAATGGGCCAAGGAAAAATCGAGCAGAAAGAATGTCTTTCAGATCACAACAGTATCAGGAAATGAGTTCC TTCTACAGTCAGATATTGACTTCATCATATTGGATTGGTTCCACGCTATCAAAAATGCAATTGACAGATTGCCAAAGGATTCAAGTTGTC CATCAAGAAACCTGGAATTATTCAAAATCCAAAGATCCTCTAGCACTGAATTGCTAAGTCACTACGACAGTGATATAAAAGAACAGAAAC CAGAGCACAGAAAATCTTTAATGTTCAGACTGCATCACAGTGCTTCCGATACAAGCGACAAAAATCGAGTTAAAAGCAGATTAAAGAAGT TTATTACCCGAAGACCTTCCCTGAAAACTCTGCAAGAAAAAGGACTTATTAAAGATCAAATTTTTGGCTCTCATCTGCACAAAGTGTGTG AACGTGAAAATTCCACAGTTCCGTGGTTTGTAAAGCAATGCATTGAAGCTGTTGAGAAAAGAGGTCTAGATGTTGATGGAATATATCGAG TTAGTGGCAATCTGGCAACAATACAGAAGTTAAGATTTATTGTCAACCAAGAAGAGAAGCTGAATTTGGACGACAGCCAGTGGGAGGACA TCCACGTTGTCACCGGAGCACTGAAGATGTTTTTCCGGGAGCTGCCTGAGCCGCTCTTCCCTTACAGTTTCTTTGAGCAGTTTGTGGAAG CGATCAAAAAGCAAGACAACAACACAAGAATTGAAGCTGTAAAATCTCTTGTACAAAAACTCCCTCCGCCAAATCGTGACACCATGAAAG TCCTCTTTGGACATCTAACTAAGATAGTGGCCAAAGCCTCCAAGAACCTCATGTCCACGCAAAGCTTGGGGATTGTATTTGGACCTACCC TTCTGCGAGCTGAAAATGAAACAGGAAACATGGCGATCCACATGGTCTACCAGAACCAGATAGCTGAGCTCATGCTGAGTGAGTACAGTA AGATCTTCGGCTCAGAGGAAGACTGACAGACAAGACAAGCTACTGAATACGTTCACATCTGTCTTGATGCCTAATATTTTTACATTTCTG TAAACATATTTCTGAAATATTTTTTGCCTTTCAAGCGACAGATGCCTCATTTTGTGAAAACTTAATGATGATTTTGTGTTTAAGTTCCAA >63577_63577_1_PDCD10-ARHGAP15_PDCD10_chr3_167437849_ENST00000392750_ARHGAP15_chr2_143986148_ENST00000295095_length(amino acids)=399AA_BP=22 MKLRPHPWFLCPSMQSCILCLMRKNWSTSWIVLSSRRIEFYKESKQQALSNMKTGHKPESVDLCGAHIEWAKEKSSRKNVFQITTVSGNE FLLQSDIDFIILDWFHAIKNAIDRLPKDSSCPSRNLELFKIQRSSSTELLSHYDSDIKEQKPEHRKSLMFRLHHSASDTSDKNRVKSRLK KFITRRPSLKTLQEKGLIKDQIFGSHLHKVCERENSTVPWFVKQCIEAVEKRGLDVDGIYRVSGNLATIQKLRFIVNQEEKLNLDDSQWE DIHVVTGALKMFFRELPEPLFPYSFFEQFVEAIKKQDNNTRIEAVKSLVQKLPPPNRDTMKVLFGHLTKIVAKASKNLMSTQSLGIVFGP -------------------------------------------------------------- >63577_63577_2_PDCD10-ARHGAP15_PDCD10_chr3_167437849_ENST00000392750_ARHGAP15_chr2_143986148_ENST00000409869_length(transcript)=3009nt_BP=514nt AAGTCAGTGCCTCAGTTGCTGGTAAGGTGTAGCTGTTCTGTCCCAGTCCCCGTAGCTCTGCACCGAGCAGAAGAGGTCTAGGGTCACGCA GGGGCCATGGCTGAGGCACTTGGGCTTGGTTCTGGCAGGCGGAGCGGACGGGGCTGAGAGCGGGGCCCTACGGGCGGAAGAGGAGCACTG