
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:PEMT-IGF2BP1 (FusionGDB2 ID:64271) |
Fusion Gene Summary for PEMT-IGF2BP1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: PEMT-IGF2BP1 | Fusion gene ID: 64271 | Hgene | Tgene | Gene symbol | PEMT | IGF2BP1 | Gene ID | 10400 | 10642 |
| Gene name | phosphatidylethanolamine N-methyltransferase | insulin like growth factor 2 mRNA binding protein 1 | |
| Synonyms | PEAMT|PEMPT|PEMT2|PLMT|PNMT | CRD-BP|CRDBP|IMP-1|IMP1|VICKZ1|ZBP1 | |
| Cytomap | 17p11.2 | 17q21.32 | |
| Type of gene | protein-coding | protein-coding | |
| Description | phosphatidylethanolamine N-methyltransferasephospholipid methyltransferase | insulin-like growth factor 2 mRNA-binding protein 1IGF-II mRNA-binding protein 1IGF2 mRNA-binding protein 1VICKZ family member 1ZBP-1coding region determinant-binding proteininsulin-like growth factor 2 mRNA binding protein 1 deltaN CRDBPzipcode-bi | |
| Modification date | 20200320 | 20200315 | |
| UniProtAcc | . | Q9NZI8 | |
| Ensembl transtripts involved in fusion gene | ENST00000255389, ENST00000395781, ENST00000395782, ENST00000395783, ENST00000435340, ENST00000484838, | ENST00000515586, ENST00000290341, ENST00000431824, | |
| Fusion gene scores | * DoF score | 11 X 7 X 6=462 | 9 X 10 X 3=270 |
| # samples | 11 | 10 | |
| ** MAII score | log2(11/462*10)=-2.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(10/270*10)=-1.43295940727611 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: PEMT [Title/Abstract] AND IGF2BP1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | PEMT(17480234)-IGF2BP1(47118740), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | PEMT-IGF2BP1 seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | IGF2BP1 | GO:0017148 | negative regulation of translation | 9891060 |
| Tgene | IGF2BP1 | GO:0070934 | CRD-mediated mRNA stabilization | 19029303 |
 Fusion gene breakpoints across PEMT (5'-gene) Fusion gene breakpoints across PEMT (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
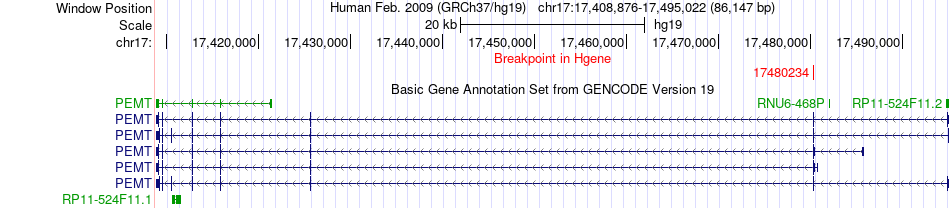 |
 Fusion gene breakpoints across IGF2BP1 (3'-gene) Fusion gene breakpoints across IGF2BP1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
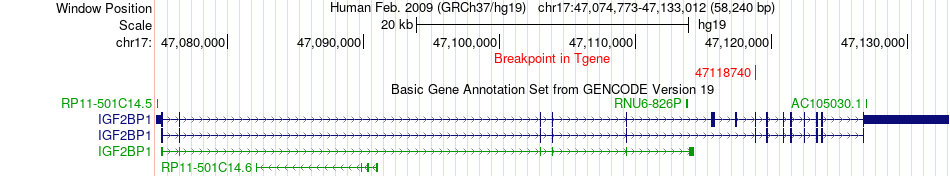 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-A8-A09Q-01A | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + |
Top |
Fusion Gene ORF analysis for PEMT-IGF2BP1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000255389 | ENST00000515586 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + |
| 5CDS-intron | ENST00000395781 | ENST00000515586 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + |
| 5CDS-intron | ENST00000395782 | ENST00000515586 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + |
| 5CDS-intron | ENST00000395783 | ENST00000515586 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + |
