
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:PDXDC1-NOB1 (FusionGDB2 ID:64103) |
Fusion Gene Summary for PDXDC1-NOB1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: PDXDC1-NOB1 | Fusion gene ID: 64103 | Hgene | Tgene | Gene symbol | PDXDC1 | NOB1 | Gene ID | 23042 | 28987 |
| Gene name | pyridoxal dependent decarboxylase domain containing 1 | NIN1 (RPN12) binding protein 1 homolog | |
| Synonyms | LP8165 | ART-4|MST158|MSTP158|NOB1P|PSMD8BP1 | |
| Cytomap | 16p13.11 | 16q22.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | pyridoxal-dependent decarboxylase domain-containing protein 1 | RNA-binding protein NOB1NIN1/PSMD8 binding protein 1 homologPSMD8 binding protein 1adenocarcinoma antigen recognized by T lymphocytes 4nin one binding proteinphosphorylation regulatory protein HP-10protein ART-4 | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000396410, ENST00000450288, ENST00000455313, ENST00000535621, ENST00000569715, ENST00000325823, ENST00000447912, ENST00000563679, | ENST00000268802, | |
| Fusion gene scores | * DoF score | 24 X 21 X 12=6048 | 4 X 2 X 3=24 |
| # samples | 27 | 4 | |
| ** MAII score | log2(27/6048*10)=-4.48542682717024 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/24*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: PDXDC1 [Title/Abstract] AND NOB1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | PDXDC1(15069077)-NOB1(69788629), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across PDXDC1 (5'-gene) Fusion gene breakpoints across PDXDC1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
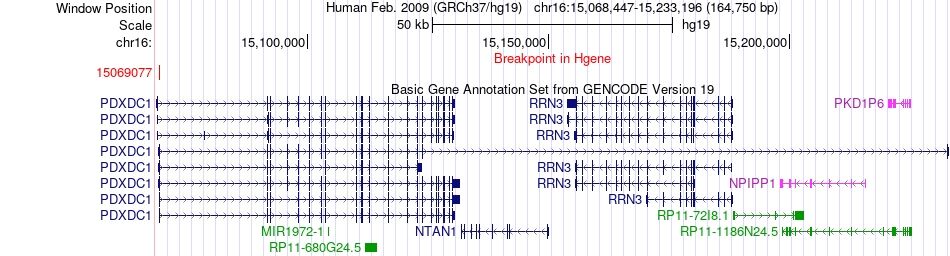 |
 Fusion gene breakpoints across NOB1 (3'-gene) Fusion gene breakpoints across NOB1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
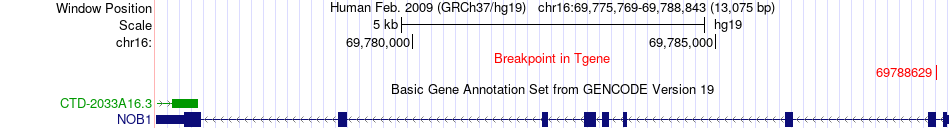 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-13-A5FT | PDXDC1 | chr16 | 15069077 | + | NOB1 | chr16 | 69788629 | - |
Top |
Fusion Gene ORF analysis for PDXDC1-NOB1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000396410 | ENST00000268802 | PDXDC1 | chr16 | 15069077 | + | NOB1 | chr16 | 69788629 | - |
| In-frame | ENST00000450288 | ENST00000268802 | PDXDC1 | chr16 | 15069077 | + | NOB1 | chr16 | 69788629 | - |