GAAGAAGGAAGAGACAAATGTTGGGGTCTACGGATTCAGTTCCAGTTTAATGATGTGGCCCTCTAAGGAGATAATTTTGCCTTAAACAGA AAGACTTGACCCTCTTAGGTGTCATACCTAAGATCAGTGTGTTTTTTGTGTCCAATTCTTTTATCACCAAAAAAGAGAAGAAATATTGCA GTGAATGAAGATTCCTCTGCATTTTAGCACTGCTTTTTCAACTGTAGTTGGCTTTTGAATGAGGATGACAATGGAAGAGATGAAGAATGA AGCTGAGACCACATCCATGGTTTCTATGCCCCTCTATGCAGTCATGTATCCTGTGTTTAATGAGGAAAAACTGGTCTACTTCCTGGATTG TTCTTTCTAGTCGAAGAATTGAATTTTACAAAGAATCCAAGCAACAGGCTCTGTCCAATATGAGCTTCTGCTTGATGTGTTCAGGAGGCA CCTGACCATGTTCATGGGATTTGGCAAATCTCCATTTTTGGTTAAACAAAACCCATTTGATGGATTGCATCATAGAGAAGAAGCTCTGCA ATTTAGAACATCCATTCATTTATTCTCTCAACAGTTAACAACTCCTACCAGGTGTCATATAATTTTTTTCTTTTTGTTTTGTTTTGTTTT TACATTAGGGGCAACAGAAATATTCAAATGAATAGTATATGATCCCTGCTCTTAAAGTACTCACAATATGGTAAAAGAGACATAATTTTG TATAGGTTTATTATAATAAAACCATGACTAAAGTGTTAAAAAATATTAGACAACATGCCATGGAACCATAGATTTAGCTATAGGTAGTCT TTTTCCAGAGAAACTATTGTTGCAAGTCATGTTCAGCTTAGGTATCTCTCCTTTTGTTCAAATATTACTGTTTGATATTATTATATCAAA TATTATTGAGCATCTCTTATGTTTAAGTCACTACGATAGGTGTTCTGAAAGATAAAAAATGAACCAGACTGAAACAAATAGAAAGTATGG CCTTAGATTGAGTATAGTGAAGAAATGGATGTTGGTAGTTGTCCTAAAATGGCATGAACAGAGACATGGCGTCGTTTTATGATAACTCAA GAGAAGAGTGTGATTGCTTCCATTTGGACAGGATTTGAAATATATTCATGGAGGAAGTGACATTTGAGCACATTTCTGAAGGATGTATAG GATTTTGATGTAAAAGGGGGCAAAAGATATTTTATGTACTAATAAGAGAGTAATTAATAGTGAACTATGGAAAATTCTTTTTTTGTTGTT TATTTTACAGATGTATATTGATCTCTTGCCTTCTTCCATGCACTGTGTACTAGTCACTGTTCTTGAAATGTTTGAATGGGGGTTCCCTGA GGCCTGGAGTGAATATTTCCTAGTCCAGAGTATTTTGTGTTTCTTCTCTAAGTTGCCTGGGAGTGAGGAACAGCTTTAGACTAAGTTGCG GAACAGCATTGTGGAACAGCAAACAGTGTAATTTGGTTAGAAAGTCAGTGTGGAACCGGTTTAGGAAAGACTTTGAACAGGATTGGGAGA AGAGATCATTTCATTCTGTAGGTAATGTGGGGTTTTTGTAACCAATATGAAATATTCTCAAATTAAGAGAGACTCAGAGTTTTCAGAGAC ATGTTTAGGGAACTGATATATATATCAGTTGTCTATAAAATTAAGTCTCTCTTAATTTTGTGTATATATAACTTAAAAACACATGCAGGA CTTGGTAAATTATCCGAGAGACATGTTCTAAAAATTTATGCAAATAGATGTGAGCAATAAGGCCTGAGCTCTTACGACAGAGAAAATTGT AAAACGGCTGTGTGTCATCATTTTGGCTCAATGCATTTAGTAAATCACAGGACATCTTTTCACTGACAAAATATTTTGATATTCTCATAT GTCTCTGACCTCCAACAGTATGGCCATTTCTAAAGACGTTCTAGTGCAAAGAGAACAAGCAGAGCTATCCATCTAAGGAGAGCGAAAAAC