| 5CDS-intron | ENST00000435340 | ENST00000515586 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + |
| Frame-shift | ENST00000255389 | ENST00000290341 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + |
| Frame-shift | ENST00000255389 | ENST00000431824 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + |
| Frame-shift | ENST00000395781 | ENST00000290341 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + |
| Frame-shift | ENST00000395781 | ENST00000431824 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + |
| Frame-shift | ENST00000395782 | ENST00000290341 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + |
| Frame-shift | ENST00000395782 | ENST00000431824 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + |
| Frame-shift | ENST00000435340 | ENST00000290341 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + |
| Frame-shift | ENST00000435340 | ENST00000431824 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + |
| In-frame | ENST00000395783 | ENST00000290341 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + |
| In-frame | ENST00000395783 | ENST00000431824 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + |
| intron-3CDS | ENST00000484838 | ENST00000290341 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + |
| intron-3CDS | ENST00000484838 | ENST00000431824 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + |
| intron-intron | ENST00000484838 | ENST00000515586 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000395783 | PEMT | chr17 | 17480234 | - | ENST00000290341 | IGF2BP1 | chr17 | 47118740 | + | 7395 | 273 | 223 | 1188 | 321 |
| ENST00000395783 | PEMT | chr17 | 17480234 | - | ENST00000431824 | IGF2BP1 | chr17 | 47118740 | + | 1189 | 273 | 223 | 1188 | 322 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000395783 | ENST00000290341 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + | 0.000912622 | 0.99908733 |
| ENST00000395783 | ENST00000431824 | PEMT | chr17 | 17480234 | - | IGF2BP1 | chr17 | 47118740 | + | 0.005048888 | 0.9949511 |
Top |
Fusion Genomic Features for PEMT-IGF2BP1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| PEMT | chr17 | 17480233 | - | IGF2BP1 | chr17 | 47118739 | + | 3.19E-14 | 1 |
| PEMT | chr17 | 17480233 | - | IGF2BP1 | chr17 | 47118739 | + | 3.19E-14 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
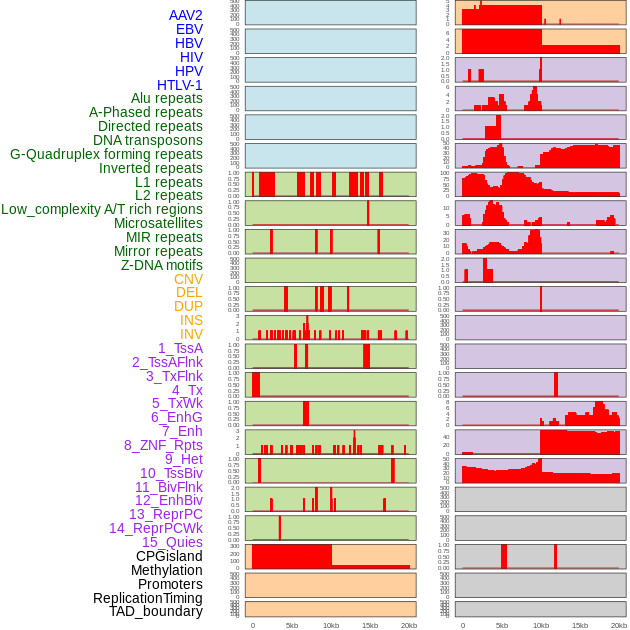 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
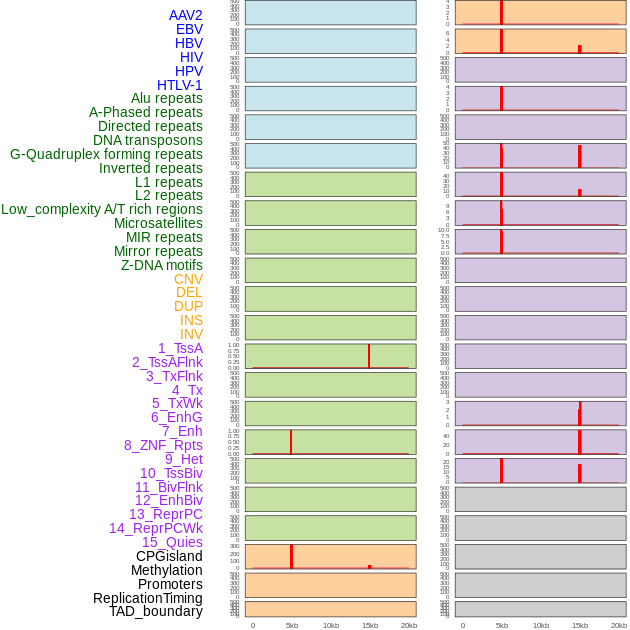 |
Top |