| In-frame | ENST00000455313 | ENST00000268802 | PDXDC1 | chr16 | 15069077 | + | NOB1 | chr16 | 69788629 | - |
| In-frame | ENST00000535621 | ENST00000268802 | PDXDC1 | chr16 | 15069077 | + | NOB1 | chr16 | 69788629 | - |
| In-frame | ENST00000569715 | ENST00000268802 | PDXDC1 | chr16 | 15069077 | + | NOB1 | chr16 | 69788629 | - |
| intron-3CDS | ENST00000325823 | ENST00000268802 | PDXDC1 | chr16 | 15069077 | + | NOB1 | chr16 | 69788629 | - |
| intron-3CDS | ENST00000447912 | ENST00000268802 | PDXDC1 | chr16 | 15069077 | + | NOB1 | chr16 | 69788629 | - |
| intron-3CDS | ENST00000563679 | ENST00000268802 | PDXDC1 | chr16 | 15069077 | + | NOB1 | chr16 | 69788629 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000535621 | PDXDC1 | chr16 | 15069077 | + | ENST00000268802 | NOB1 | chr16 | 69788629 | - | 1850 | 209 | 50 | 1384 | 444 |
| ENST00000455313 | PDXDC1 | chr16 | 15069077 | + | ENST00000268802 | NOB1 | chr16 | 69788629 | - | 1782 | 141 | 120 | 1316 | 398 |
| ENST00000396410 | PDXDC1 | chr16 | 15069077 | + | ENST00000268802 | NOB1 | chr16 | 69788629 | - | 1759 | 118 | 97 | 1293 | 398 |
| ENST00000569715 | PDXDC1 | chr16 | 15069077 | + | ENST00000268802 | NOB1 | chr16 | 69788629 | - | 1739 | 98 | 77 | 1273 | 398 |
| ENST00000450288 | PDXDC1 | chr16 | 15069077 | + | ENST00000268802 | NOB1 | chr16 | 69788629 | - | 1733 | 92 | 71 | 1267 | 398 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000535621 | ENST00000268802 | PDXDC1 | chr16 | 15069077 | + | NOB1 | chr16 | 69788629 | - | 0.004933318 | 0.99506664 |
| ENST00000455313 | ENST00000268802 | PDXDC1 | chr16 | 15069077 | + | NOB1 | chr16 | 69788629 | - | 0.004121795 | 0.9958782 |
| ENST00000396410 | ENST00000268802 | PDXDC1 | chr16 | 15069077 | + | NOB1 | chr16 | 69788629 | - | 0.004088013 | 0.99591196 |
| ENST00000569715 | ENST00000268802 | PDXDC1 | chr16 | 15069077 | + | NOB1 | chr16 | 69788629 | - | 0.003916046 | 0.996084 |
| ENST00000450288 | ENST00000268802 | PDXDC1 | chr16 | 15069077 | + | NOB1 | chr16 | 69788629 | - | 0.003905883 | 0.99609417 |
Top |
Fusion Genomic Features for PDXDC1-NOB1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