ATGCTGTCCCAAGAACTAGGTGTGTCACCATCTCAGTGAGCACCCGTTGCAAACCTACTGGTGCTTCCATCTGACTCCAGAACATCTGTT TAGTGTCACACCCTACTTAAACAAATGCATTGTGTGAAGTCTTTCCACCAACAATCAGACGCTGTGGAACTCATTGTCACCAATTGGGTG GATGAGAGCCACCGTTTCTATTATATTCATCACACACTGCTCAGAAAATTTAGTGTCACTTTTGATATAAATTCAAAAACATTGTAAACA TTTTAGGGAATATTTTCCCCCTAAAATGGTGTGGTTCTAAAGCCAGTATATTCAGTAGAAAGCCTATATGTTGGTTGTGTAAGGGACTTT TTTGTTACTCTTATGTTTTTTATTTTACATTGTACTAATATGTATTTTTAATCAGAGCCAACAACTGGCAATTTTTACTTTTTAAAAATT ATATTTTTGGGTTTTACATTTAAGTCTTTAATCCATCTTGAGTTAATTTTTGTATATGGTGTAAGGAAGGGGTCCAGTTTCAATTTTCTG CATATGGTTACCCAGTTCCCCACATTTATTAAATAAACTATAAAAACTCTAGAATGAAATCTAGGCAATACCATTCAGGACATAGGCACA GACAAAGATTTTATGATGAAAATGCCAAAAGCAATTGTAATAAAAGCAAATGGTGACAAATGGGGTCTAATTAAACTAAAGAGCTTCCGC ACAGCAAAAGTAACTATCATCAGAGTGAACAGACAACTTTCAGAATGGGAGACAATTTTTGCAATCTATTCAGCTGACAAAATTCTAATA >63577_63577_2_PDCD10-ARHGAP15_PDCD10_chr3_167437849_ENST00000392750_ARHGAP15_chr2_143986148_ENST00000409869_length(amino acids)=77AA_BP= -------------------------------------------------------------- >63577_63577_3_PDCD10-ARHGAP15_PDCD10_chr3_167437849_ENST00000461494_ARHGAP15_chr2_143986148_ENST00000295095_length(transcript)=1637nt_BP=325nt AGGTTGGCCCCTGGTGCCCCGGGAGAGGGGACAGGGCGGATGTGCCTGGGGTGGGGTCCTTTGAAGGTGTGGCTTCGGACCGAGAGGTGA GGCTGCTGTTTTCCCCCTGCTTTATCAGTGTGTTTTTTGTGTCCAATTCTTTTATCACCAAAAAAGAGAAGAAATATTGCAGTGAATGAA GATTCCTCTGCATTTTAGCACTGCTTTTTCAACTGTAGTTGGCTTTTGAATGAGGATGACAATGGAAGAGATGAAGAATGAAGCTGAGAC CACATCCATGGTTTCTATGCCCCTCTATGCAGTCATGTATCCTGTGTTTAATGAGGAAAAACTGGTCTACTTCCTGGATTGTTCTTTCTA GTCGAAGAATTGAATTTTACAAAGAATCCAAGCAACAGGCTCTGTCCAATATGAAAACTGGGCACAAACCAGAAAGTGTGGATTTGTGTG GAGCACACATTGAATGGGCCAAGGAAAAATCGAGCAGAAAGAATGTCTTTCAGATCACAACAGTATCAGGAAATGAGTTCCTTCTACAGT CAGATATTGACTTCATCATATTGGATTGGTTCCACGCTATCAAAAATGCAATTGACAGATTGCCAAAGGATTCAAGTTGTCCATCAAGAA ACCTGGAATTATTCAAAATCCAAAGATCCTCTAGCACTGAATTGCTAAGTCACTACGACAGTGATATAAAAGAACAGAAACCAGAGCACA GAAAATCTTTAATGTTCAGACTGCATCACAGTGCTTCCGATACAAGCGACAAAAATCGAGTTAAAAGCAGATTAAAGAAGTTTATTACCC