Fusion Protein Features for PEMT-IGF2BP1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:17480234/chr17:47118740) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | IGF2BP1 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: RNA-binding factor that recruits target transcripts to cytoplasmic protein-RNA complexes (mRNPs). This transcript 'caging' into mRNPs allows mRNA transport and transient storage. It also modulates the rate and location at which target transcripts encounter the translational apparatus and shields them from endonuclease attacks or microRNA-mediated degradation. Plays a direct role in the transport and translation of transcripts required for axonal regeneration in adult sensory neurons (By similarity). Regulates localized beta-actin/ACTB mRNA translation, a crucial process for cell polarity, cell migration and neurite outgrowth. Co-transcriptionally associates with the ACTB mRNA in the nucleus. This binding involves a conserved 54-nucleotide element in the ACTB mRNA 3'-UTR, known as the 'zipcode'. The RNP thus formed is exported to the cytoplasm, binds to a motor protein and is transported along the cytoskeleton to the cell periphery. During transport, prevents ACTB mRNA from being translated into protein. When the RNP complex reaches its destination near the plasma membrane, IGF2BP1 is phosphorylated. This releases the mRNA, allowing ribosomal 40S and 60S subunits to assemble and initiate ACTB protein synthesis. Monomeric ACTB then assembles into the subcortical actin cytoskeleton (By similarity). During neuronal development, key regulator of neurite outgrowth, growth cone guidance and neuronal cell migration, presumably through the spatiotemporal fine tuning of protein synthesis, such as that of ACTB (By similarity). May regulate mRNA transport to activated synapses (By similarity). Binds to and stabilizes ABCB1/MDR-1 mRNA (By similarity). During interstinal wound repair, interacts with and stabilizes PTGS2 transcript. PTGS2 mRNA stabilization may be crucial for colonic mucosal wound healing (By similarity). Binds to the 3'-UTR of IGF2 mRNA by a mechanism of cooperative and sequential dimerization and regulates IGF2 mRNA subcellular localization and translation. Binds to MYC mRNA, in the coding region instability determinant (CRD) of the open reading frame (ORF), hence prevents MYC cleavage by endonucleases and possibly microRNA targeting to MYC-CRD. Binds to the 3'-UTR of CD44 mRNA and stabilizes it, hence promotes cell adhesion and invadopodia formation in cancer cells. Binds to the oncofetal H19 transcript and to the neuron-specific TAU mRNA and regulates their localizations. Binds to and stabilizes BTRC/FBW1A mRNA. Binds to the adenine-rich autoregulatory sequence (ARS) located in PABPC1 mRNA and represses its translation. PABPC1 mRNA-binding is stimulated by PABPC1 protein. Prevents BTRC/FBW1A mRNA degradation by disrupting microRNA-dependent interaction with AGO2. Promotes the directed movement of tumor-derived cells by fine-tuning intracellular signaling networks. Binds to MAPK4 3'-UTR and inhibits its translation. Interacts with PTEN transcript open reading frame (ORF) and prevents mRNA