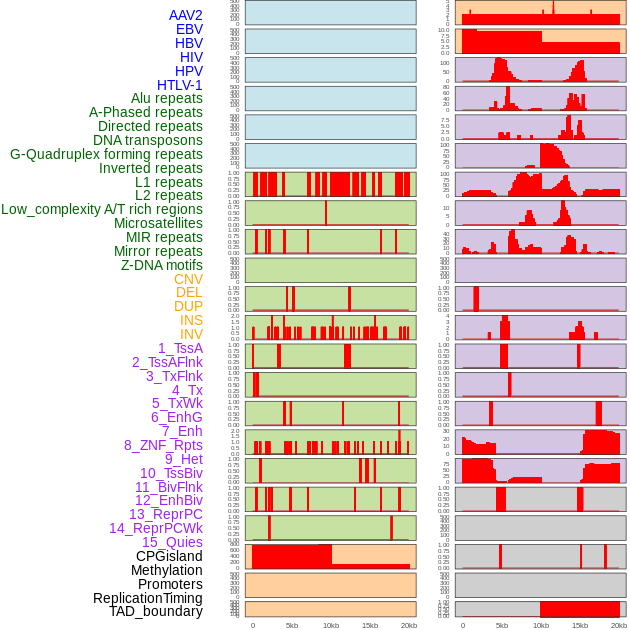 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for PDXDC1-NOB1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:15069077/chr16:69788629) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | NOB1 | chr16:15069077 | chr16:69788629 | ENST00000268802 | 0 | 9 | 185_191 | 21 | 413.0 | Compositional bias | Poly-Glu | |
| Tgene | NOB1 | chr16:15069077 | chr16:69788629 | ENST00000268802 | 0 | 9 | 260_332 | 21 | 413.0 | Zinc finger | NOB1 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | NOB1 | chr16:15069077 | chr16:69788629 | ENST00000268802 | 0 | 9 | 5_108 | 21 | 413.0 | Domain | PINc |
Top |
Fusion Gene Sequence for PDXDC1-NOB1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >64103_64103_1_PDXDC1-NOB1_PDXDC1_chr16_15069077_ENST00000396410_NOB1_chr16_69788629_ENST00000268802_length(transcript)=1759nt_BP=118nt CGGGGGACGTCAGCGCTGCCAGCGTGGAAGGAGCTGCGGGGCGCGGGAGGAGGAAGTAGAGCCCGGGACCGCCAGGCCACCACCGGCCGC CTCAGCCATGGACGCGTCCCTGGAGAAGGACATCGGGAAGAACATTTACACCATCCGGGAGGTGGTCACTGAGATTCGGGACAAGGCCAC ACGCAGGCGGCTCGCTGTCCTGCCCTACGAGCTGCGGTTCAAGGAGCCCTTACCGGAATACGTGCGGCTGGTGACTGAGTTTTCAAAGAA AACAGGAGACTACCCCAGCCTCTCTGCCACGGACATCCAAGTGCTTGCACTCACATACCAGTTGGAAGCAGAGTTTGTTGGGGTGTCTCA CCTAAAACAAGAACCACAGAAGGTTAAGGTGAGCTCATCGATTCAGCACCCAGAAACACCTCTGCACATTTCTGGTTTCCATCTGCCCTA CAAGCCTAAACCCCCACAAGAAACAGAAAAAGGACACTCAGCTTGTGAGCCTGAGAACCTGGAATTTAGTTCCTTCATGTTCTGGAGAAA CCCTTTGCCCAACATCGATCATGAACTGCAGGAGCTGCTGATTGACAGAGGTGAGGACGTTCCAAGTGAGGAGGAGGAGGAGGAAGAAAA CGGGTTTGAAGACAGAAAAGATGACAGCGATGACGACGGGGGTGGCTGGATAACCCCCAGTAACATCAAGCAGATCCAGCAGGAGCTGGA GCAGTGTGACGTCCCCGAGGACGTGCGGGTTGGCTGCCTGACCACAGACTTCGCCATGCAGAATGTTCTGCTGCAGATGGGGCTGCACGT GCTGGCGGTGAACGGCATGCTGATTCGTGAGGCCCGGAGCTACATCTTGCGCTGCCATGGCTGTTTCAAGACAACGTCTGACATGAGCCG AGTGTTCTGCTCACACTGTGGGAACAAGACCCTGAAGAAAGTGTCCGTGACCGTCAGCGACGACGGCACCCTGCACATGCACTTCTCCCG CAACCCCAAGGTGCTGAACCCCCGCGGCCTCCGGTACTCGCTTCCCACTCCCAAAGGGGGCAAATACGCCATCAACCCCCATCTCACCGA GGATCAGCGCTTCCCTCAGCTGCGACTCTCCCAAAAGGCCAGGCAGAAAACCAACGTGTTCGCCCCTGACTACATCGCCGGGGTGTCACC CTTTGTCGAGAATGACATCTCCAGCCGCTCAGCTACCCTGCAGGTCCGGGACAGCACCTTGGGAGCTGGGCGGAGACGCTTAAATCCCAA CGCTTCCAGAAAGAAGTTTGTGAAGAAAAGGTGAAGAGCGAGTTCCCGCAGGCAAATTGGATGGGCGTCTGGCCGCCGTGGAGTTCCGGT