GAAGACCTTCCCTGAAAACTCTGCAAGAAAAAGGACTTATTAAAGATCAAATTTTTGGCTCTCATCTGCACAAAGTGTGTGAACGTGAAA ATTCCACAGTTCCGTGGTTTGTAAAGCAATGCATTGAAGCTGTTGAGAAAAGAGGTCTAGATGTTGATGGAATATATCGAGTTAGTGGCA ATCTGGCAACAATACAGAAGTTAAGATTTATTGTCAACCAAGAAGAGAAGCTGAATTTGGACGACAGCCAGTGGGAGGACATCCACGTTG TCACCGGAGCACTGAAGATGTTTTTCCGGGAGCTGCCTGAGCCGCTCTTCCCTTACAGTTTCTTTGAGCAGTTTGTGGAAGCGATCAAAA AGCAAGACAACAACACAAGAATTGAAGCTGTAAAATCTCTTGTACAAAAACTCCCTCCGCCAAATCGTGACACCATGAAAGTCCTCTTTG GACATCTAACTAAGATAGTGGCCAAAGCCTCCAAGAACCTCATGTCCACGCAAAGCTTGGGGATTGTATTTGGACCTACCCTTCTGCGAG CTGAAAATGAAACAGGAAACATGGCGATCCACATGGTCTACCAGAACCAGATAGCTGAGCTCATGCTGAGTGAGTACAGTAAGATCTTCG GCTCAGAGGAAGACTGACAGACAAGACAAGCTACTGAATACGTTCACATCTGTCTTGATGCCTAATATTTTTACATTTCTGTAAACATAT TTCTGAAATATTTTTTGCCTTTCAAGCGACAGATGCCTCATTTTGTGAAAACTTAATGATGATTTTGTGTTTAAGTTCCAAACATTTGAA >63577_63577_3_PDCD10-ARHGAP15_PDCD10_chr3_167437849_ENST00000461494_ARHGAP15_chr2_143986148_ENST00000295095_length(amino acids)=399AA_BP=22 MKLRPHPWFLCPSMQSCILCLMRKNWSTSWIVLSSRRIEFYKESKQQALSNMKTGHKPESVDLCGAHIEWAKEKSSRKNVFQITTVSGNE FLLQSDIDFIILDWFHAIKNAIDRLPKDSSCPSRNLELFKIQRSSSTELLSHYDSDIKEQKPEHRKSLMFRLHHSASDTSDKNRVKSRLK KFITRRPSLKTLQEKGLIKDQIFGSHLHKVCERENSTVPWFVKQCIEAVEKRGLDVDGIYRVSGNLATIQKLRFIVNQEEKLNLDDSQWE DIHVVTGALKMFFRELPEPLFPYSFFEQFVEAIKKQDNNTRIEAVKSLVQKLPPPNRDTMKVLFGHLTKIVAKASKNLMSTQSLGIVFGP -------------------------------------------------------------- >63577_63577_4_PDCD10-ARHGAP15_PDCD10_chr3_167437849_ENST00000461494_ARHGAP15_chr2_143986148_ENST00000409869_length(transcript)=2820nt_BP=325nt AGGTTGGCCCCTGGTGCCCCGGGAGAGGGGACAGGGCGGATGTGCCTGGGGTGGGGTCCTTTGAAGGTGTGGCTTCGGACCGAGAGGTGA GGCTGCTGTTTTCCCCCTGCTTTATCAGTGTGTTTTTTGTGTCCAATTCTTTTATCACCAAAAAAGAGAAGAAATATTGCAGTGAATGAA GATTCCTCTGCATTTTAGCACTGCTTTTTCAACTGTAGTTGGCTTTTGAATGAGGATGACAATGGAAGAGATGAAGAATGAAGCTGAGAC CACATCCATGGTTTCTATGCCCCTCTATGCAGTCATGTATCCTGTGTTTAATGAGGAAAAACTGGTCTACTTCCTGGATTGTTCTTTCTA GTCGAAGAATTGAATTTTACAAAGAATCCAAGCAACAGGCTCTGTCCAATATGAGCTTCTGCTTGATGTGTTCAGGAGGCACCTGACCAT