decay. This combined action on MAPK4 (down-regulation) and PTEN (up-regulation) antagonizes HSPB1 phosphorylation, consequently it prevents G-actin sequestration by phosphorylated HSPB1, allowing F-actin polymerization. Hence enhances the velocity of cell migration and stimulates directed cell migration by PTEN-modulated polarization. Interacts with Hepatitis C virus (HCV) 5'-UTR and 3'-UTR and specifically enhances translation at the HCV IRES, but not 5'-cap-dependent translation, possibly by recruiting eIF3. Interacts with HIV-1 GAG protein and blocks the formation of infectious HIV-1 particles. Reduces HIV-1 assembly by inhibiting viral RNA packaging, as well as assembly and processing of GAG protein on cellular membranes. During cellular stress, such as oxidative stress or heat shock, stabilizes target mRNAs that are recruited to stress granules, including CD44, IGF2, MAPK4, MYC, PTEN, RAPGEF2 and RPS6KA5 transcripts. {ECO:0000250, ECO:0000269|PubMed:10875929, ECO:0000269|PubMed:16356927, ECO:0000269|PubMed:16541107, ECO:0000269|PubMed:16778892, ECO:0000269|PubMed:17101699, ECO:0000269|PubMed:17255263, ECO:0000269|PubMed:17893325, ECO:0000269|PubMed:18385235, ECO:0000269|PubMed:19029303, ECO:0000269|PubMed:19541769, ECO:0000269|PubMed:19647520, ECO:0000269|PubMed:20080952, ECO:0000269|PubMed:22279049, ECO:0000269|PubMed:8132663, ECO:0000269|PubMed:9891060}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000255389 | - | 2 | 7 | 13_33 | 68 | 237.0 | Intramembrane | Helical |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395781 | - | 2 | 8 | 13_33 | 68 | 233.0 | Intramembrane | Helical |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395782 | - | 2 | 7 | 13_33 | 31 | 200.0 | Intramembrane | Helical |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395783 | - | 2 | 7 | 13_33 | 31 | 200.0 | Intramembrane | Helical |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000255389 | - | 2 | 7 | 1_12 | 68 | 237.0 | Topological domain | Lumenal |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000255389 | - | 2 | 7 | 34_45 | 68 | 237.0 | Topological domain | Lumenal |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395781 | - | 2 | 8 | 1_12 | 68 | 233.0 | Topological domain | Lumenal |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395781 | - | 2 | 8 | 34_45 | 68 | 233.0 | Topological domain | Lumenal |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395782 | - | 2 | 7 | 1_12 | 31 | 200.0 | Topological domain | Lumenal |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395783 | - | 2 | 7 | 1_12 | 31 | 200.0 | Topological domain | Lumenal |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000255389 | - | 2 | 7 | 46_66 | 68 | 237.0 | Transmembrane | Helical |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395781 | - | 2 | 8 | 46_66 | 68 | 233.0 | Transmembrane | Helical |