GACCCATTTCCCCAGCCGTGTCGTCTCCAGGACCACCCGATGGAAATAACAGGCGGGCTTCACGGTGCGGCTCTGTCCGCCCATGCCCCG CTGGGTCTGCAGGGAACTGGACTGTCCCATGGCCTGTGAGCACCGGAGCGCCTGGCTGCCTGCCAAGGAAGTGCAATTGCATAAAAACAG AAAGAACAACGCCCTGGAGCCAATCTTCAAGAAAGGAATTTCCAAAGGATAATATTTTTCTAATAAATGCGGCTGCAACCTCCTGTGCAT TTAATTAAATAGGCCAAATTTTTGCTGCTTAGGTCATCTCAAGGCTGATACTTGAGCTGTGTGCCCAGAGATCATGCATTTAGATTTATA >64103_64103_1_PDXDC1-NOB1_PDXDC1_chr16_15069077_ENST00000396410_NOB1_chr16_69788629_ENST00000268802_length(amino acids)=398AA_BP=7 MDASLEKDIGKNIYTIREVVTEIRDKATRRRLAVLPYELRFKEPLPEYVRLVTEFSKKTGDYPSLSATDIQVLALTYQLEAEFVGVSHLK QEPQKVKVSSSIQHPETPLHISGFHLPYKPKPPQETEKGHSACEPENLEFSSFMFWRNPLPNIDHELQELLIDRGEDVPSEEEEEEENGF EDRKDDSDDDGGGWITPSNIKQIQQELEQCDVPEDVRVGCLTTDFAMQNVLLQMGLHVLAVNGMLIREARSYILRCHGCFKTTSDMSRVF CSHCGNKTLKKVSVTVSDDGTLHMHFSRNPKVLNPRGLRYSLPTPKGGKYAINPHLTEDQRFPQLRLSQKARQKTNVFAPDYIAGVSPFV -------------------------------------------------------------- >64103_64103_2_PDXDC1-NOB1_PDXDC1_chr16_15069077_ENST00000450288_NOB1_chr16_69788629_ENST00000268802_length(transcript)=1733nt_BP=92nt GAAGGAGCTGCGGGGCGCGGGAGGAGGAAGTAGAGCCCGGGACCGCCAGGCCACCACCGGCCGCCTCAGCCATGGACGCGTCCCTGGAGA AGGACATCGGGAAGAACATTTACACCATCCGGGAGGTGGTCACTGAGATTCGGGACAAGGCCACACGCAGGCGGCTCGCTGTCCTGCCCT ACGAGCTGCGGTTCAAGGAGCCCTTACCGGAATACGTGCGGCTGGTGACTGAGTTTTCAAAGAAAACAGGAGACTACCCCAGCCTCTCTG CCACGGACATCCAAGTGCTTGCACTCACATACCAGTTGGAAGCAGAGTTTGTTGGGGTGTCTCACCTAAAACAAGAACCACAGAAGGTTA AGGTGAGCTCATCGATTCAGCACCCAGAAACACCTCTGCACATTTCTGGTTTCCATCTGCCCTACAAGCCTAAACCCCCACAAGAAACAG AAAAAGGACACTCAGCTTGTGAGCCTGAGAACCTGGAATTTAGTTCCTTCATGTTCTGGAGAAACCCTTTGCCCAACATCGATCATGAAC TGCAGGAGCTGCTGATTGACAGAGGTGAGGACGTTCCAAGTGAGGAGGAGGAGGAGGAAGAAAACGGGTTTGAAGACAGAAAAGATGACA GCGATGACGACGGGGGTGGCTGGATAACCCCCAGTAACATCAAGCAGATCCAGCAGGAGCTGGAGCAGTGTGACGTCCCCGAGGACGTGC GGGTTGGCTGCCTGACCACAGACTTCGCCATGCAGAATGTTCTGCTGCAGATGGGGCTGCACGTGCTGGCGGTGAACGGCATGCTGATTC GTGAGGCCCGGAGCTACATCTTGCGCTGCCATGGCTGTTTCAAGACAACGTCTGACATGAGCCGAGTGTTCTGCTCACACTGTGGGAACA AGACCCTGAAGAAAGTGTCCGTGACCGTCAGCGACGACGGCACCCTGCACATGCACTTCTCCCGCAACCCCAAGGTGCTGAACCCCCGCG GCCTCCGGTACTCGCTTCCCACTCCCAAAGGGGGCAAATACGCCATCAACCCCCATCTCACCGAGGATCAGCGCTTCCCTCAGCTGCGAC