GTTCATGGGATTTGGCAAATCTCCATTTTTGGTTAAACAAAACCCATTTGATGGATTGCATCATAGAGAAGAAGCTCTGCAATTTAGAAC ATCCATTCATTTATTCTCTCAACAGTTAACAACTCCTACCAGGTGTCATATAATTTTTTTCTTTTTGTTTTGTTTTGTTTTTACATTAGG GGCAACAGAAATATTCAAATGAATAGTATATGATCCCTGCTCTTAAAGTACTCACAATATGGTAAAAGAGACATAATTTTGTATAGGTTT ATTATAATAAAACCATGACTAAAGTGTTAAAAAATATTAGACAACATGCCATGGAACCATAGATTTAGCTATAGGTAGTCTTTTTCCAGA GAAACTATTGTTGCAAGTCATGTTCAGCTTAGGTATCTCTCCTTTTGTTCAAATATTACTGTTTGATATTATTATATCAAATATTATTGA GCATCTCTTATGTTTAAGTCACTACGATAGGTGTTCTGAAAGATAAAAAATGAACCAGACTGAAACAAATAGAAAGTATGGCCTTAGATT GAGTATAGTGAAGAAATGGATGTTGGTAGTTGTCCTAAAATGGCATGAACAGAGACATGGCGTCGTTTTATGATAACTCAAGAGAAGAGT GTGATTGCTTCCATTTGGACAGGATTTGAAATATATTCATGGAGGAAGTGACATTTGAGCACATTTCTGAAGGATGTATAGGATTTTGAT GTAAAAGGGGGCAAAAGATATTTTATGTACTAATAAGAGAGTAATTAATAGTGAACTATGGAAAATTCTTTTTTTGTTGTTTATTTTACA GATGTATATTGATCTCTTGCCTTCTTCCATGCACTGTGTACTAGTCACTGTTCTTGAAATGTTTGAATGGGGGTTCCCTGAGGCCTGGAG TGAATATTTCCTAGTCCAGAGTATTTTGTGTTTCTTCTCTAAGTTGCCTGGGAGTGAGGAACAGCTTTAGACTAAGTTGCGGAACAGCAT TGTGGAACAGCAAACAGTGTAATTTGGTTAGAAAGTCAGTGTGGAACCGGTTTAGGAAAGACTTTGAACAGGATTGGGAGAAGAGATCAT TTCATTCTGTAGGTAATGTGGGGTTTTTGTAACCAATATGAAATATTCTCAAATTAAGAGAGACTCAGAGTTTTCAGAGACATGTTTAGG GAACTGATATATATATCAGTTGTCTATAAAATTAAGTCTCTCTTAATTTTGTGTATATATAACTTAAAAACACATGCAGGACTTGGTAAA TTATCCGAGAGACATGTTCTAAAAATTTATGCAAATAGATGTGAGCAATAAGGCCTGAGCTCTTACGACAGAGAAAATTGTAAAACGGCT GTGTGTCATCATTTTGGCTCAATGCATTTAGTAAATCACAGGACATCTTTTCACTGACAAAATATTTTGATATTCTCATATGTCTCTGAC CTCCAACAGTATGGCCATTTCTAAAGACGTTCTAGTGCAAAGAGAACAAGCAGAGCTATCCATCTAAGGAGAGCGAAAAACATGCTGTCC CAAGAACTAGGTGTGTCACCATCTCAGTGAGCACCCGTTGCAAACCTACTGGTGCTTCCATCTGACTCCAGAACATCTGTTTAGTGTCAC ACCCTACTTAAACAAATGCATTGTGTGAAGTCTTTCCACCAACAATCAGACGCTGTGGAACTCATTGTCACCAATTGGGTGGATGAGAGC CACCGTTTCTATTATATTCATCACACACTGCTCAGAAAATTTAGTGTCACTTTTGATATAAATTCAAAAACATTGTAAACATTTTAGGGA ATATTTTCCCCCTAAAATGGTGTGGTTCTAAAGCCAGTATATTCAGTAGAAAGCCTATATGTTGGTTGTGTAAGGGACTTTTTTGTTACT CTTATGTTTTTTATTTTACATTGTACTAATATGTATTTTTAATCAGAGCCAACAACTGGCAATTTTTACTTTTTAAAAATTATATTTTTG GGTTTTACATTTAAGTCTTTAATCCATCTTGAGTTAATTTTTGTATATGGTGTAAGGAAGGGGTCCAGTTTCAATTTTCTGCATATGGTT