| Tgene | IGF2BP1 | chr17:17480234 | chr17:47118740 | ENST00000290341 | 6 | 15 | 276_343 | 272 | 578.0 | Domain | KH 2 | |
| Tgene | IGF2BP1 | chr17:17480234 | chr17:47118740 | ENST00000290341 | 6 | 15 | 405_470 | 272 | 578.0 | Domain | KH 3 | |
| Tgene | IGF2BP1 | chr17:17480234 | chr17:47118740 | ENST00000290341 | 6 | 15 | 487_553 | 272 | 578.0 | Domain | KH 4 | |
| Tgene | IGF2BP1 | chr17:17480234 | chr17:47118740 | ENST00000431824 | 4 | 13 | 195_260 | 133 | 439.0 | Domain | KH 1 | |
| Tgene | IGF2BP1 | chr17:17480234 | chr17:47118740 | ENST00000431824 | 4 | 13 | 276_343 | 133 | 439.0 | Domain | KH 2 | |
| Tgene | IGF2BP1 | chr17:17480234 | chr17:47118740 | ENST00000431824 | 4 | 13 | 405_470 | 133 | 439.0 | Domain | KH 3 | |
| Tgene | IGF2BP1 | chr17:17480234 | chr17:47118740 | ENST00000431824 | 4 | 13 | 487_553 | 133 | 439.0 | Domain | KH 4 | |
| Tgene | IGF2BP1 | chr17:17480234 | chr17:47118740 | ENST00000290341 | 6 | 15 | 310_324 | 272 | 578.0 | Region | Note=Sufficient for nuclear export | |
| Tgene | IGF2BP1 | chr17:17480234 | chr17:47118740 | ENST00000290341 | 6 | 15 | 485_495 | 272 | 578.0 | Region | Note=Sufficient for nuclear export | |
| Tgene | IGF2BP1 | chr17:17480234 | chr17:47118740 | ENST00000431824 | 4 | 13 | 310_324 | 133 | 439.0 | Region | Note=Sufficient for nuclear export | |
| Tgene | IGF2BP1 | chr17:17480234 | chr17:47118740 | ENST00000431824 | 4 | 13 | 485_495 | 133 | 439.0 | Region | Note=Sufficient for nuclear export |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000255389 | - | 2 | 7 | 180_181 | 68 | 237.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000255389 | - | 2 | 7 | 98_100 | 68 | 237.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395781 | - | 2 | 8 | 180_181 | 68 | 233.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395781 | - | 2 | 8 | 98_100 | 68 | 233.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395782 | - | 2 | 7 | 180_181 | 31 | 200.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395782 | - | 2 | 7 | 98_100 | 31 | 200.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395783 | - | 2 | 7 | 180_181 | 31 | 200.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395783 | - | 2 | 7 | 98_100 | 31 | 200.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000255389 | - | 2 | 7 | 115_157 | 68 | 237.0 | Topological domain | Lumenal |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000255389 | - | 2 | 7 | 179_199 | 68 | 237.0 | Topological domain | Cytoplasmic |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000255389 | - | 2 | 7 | 67_93 | 68 | 237.0 | Topological domain | Cytoplasmic |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395781 | - | 2 | 8 | 115_157 | 68 | 233.0 | Topological domain | Lumenal |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395781 | - | 2 | 8 | 179_199 | 68 | 233.0 | Topological