TCTCCCAAAAGGCCAGGCAGAAAACCAACGTGTTCGCCCCTGACTACATCGCCGGGGTGTCACCCTTTGTCGAGAATGACATCTCCAGCC GCTCAGCTACCCTGCAGGTCCGGGACAGCACCTTGGGAGCTGGGCGGAGACGCTTAAATCCCAACGCTTCCAGAAAGAAGTTTGTGAAGA AAAGGTGAAGAGCGAGTTCCCGCAGGCAAATTGGATGGGCGTCTGGCCGCCGTGGAGTTCCGGTGACCCATTTCCCCAGCCGTGTCGTCT CCAGGACCACCCGATGGAAATAACAGGCGGGCTTCACGGTGCGGCTCTGTCCGCCCATGCCCCGCTGGGTCTGCAGGGAACTGGACTGTC CCATGGCCTGTGAGCACCGGAGCGCCTGGCTGCCTGCCAAGGAAGTGCAATTGCATAAAAACAGAAAGAACAACGCCCTGGAGCCAATCT TCAAGAAAGGAATTTCCAAAGGATAATATTTTTCTAATAAATGCGGCTGCAACCTCCTGTGCATTTAATTAAATAGGCCAAATTTTTGCT GCTTAGGTCATCTCAAGGCTGATACTTGAGCTGTGTGCCCAGAGATCATGCATTTAGATTTATATTTTTGCCAGAAAATACAAGGTTATA >64103_64103_2_PDXDC1-NOB1_PDXDC1_chr16_15069077_ENST00000450288_NOB1_chr16_69788629_ENST00000268802_length(amino acids)=398AA_BP=7 MDASLEKDIGKNIYTIREVVTEIRDKATRRRLAVLPYELRFKEPLPEYVRLVTEFSKKTGDYPSLSATDIQVLALTYQLEAEFVGVSHLK QEPQKVKVSSSIQHPETPLHISGFHLPYKPKPPQETEKGHSACEPENLEFSSFMFWRNPLPNIDHELQELLIDRGEDVPSEEEEEEENGF EDRKDDSDDDGGGWITPSNIKQIQQELEQCDVPEDVRVGCLTTDFAMQNVLLQMGLHVLAVNGMLIREARSYILRCHGCFKTTSDMSRVF CSHCGNKTLKKVSVTVSDDGTLHMHFSRNPKVLNPRGLRYSLPTPKGGKYAINPHLTEDQRFPQLRLSQKARQKTNVFAPDYIAGVSPFV -------------------------------------------------------------- >64103_64103_3_PDXDC1-NOB1_PDXDC1_chr16_15069077_ENST00000455313_NOB1_chr16_69788629_ENST00000268802_length(transcript)=1782nt_BP=141nt CTCCGCCCCGCCGGCCGCGGGCGCGGGGGACGTCAGCGCTGCCAGCGTGGAAGGAGCTGCGGGGCGCGGGAGGAGGAAGTAGAGCCCGGG ACCGCCAGGCCACCACCGGCCGCCTCAGCCATGGACGCGTCCCTGGAGAAGGACATCGGGAAGAACATTTACACCATCCGGGAGGTGGTC ACTGAGATTCGGGACAAGGCCACACGCAGGCGGCTCGCTGTCCTGCCCTACGAGCTGCGGTTCAAGGAGCCCTTACCGGAATACGTGCGG CTGGTGACTGAGTTTTCAAAGAAAACAGGAGACTACCCCAGCCTCTCTGCCACGGACATCCAAGTGCTTGCACTCACATACCAGTTGGAA GCAGAGTTTGTTGGGGTGTCTCACCTAAAACAAGAACCACAGAAGGTTAAGGTGAGCTCATCGATTCAGCACCCAGAAACACCTCTGCAC ATTTCTGGTTTCCATCTGCCCTACAAGCCTAAACCCCCACAAGAAACAGAAAAAGGACACTCAGCTTGTGAGCCTGAGAACCTGGAATTT AGTTCCTTCATGTTCTGGAGAAACCCTTTGCCCAACATCGATCATGAACTGCAGGAGCTGCTGATTGACAGAGGTGAGGACGTTCCAAGT GAGGAGGAGGAGGAGGAAGAAAACGGGTTTGAAGACAGAAAAGATGACAGCGATGACGACGGGGGTGGCTGGATAACCCCCAGTAACATC AAGCAGATCCAGCAGGAGCTGGAGCAGTGTGACGTCCCCGAGGACGTGCGGGTTGGCTGCCTGACCACAGACTTCGCCATGCAGAATGTT