ACCCAGTTCCCCACATTTATTAAATAAACTATAAAAACTCTAGAATGAAATCTAGGCAATACCATTCAGGACATAGGCACAGACAAAGAT TTTATGATGAAAATGCCAAAAGCAATTGTAATAAAAGCAAATGGTGACAAATGGGGTCTAATTAAACTAAAGAGCTTCCGCACAGCAAAA GTAACTATCATCAGAGTGAACAGACAACTTTCAGAATGGGAGACAATTTTTGCAATCTATTCAGCTGACAAAATTCTAATATCCAGAGTC >63577_63577_4_PDCD10-ARHGAP15_PDCD10_chr3_167437849_ENST00000461494_ARHGAP15_chr2_143986148_ENST00000409869_length(amino acids)=77AA_BP= -------------------------------------------------------------- >63577_63577_5_PDCD10-ARHGAP15_PDCD10_chr3_167437849_ENST00000487947_ARHGAP15_chr2_143986148_ENST00000295095_length(transcript)=1548nt_BP=236nt GAAGGTGTGGCTTCGGACCGAGAGATCAGTGTGTTTTTTGTGTCCAATTCTTTTATCACCAAAAAAGAGAAGAAATATTGCAGTGAATGA AGATTCCTCTGCATTTTAGCACTGCTTTTTCAACTGTAGTTGGCTTTTGAATGAGGATGACAATGGAAGAGATGAAGAATGAAGCTGAGA CCACATCCATGGTTTCTATGCCCCTCTATGCAGTCATGTATCCTGTGTTTAATGAGGAAAAACTGGTCTACTTCCTGGATTGTTCTTTCT AGTCGAAGAATTGAATTTTACAAAGAATCCAAGCAACAGGCTCTGTCCAATATGAAAACTGGGCACAAACCAGAAAGTGTGGATTTGTGT GGAGCACACATTGAATGGGCCAAGGAAAAATCGAGCAGAAAGAATGTCTTTCAGATCACAACAGTATCAGGAAATGAGTTCCTTCTACAG TCAGATATTGACTTCATCATATTGGATTGGTTCCACGCTATCAAAAATGCAATTGACAGATTGCCAAAGGATTCAAGTTGTCCATCAAGA AACCTGGAATTATTCAAAATCCAAAGATCCTCTAGCACTGAATTGCTAAGTCACTACGACAGTGATATAAAAGAACAGAAACCAGAGCAC AGAAAATCTTTAATGTTCAGACTGCATCACAGTGCTTCCGATACAAGCGACAAAAATCGAGTTAAAAGCAGATTAAAGAAGTTTATTACC CGAAGACCTTCCCTGAAAACTCTGCAAGAAAAAGGACTTATTAAAGATCAAATTTTTGGCTCTCATCTGCACAAAGTGTGTGAACGTGAA AATTCCACAGTTCCGTGGTTTGTAAAGCAATGCATTGAAGCTGTTGAGAAAAGAGGTCTAGATGTTGATGGAATATATCGAGTTAGTGGC AATCTGGCAACAATACAGAAGTTAAGATTTATTGTCAACCAAGAAGAGAAGCTGAATTTGGACGACAGCCAGTGGGAGGACATCCACGTT GTCACCGGAGCACTGAAGATGTTTTTCCGGGAGCTGCCTGAGCCGCTCTTCCCTTACAGTTTCTTTGAGCAGTTTGTGGAAGCGATCAAA AAGCAAGACAACAACACAAGAATTGAAGCTGTAAAATCTCTTGTACAAAAACTCCCTCCGCCAAATCGTGACACCATGAAAGTCCTCTTT GGACATCTAACTAAGATAGTGGCCAAAGCCTCCAAGAACCTCATGTCCACGCAAAGCTTGGGGATTGTATTTGGACCTACCCTTCTGCGA GCTGAAAATGAAACAGGAAACATGGCGATCCACATGGTCTACCAGAACCAGATAGCTGAGCTCATGCTGAGTGAGTACAGTAAGATCTTC