domain | Cytoplasmic |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395781 | - | 2 | 8 | 67_93 | 68 | 233.0 | Topological domain | Cytoplasmic |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395782 | - | 2 | 7 | 115_157 | 31 | 200.0 | Topological domain | Lumenal |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395782 | - | 2 | 7 | 179_199 | 31 | 200.0 | Topological domain | Cytoplasmic |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395782 | - | 2 | 7 | 34_45 | 31 | 200.0 | Topological domain | Lumenal |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395782 | - | 2 | 7 | 67_93 | 31 | 200.0 | Topological domain | Cytoplasmic |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395783 | - | 2 | 7 | 115_157 | 31 | 200.0 | Topological domain | Lumenal |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395783 | - | 2 | 7 | 179_199 | 31 | 200.0 | Topological domain | Cytoplasmic |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395783 | - | 2 | 7 | 34_45 | 31 | 200.0 | Topological domain | Lumenal |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395783 | - | 2 | 7 | 67_93 | 31 | 200.0 | Topological domain | Cytoplasmic |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000255389 | - | 2 | 7 | 158_178 | 68 | 237.0 | Transmembrane | Helical |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000255389 | - | 2 | 7 | 94_114 | 68 | 237.0 | Transmembrane | Helical |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395781 | - | 2 | 8 | 158_178 | 68 | 233.0 | Transmembrane | Helical |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395781 | - | 2 | 8 | 94_114 | 68 | 233.0 | Transmembrane | Helical |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395782 | - | 2 | 7 | 158_178 | 31 | 200.0 | Transmembrane | Helical |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395782 | - | 2 | 7 | 46_66 | 31 | 200.0 | Transmembrane | Helical |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395782 | - | 2 | 7 | 94_114 | 31 | 200.0 | Transmembrane | Helical |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395783 | - | 2 | 7 | 158_178 | 31 | 200.0 | Transmembrane | Helical |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395783 | - | 2 | 7 | 46_66 | 31 | 200.0 | Transmembrane | Helical |
| Hgene | PEMT | chr17:17480234 | chr17:47118740 | ENST00000395783 | - | 2 | 7 | 94_114 | 31 | 200.0 | Transmembrane | Helical |
| Tgene | IGF2BP1 | chr17:17480234 | chr17:47118740 | ENST00000290341 | 6 | 15 | 195_260 | 272 | 578.0 | Domain | KH 1 | |
| Tgene | IGF2BP1 | chr17:17480234 | chr17:47118740 | ENST00000290341 | 6 | 15 | 2_75 | 272 | 578.0 | Domain | RRM 1 | |
| Tgene | IGF2BP1 | chr17:17480234 | chr17:47118740 | ENST00000290341 | 6 | 15 | 81_156 | 272 | 578.0 | Domain | RRM 2 | |
| Tgene | IGF2BP1 | chr17:17480234 | chr17:47118740 | ENST00000431824 | 4 | 13 | 2_75 | 133 | 439.0 | Domain | RRM 1 | |