CTGCTGCAGATGGGGCTGCACGTGCTGGCGGTGAACGGCATGCTGATTCGTGAGGCCCGGAGCTACATCTTGCGCTGCCATGGCTGTTTC AAGACAACGTCTGACATGAGCCGAGTGTTCTGCTCACACTGTGGGAACAAGACCCTGAAGAAAGTGTCCGTGACCGTCAGCGACGACGGC ACCCTGCACATGCACTTCTCCCGCAACCCCAAGGTGCTGAACCCCCGCGGCCTCCGGTACTCGCTTCCCACTCCCAAAGGGGGCAAATAC GCCATCAACCCCCATCTCACCGAGGATCAGCGCTTCCCTCAGCTGCGACTCTCCCAAAAGGCCAGGCAGAAAACCAACGTGTTCGCCCCT GACTACATCGCCGGGGTGTCACCCTTTGTCGAGAATGACATCTCCAGCCGCTCAGCTACCCTGCAGGTCCGGGACAGCACCTTGGGAGCT GGGCGGAGACGCTTAAATCCCAACGCTTCCAGAAAGAAGTTTGTGAAGAAAAGGTGAAGAGCGAGTTCCCGCAGGCAAATTGGATGGGCG TCTGGCCGCCGTGGAGTTCCGGTGACCCATTTCCCCAGCCGTGTCGTCTCCAGGACCACCCGATGGAAATAACAGGCGGGCTTCACGGTG CGGCTCTGTCCGCCCATGCCCCGCTGGGTCTGCAGGGAACTGGACTGTCCCATGGCCTGTGAGCACCGGAGCGCCTGGCTGCCTGCCAAG GAAGTGCAATTGCATAAAAACAGAAAGAACAACGCCCTGGAGCCAATCTTCAAGAAAGGAATTTCCAAAGGATAATATTTTTCTAATAAA TGCGGCTGCAACCTCCTGTGCATTTAATTAAATAGGCCAAATTTTTGCTGCTTAGGTCATCTCAAGGCTGATACTTGAGCTGTGTGCCCA >64103_64103_3_PDXDC1-NOB1_PDXDC1_chr16_15069077_ENST00000455313_NOB1_chr16_69788629_ENST00000268802_length(amino acids)=398AA_BP=7 MDASLEKDIGKNIYTIREVVTEIRDKATRRRLAVLPYELRFKEPLPEYVRLVTEFSKKTGDYPSLSATDIQVLALTYQLEAEFVGVSHLK QEPQKVKVSSSIQHPETPLHISGFHLPYKPKPPQETEKGHSACEPENLEFSSFMFWRNPLPNIDHELQELLIDRGEDVPSEEEEEEENGF EDRKDDSDDDGGGWITPSNIKQIQQELEQCDVPEDVRVGCLTTDFAMQNVLLQMGLHVLAVNGMLIREARSYILRCHGCFKTTSDMSRVF CSHCGNKTLKKVSVTVSDDGTLHMHFSRNPKVLNPRGLRYSLPTPKGGKYAINPHLTEDQRFPQLRLSQKARQKTNVFAPDYIAGVSPFV -------------------------------------------------------------- >64103_64103_4_PDXDC1-NOB1_PDXDC1_chr16_15069077_ENST00000535621_NOB1_chr16_69788629_ENST00000268802_length(transcript)=1850nt_BP=209nt CCCCGCCCCGCCGCCTCTCAACCATCAGGTTCGGCAGCCCGCGGCGCCGCCTGGCAGCTCCTCCTCTTCTCCGCCCCGCCGGCCGCGGGC GCGGGGGACGTCAGCGCTGCCAGCGTGGAAGGAGCTGCGGGGCGCGGGAGGAGGAAGTAGAGCCCGGGACCGCCAGGCCACCACCGGCCG CCTCAGCCATGGACGCGTCCCTGGAGAAGGACATCGGGAAGAACATTTACACCATCCGGGAGGTGGTCACTGAGATTCGGGACAAGGCCA CACGCAGGCGGCTCGCTGTCCTGCCCTACGAGCTGCGGTTCAAGGAGCCCTTACCGGAATACGTGCGGCTGGTGACTGAGTTTTCAAAGA AAACAGGAGACTACCCCAGCCTCTCTGCCACGGACATCCAAGTGCTTGCACTCACATACCAGTTGGAAGCAGAGTTTGTTGGGGTGTCTC ACCTAAAACAAGAACCACAGAAGGTTAAGGTGAGCTCATCGATTCAGCACCCAGAAACACCTCTGCACATTTCTGGTTTCCATCTGCCCT ACAAGCCTAAACCCCCACAAGAAACAGAAAAAGGACACTCAGCTTGTGAGCCTGAGAACCTGGAATTTAGTTCCTTCATGTTCTGGAGAA