GGCTCAGAGGAAGACTGACAGACAAGACAAGCTACTGAATACGTTCACATCTGTCTTGATGCCTAATATTTTTACATTTCTGTAAACATA TTTCTGAAATATTTTTTGCCTTTCAAGCGACAGATGCCTCATTTTGTGAAAACTTAATGATGATTTTGTGTTTAAGTTCCAAACATTTGA >63577_63577_5_PDCD10-ARHGAP15_PDCD10_chr3_167437849_ENST00000487947_ARHGAP15_chr2_143986148_ENST00000295095_length(amino acids)=399AA_BP=22 MKLRPHPWFLCPSMQSCILCLMRKNWSTSWIVLSSRRIEFYKESKQQALSNMKTGHKPESVDLCGAHIEWAKEKSSRKNVFQITTVSGNE FLLQSDIDFIILDWFHAIKNAIDRLPKDSSCPSRNLELFKIQRSSSTELLSHYDSDIKEQKPEHRKSLMFRLHHSASDTSDKNRVKSRLK KFITRRPSLKTLQEKGLIKDQIFGSHLHKVCERENSTVPWFVKQCIEAVEKRGLDVDGIYRVSGNLATIQKLRFIVNQEEKLNLDDSQWE DIHVVTGALKMFFRELPEPLFPYSFFEQFVEAIKKQDNNTRIEAVKSLVQKLPPPNRDTMKVLFGHLTKIVAKASKNLMSTQSLGIVFGP -------------------------------------------------------------- >63577_63577_6_PDCD10-ARHGAP15_PDCD10_chr3_167437849_ENST00000487947_ARHGAP15_chr2_143986148_ENST00000409869_length(transcript)=2731nt_BP=236nt GAAGGTGTGGCTTCGGACCGAGAGATCAGTGTGTTTTTTGTGTCCAATTCTTTTATCACCAAAAAAGAGAAGAAATATTGCAGTGAATGA AGATTCCTCTGCATTTTAGCACTGCTTTTTCAACTGTAGTTGGCTTTTGAATGAGGATGACAATGGAAGAGATGAAGAATGAAGCTGAGA CCACATCCATGGTTTCTATGCCCCTCTATGCAGTCATGTATCCTGTGTTTAATGAGGAAAAACTGGTCTACTTCCTGGATTGTTCTTTCT AGTCGAAGAATTGAATTTTACAAAGAATCCAAGCAACAGGCTCTGTCCAATATGAGCTTCTGCTTGATGTGTTCAGGAGGCACCTGACCA TGTTCATGGGATTTGGCAAATCTCCATTTTTGGTTAAACAAAACCCATTTGATGGATTGCATCATAGAGAAGAAGCTCTGCAATTTAGAA CATCCATTCATTTATTCTCTCAACAGTTAACAACTCCTACCAGGTGTCATATAATTTTTTTCTTTTTGTTTTGTTTTGTTTTTACATTAG GGGCAACAGAAATATTCAAATGAATAGTATATGATCCCTGCTCTTAAAGTACTCACAATATGGTAAAAGAGACATAATTTTGTATAGGTT TATTATAATAAAACCATGACTAAAGTGTTAAAAAATATTAGACAACATGCCATGGAACCATAGATTTAGCTATAGGTAGTCTTTTTCCAG AGAAACTATTGTTGCAAGTCATGTTCAGCTTAGGTATCTCTCCTTTTGTTCAAATATTACTGTTTGATATTATTATATCAAATATTATTG AGCATCTCTTATGTTTAAGTCACTACGATAGGTGTTCTGAAAGATAAAAAATGAACCAGACTGAAACAAATAGAAAGTATGGCCTTAGAT TGAGTATAGTGAAGAAATGGATGTTGGTAGTTGTCCTAAAATGGCATGAACAGAGACATGGCGTCGTTTTATGATAACTCAAGAGAAGAG TGTGATTGCTTCCATTTGGACAGGATTTGAAATATATTCATGGAGGAAGTGACATTTGAGCACATTTCTGAAGGATGTATAGGATTTTGA