| Tgene | IGF2BP1 | chr17:17480234 | chr17:47118740 | ENST00000431824 | 4 | 13 | 81_156 | 133 | 439.0 | Domain | RRM 2 |
Top |
Fusion Gene Sequence for PEMT-IGF2BP1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >64271_64271_1_PEMT-IGF2BP1_PEMT_chr17_17480234_ENST00000395783_IGF2BP1_chr17_47118740_ENST00000290341_length(transcript)=7395nt_BP=273nt CTGACCACAGAGCGCTGCTCCCGAGAACCCTGCACCCCTCAATGGAGTAAATTACCATAAAGCCTCTTCCTTACCCATGCTTTGGGGTGT TAACAGCTGAGGCTATTCGTCGGTGACCTGTGGGACTCGAGCTATTCCTGCAGCTCAGCAGACCTCCTGGCCGTGGCAGACTTCTGCGTT ATGACCCGGCTGCTGGGCTACGTGGACCCCCTGGATCCCAGCTTTGTGGCTGCCGTCATCACCATCACCTTCAATCCGCTCTACTGGAAT GTGGGCTGACGAGGTTCCCCTGAAGATCCTGGCCCATAATAACTTTGTAGGGCGTCTCATTGGCAAGGAAGGACGGAACCTGAAGAAGGT AGAGCAAGATACCGAGACAAAAATCACCATCTCCTCGTTGCAAGACCTTACCCTTTACAACCCTGAGAGGACCATCACTGTGAAGGGGGC CATCGAGAATTGTTGCAGGGCCGAGCAGGAAATAATGAAGAAAGTTCGGGAGGCCTATGAGAATGATGTGGCTGCCATGAGCCTGCAGTC TCACCTGATCCCTGGCCTGAACCTGGCTGCTGTAGGTCTTTTCCCAGCTTCATCCAGCGCAGTCCCGCCGCCTCCCAGCAGCGTTACTGG GGCTGCTCCCTATAGCTCCTTTATGCAGGCTCCCGAGCAGGAGATGGTGCAGGTGTTTATCCCCGCCCAGGCAGTGGGCGCCATCATCGG CAAGAAGGGGCAGCACATCAAACAGCTCTCCCGGTTTGCCAGCGCCTCCATCAAGATTGCACCACCCGAAACACCTGACTCCAAAGTTCG TATGGTTATCATCACTGGACCGCCAGAGGCCCAATTCAAGGCTCAGGGAAGAATCTATGGCAAACTCAAGGAGGAGAACTTCTTTGGTCC CAAGGAGGAAGTGAAGCTGGAGACCCACATACGTGTGCCAGCATCAGCAGCTGGCCGGGTCATTGGCAAAGGTGGAAAAACGGTGAACGA GTTGCAGAATTTGACGGCAGCTGAGGTGGTAGTACCAAGAGACCAGACCCCTGATGAGAACGACCAGGTCATCGTGAAAATCATCGGACA TTTCTATGCCAGTCAGATGGCTCAACGGAAGATCCGAGACATCCTGGCCCAGGTTAAGCAGCAGCATCAGAAGGGACAGAGTAACCAGGC CCAGGCACGGAGGAAGTGACCAGCCCCTCCCTGTCCCTTCGAGTCCAGGACAACAACGGGCAGAAATCGAGAGTGTGCTCTCCCCGGCAG GCCTGAGAATGAGTGGGAATCCGGGACACCTGGGCCGGGCTGTAGATCAGGTTTGCCCACTTGATTGAGAAAGATGTTCCAGTGAGGAAC CCTGATCTCTCAGCCCCAAACACCCACCCAATTGGCCCAACACTGTCTGCCCCTCGGGGTGTCAGAAATTCTAGCGCAAGGCACTTTTAA ACGTGGATTGTTTAAAGAAGCTCTCCAGGCCCCACCAAGAGGGTGGATCACACCTCAGTGGGAAGAAAAATAAAATTTCCTTCAGGTTTT AAAAACATGCAGAGAGGTGTTTTAATCAGCCTTAAAGGATGGTTCATTTCTTGACCTTAATGTTTTTCCAATCTTCTTCCCCCTACTTGG GTAATTGATTAAAATACCTCCATTTACGGCCTCTTTCTATATTTACACTAATTTTTTTATCTTTATTGCTACCAGAAAAAAATGCGAACG AATGCATTGCTTTGCTTACAGTATTGACTCAAGGGAAAAGAACTGTCAGTATCTGTAGATTAATTCCAATCACTCCCTAACCAATAGGTA CAATACGGAATGAAGAAGAGGGGAAAATGGGGAGAAAGATGGTTAAAATACATAATAATCCACGTTTAAAAGGAGCGCACTTGTGGCTGA TCTATGCCAGATCACCATCTTCAAATTGGCACAACTGAAATTTCCCCACTCTGTTGGGGCTTCCCCACCACATTCATGTCCCTCTCCCGT GTAGGTTTCACATTATGTCCAGGTGCACATAGGTGGTATTGAATGCTCAGCAGGGTAGGGGCTGACCACTGTCCCTGATTCCCATCGTTC TCAGGCGGATTTTATATTTTTTTAAAGTCTATTTTAATGATTGGATATGAGCACTGGGAAGGGGACGCTAACTCCCCTTGATAAAGTCTC GGTTCCATGGAGGACTTGAGTGGCCCCAAAGGCTGCCACGGTGCCCTCACCCCAGCCCATGTGCTCCCATAAGGGCTGGTTCCTAGAGGC AGGGGTTGTGGGGCACTCCCAGCCACGGCACTGTTACCTTGGTGGTGGGACTTGGAACCCAACCCTGAGCTCCCGATAAAGCTAAAGTCC ATCATCTGGCAAATTCAGTAAATTGGAGAGTACTTGCTTCTGTTTGTATCTGAGAGGAATTTTTAACTGACGGCTTCTGTCTCCATGAAT