ACCCTTTGCCCAACATCGATCATGAACTGCAGGAGCTGCTGATTGACAGAGGTGAGGACGTTCCAAGTGAGGAGGAGGAGGAGGAAGAAA ACGGGTTTGAAGACAGAAAAGATGACAGCGATGACGACGGGGGTGGCTGGATAACCCCCAGTAACATCAAGCAGATCCAGCAGGAGCTGG AGCAGTGTGACGTCCCCGAGGACGTGCGGGTTGGCTGCCTGACCACAGACTTCGCCATGCAGAATGTTCTGCTGCAGATGGGGCTGCACG TGCTGGCGGTGAACGGCATGCTGATTCGTGAGGCCCGGAGCTACATCTTGCGCTGCCATGGCTGTTTCAAGACAACGTCTGACATGAGCC GAGTGTTCTGCTCACACTGTGGGAACAAGACCCTGAAGAAAGTGTCCGTGACCGTCAGCGACGACGGCACCCTGCACATGCACTTCTCCC GCAACCCCAAGGTGCTGAACCCCCGCGGCCTCCGGTACTCGCTTCCCACTCCCAAAGGGGGCAAATACGCCATCAACCCCCATCTCACCG AGGATCAGCGCTTCCCTCAGCTGCGACTCTCCCAAAAGGCCAGGCAGAAAACCAACGTGTTCGCCCCTGACTACATCGCCGGGGTGTCAC CCTTTGTCGAGAATGACATCTCCAGCCGCTCAGCTACCCTGCAGGTCCGGGACAGCACCTTGGGAGCTGGGCGGAGACGCTTAAATCCCA ACGCTTCCAGAAAGAAGTTTGTGAAGAAAAGGTGAAGAGCGAGTTCCCGCAGGCAAATTGGATGGGCGTCTGGCCGCCGTGGAGTTCCGG TGACCCATTTCCCCAGCCGTGTCGTCTCCAGGACCACCCGATGGAAATAACAGGCGGGCTTCACGGTGCGGCTCTGTCCGCCCATGCCCC GCTGGGTCTGCAGGGAACTGGACTGTCCCATGGCCTGTGAGCACCGGAGCGCCTGGCTGCCTGCCAAGGAAGTGCAATTGCATAAAAACA GAAAGAACAACGCCCTGGAGCCAATCTTCAAGAAAGGAATTTCCAAAGGATAATATTTTTCTAATAAATGCGGCTGCAACCTCCTGTGCA TTTAATTAAATAGGCCAAATTTTTGCTGCTTAGGTCATCTCAAGGCTGATACTTGAGCTGTGTGCCCAGAGATCATGCATTTAGATTTAT >64103_64103_4_PDXDC1-NOB1_PDXDC1_chr16_15069077_ENST00000535621_NOB1_chr16_69788629_ENST00000268802_length(amino acids)=444AA_BP=1 MAAPPLLRPAGRGRGGRQRCQRGRSCGAREEEVEPGTARPPPAASAMDASLEKDIGKNIYTIREVVTEIRDKATRRRLAVLPYELRFKEP LPEYVRLVTEFSKKTGDYPSLSATDIQVLALTYQLEAEFVGVSHLKQEPQKVKVSSSIQHPETPLHISGFHLPYKPKPPQETEKGHSACE PENLEFSSFMFWRNPLPNIDHELQELLIDRGEDVPSEEEEEEENGFEDRKDDSDDDGGGWITPSNIKQIQQELEQCDVPEDVRVGCLTTD FAMQNVLLQMGLHVLAVNGMLIREARSYILRCHGCFKTTSDMSRVFCSHCGNKTLKKVSVTVSDDGTLHMHFSRNPKVLNPRGLRYSLPT -------------------------------------------------------------- >64103_64103_5_PDXDC1-NOB1_PDXDC1_chr16_15069077_ENST00000569715_NOB1_chr16_69788629_ENST00000268802_length(transcript)=1739nt_BP=98nt AGCGTGGAAGGAGCTGCGGGGCGCGGGAGGAGGAAGTAGAGCCCGGGACCGCCAGGCCACCACCGGCCGCCTCAGCCATGGACGCGTCCC TGGAGAAGGACATCGGGAAGAACATTTACACCATCCGGGAGGTGGTCACTGAGATTCGGGACAAGGCCACACGCAGGCGGCTCGCTGTCC TGCCCTACGAGCTGCGGTTCAAGGAGCCCTTACCGGAATACGTGCGGCTGGTGACTGAGTTTTCAAAGAAAACAGGAGACTACCCCAGCC