TGTAAAAGGGGGCAAAAGATATTTTATGTACTAATAAGAGAGTAATTAATAGTGAACTATGGAAAATTCTTTTTTTGTTGTTTATTTTAC AGATGTATATTGATCTCTTGCCTTCTTCCATGCACTGTGTACTAGTCACTGTTCTTGAAATGTTTGAATGGGGGTTCCCTGAGGCCTGGA GTGAATATTTCCTAGTCCAGAGTATTTTGTGTTTCTTCTCTAAGTTGCCTGGGAGTGAGGAACAGCTTTAGACTAAGTTGCGGAACAGCA TTGTGGAACAGCAAACAGTGTAATTTGGTTAGAAAGTCAGTGTGGAACCGGTTTAGGAAAGACTTTGAACAGGATTGGGAGAAGAGATCA TTTCATTCTGTAGGTAATGTGGGGTTTTTGTAACCAATATGAAATATTCTCAAATTAAGAGAGACTCAGAGTTTTCAGAGACATGTTTAG GGAACTGATATATATATCAGTTGTCTATAAAATTAAGTCTCTCTTAATTTTGTGTATATATAACTTAAAAACACATGCAGGACTTGGTAA ATTATCCGAGAGACATGTTCTAAAAATTTATGCAAATAGATGTGAGCAATAAGGCCTGAGCTCTTACGACAGAGAAAATTGTAAAACGGC TGTGTGTCATCATTTTGGCTCAATGCATTTAGTAAATCACAGGACATCTTTTCACTGACAAAATATTTTGATATTCTCATATGTCTCTGA CCTCCAACAGTATGGCCATTTCTAAAGACGTTCTAGTGCAAAGAGAACAAGCAGAGCTATCCATCTAAGGAGAGCGAAAAACATGCTGTC CCAAGAACTAGGTGTGTCACCATCTCAGTGAGCACCCGTTGCAAACCTACTGGTGCTTCCATCTGACTCCAGAACATCTGTTTAGTGTCA CACCCTACTTAAACAAATGCATTGTGTGAAGTCTTTCCACCAACAATCAGACGCTGTGGAACTCATTGTCACCAATTGGGTGGATGAGAG CCACCGTTTCTATTATATTCATCACACACTGCTCAGAAAATTTAGTGTCACTTTTGATATAAATTCAAAAACATTGTAAACATTTTAGGG AATATTTTCCCCCTAAAATGGTGTGGTTCTAAAGCCAGTATATTCAGTAGAAAGCCTATATGTTGGTTGTGTAAGGGACTTTTTTGTTAC TCTTATGTTTTTTATTTTACATTGTACTAATATGTATTTTTAATCAGAGCCAACAACTGGCAATTTTTACTTTTTAAAAATTATATTTTT GGGTTTTACATTTAAGTCTTTAATCCATCTTGAGTTAATTTTTGTATATGGTGTAAGGAAGGGGTCCAGTTTCAATTTTCTGCATATGGT TACCCAGTTCCCCACATTTATTAAATAAACTATAAAAACTCTAGAATGAAATCTAGGCAATACCATTCAGGACATAGGCACAGACAAAGA TTTTATGATGAAAATGCCAAAAGCAATTGTAATAAAAGCAAATGGTGACAAATGGGGTCTAATTAAACTAAAGAGCTTCCGCACAGCAAA AGTAACTATCATCAGAGTGAACAGACAACTTTCAGAATGGGAGACAATTTTTGCAATCTATTCAGCTGACAAAATTCTAATATCCAGAGT >63577_63577_6_PDCD10-ARHGAP15_PDCD10_chr3_167437849_ENST00000487947_ARHGAP15_chr2_143986148_ENST00000409869_length(amino acids)=77AA_BP= -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for PDCD10-ARHGAP15 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for PDCD10-ARHGAP15 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for PDCD10-ARHGAP15 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