TCTCTGCCACGGACATCCAAGTGCTTGCACTCACATACCAGTTGGAAGCAGAGTTTGTTGGGGTGTCTCACCTAAAACAAGAACCACAGA AGGTTAAGGTGAGCTCATCGATTCAGCACCCAGAAACACCTCTGCACATTTCTGGTTTCCATCTGCCCTACAAGCCTAAACCCCCACAAG AAACAGAAAAAGGACACTCAGCTTGTGAGCCTGAGAACCTGGAATTTAGTTCCTTCATGTTCTGGAGAAACCCTTTGCCCAACATCGATC ATGAACTGCAGGAGCTGCTGATTGACAGAGGTGAGGACGTTCCAAGTGAGGAGGAGGAGGAGGAAGAAAACGGGTTTGAAGACAGAAAAG ATGACAGCGATGACGACGGGGGTGGCTGGATAACCCCCAGTAACATCAAGCAGATCCAGCAGGAGCTGGAGCAGTGTGACGTCCCCGAGG ACGTGCGGGTTGGCTGCCTGACCACAGACTTCGCCATGCAGAATGTTCTGCTGCAGATGGGGCTGCACGTGCTGGCGGTGAACGGCATGC TGATTCGTGAGGCCCGGAGCTACATCTTGCGCTGCCATGGCTGTTTCAAGACAACGTCTGACATGAGCCGAGTGTTCTGCTCACACTGTG GGAACAAGACCCTGAAGAAAGTGTCCGTGACCGTCAGCGACGACGGCACCCTGCACATGCACTTCTCCCGCAACCCCAAGGTGCTGAACC CCCGCGGCCTCCGGTACTCGCTTCCCACTCCCAAAGGGGGCAAATACGCCATCAACCCCCATCTCACCGAGGATCAGCGCTTCCCTCAGC TGCGACTCTCCCAAAAGGCCAGGCAGAAAACCAACGTGTTCGCCCCTGACTACATCGCCGGGGTGTCACCCTTTGTCGAGAATGACATCT CCAGCCGCTCAGCTACCCTGCAGGTCCGGGACAGCACCTTGGGAGCTGGGCGGAGACGCTTAAATCCCAACGCTTCCAGAAAGAAGTTTG TGAAGAAAAGGTGAAGAGCGAGTTCCCGCAGGCAAATTGGATGGGCGTCTGGCCGCCGTGGAGTTCCGGTGACCCATTTCCCCAGCCGTG TCGTCTCCAGGACCACCCGATGGAAATAACAGGCGGGCTTCACGGTGCGGCTCTGTCCGCCCATGCCCCGCTGGGTCTGCAGGGAACTGG ACTGTCCCATGGCCTGTGAGCACCGGAGCGCCTGGCTGCCTGCCAAGGAAGTGCAATTGCATAAAAACAGAAAGAACAACGCCCTGGAGC CAATCTTCAAGAAAGGAATTTCCAAAGGATAATATTTTTCTAATAAATGCGGCTGCAACCTCCTGTGCATTTAATTAAATAGGCCAAATT TTTGCTGCTTAGGTCATCTCAAGGCTGATACTTGAGCTGTGTGCCCAGAGATCATGCATTTAGATTTATATTTTTGCCAGAAAATACAAG >64103_64103_5_PDXDC1-NOB1_PDXDC1_chr16_15069077_ENST00000569715_NOB1_chr16_69788629_ENST00000268802_length(amino acids)=398AA_BP=7 MDASLEKDIGKNIYTIREVVTEIRDKATRRRLAVLPYELRFKEPLPEYVRLVTEFSKKTGDYPSLSATDIQVLALTYQLEAEFVGVSHLK QEPQKVKVSSSIQHPETPLHISGFHLPYKPKPPQETEKGHSACEPENLEFSSFMFWRNPLPNIDHELQELLIDRGEDVPSEEEEEEENGF EDRKDDSDDDGGGWITPSNIKQIQQELEQCDVPEDVRVGCLTTDFAMQNVLLQMGLHVLAVNGMLIREARSYILRCHGCFKTTSDMSRVF CSHCGNKTLKKVSVTVSDDGTLHMHFSRNPKVLNPRGLRYSLPTPKGGKYAINPHLTEDQRFPQLRLSQKARQKTNVFAPDYIAGVSPFV -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for PDXDC1-NOB1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for PDXDC1-NOB1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for PDXDC1-NOB1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