CATTATCAGCATGATGAAAGGTGTGTCTAAAAAACAATTCAGAATACCAGCAGCATTGTACAGCAAGGGGTAAATAAGCTTAATTTATTA ATTTACCAGGCTTAATTAAGATCCCATGGAGTGTTTAGCCCTTGTGGGAGACAGAAGCCATCAGTTAAATGAGGTTAGGCCTCTCCTCCT AATATACTGATTGACAATGCATATTAGCCAGGTAATGCACTTTAGCTACCCTGGACAATGCTATCAAGTGTGCTGGGAAGGGAGGAAGGC CTCTCTACATATGGAAAAGCCCATGCGTGGAGTTCCCCTCCTTTCAACATTGCAACAACAGTAACAACAAGACAACCGCAACATGTGGGC GTAGTCAGGCAATGCTGTGTGCGAAGTAAACTACCTCAAGGTATGAAGTTACCTCAGCAATTATTTTCCTTTTTGTTCCCCCCAACCCCA TTAAAAAAATTTTTTTTTGATTTTTGTTTTTTTGCAGCTTGCTGATATTTTATATAAAAAAGAAAAGCAAAGCAAAAGAGAAGCTGATAG TCTTGAATATTTTATTTTTTTAATGAAAAGAAAAAACAAGAAAGTTATGTTTCATAATTTCTTACAACATGAGCCAGTAACCCTTTAGGA ACTCTCTATGGAGAACAGGCCTGGTGGGAAAGGCTTTGGGGGCTGCCCCCTTAGGAGGAGGCTAGTGCTAAGAGGGAAGGCCCAGGTTTG AGAGAGCCCAGAGGGGCAGAGCCCAGAGCCTTGTTTGGCCCTGATCTCTGACTTCTAGAGCCCCAGCTGCTGGCGGCTGCTGGAATATCC TACCTGATAGGATTAAAAGGCCTAGTGGAGCTGGGGGCTCTCAGTGGTTAAACAATGCCCAACAACCAACCAGCTGGCCCTTGGTCTCCT CTCTTTCCTCCTTTGGTTAAAGAGCATCTCAGCCAGCTTTTCCCACCAGTGGTGCTGTTGAGATATTTTAAAATATTGCCTCCGTTTTAT CGAGGAGAGAAATAATAACTAAAAAATATACCCTTTAAAAAAACCTATATTTCTCTGTCTAAAAATATGGGAGCTGAGATTCCGTTCGTG GAAAAAAGACAAGGCCACCCTCTCGCCCTCAGAGAGGTCCACCTGGTTTGTCATTGCAATGCTTTTCATTTTTTTTTTTTGTTATTGTTT CATTTCAGTTCCGTCTTGCTATTCTTCCTAATCTATATCCATAGATCTAAGGGGCAAACAGATACTAGTTAACTGCCCCCACCTCTGTCT CCCTGTCTTCTTTAGATCGGTCTGATTGATTTTAAAAGTGGACCCAAACTTAGGGAATTCTTGATTTAGGGTGGCTGGTGGCAAGGAGGG GCAGGGGATATGGGGACGTGACTGGGACAGGTTCCTGCCTTATCATTTTCTCCCTAGGACATTCCCTTGTAGCCCCCAGAATTGTCTGGC CCAAATTGAATAGAAGCAGAAAAACATTTAGGGATAACATCAGGCCAGTAGAATTAAGCCTCTCCACCTGTCCCAACCATAAAAAGGGTC TCCCAGCTTTCCATCTCTGGCTCTATATGCTTTATCCCAAAACAAAGCAGATAACGTTCAGACGTCGGCCATTTAGTAATTTAAAGCGAA TTTCCAGCAGCAAGCATGCTTTGATATCTGGTTCAGACTATCATCAGGAAGAAAAAAAAATCCCACAGTACCTGAAATGTGATTGTTGCA GTGTTCAGTTTCCTTGGGGGCCTGCTCCCTTCACACCTTGAGCCCAAGTCCTTTTCCGTTGGCTGATTCAGCTCCCAGAAGAGACGAGGA AGTGTGTGGCAAGGGACTGGAAAACTTCACTTGCTTGGATTAGGCAAGGCTCCACTCATTGTTGATATTTGCCCAGCAGGAAAATCATGT AAGTTATACCACCAGAAAGCAAAAGGAGCATGGTTTGGTGGTTAAGGTTTAGTGGGATGAAGGACCTGTCTTGGTGGGCCGGGCCCTCTT GTGCCCCGTAGGCTAGGTCTTAGGGCAACTCCTTGCCCTCCTGCTCAGCACCTCCATTTCCCCATCCTTGGTGAGATAACAAGCTATCGC GAAAAGCACTTGGGAGATTTGGATGATTTGAGAAGAGTGACTTAAAAAAAATGCTTCTGTGCTCTAAGATATATATGTGTGTGTGTGTGC TACATATATATTTTTAAGAAAGGACCATCTCTTTAGGATATATTTTTAAATTCTTTGAAACACATAACCAAAATGGTTTGATTCACTGAC TGACTTTGAAGCTGCATCTGCCAGTTACACCCCAAATGGCTTTAATCCCCTCTCGGGTCTGGTTGCCTTTTGCAGTTTGGGTTGTGGACT CAGCTCCTGTGAGGGGTCTGGTTAGGAGAGAGCCATTTTTAAGGACAGGGAGTTTTATAGCCCTTTTCTACTTTCCTCCCCTCCTCCCAG TCCTTATCAATCTTTTTTCCTTTTTCCTGACCCCCTCCTTCTGGAGGCAGTTGGGAGCTATCCTTGTTTATGCCTCACTATTGGCAGAAA AGACCCCATTTAAAACCCAGAGAACACTGGAGGGGGATGCTCTAGTTGGTTCTGTGTCCATTTTCCTCTGTGCCAAAGACAGACAGACAG AGGCTGAGAGAGGCTGTTCCTGAATCAAAGCAATAGCCAGCTTTCGACACATACCTGGCTGTCTGAGGAGGAAGGCCTCCTGGAAACTGG GAGCTAAGGGCGAGGCCCTTCCCTTCAGAGGCTCCTGGGGGATTAGGGTGTGGTGTTTGCCAAGCCAAGGGGTAGGGAGCCGAGAAATTG GTCTGTCGGCTCCTGGTTGCACTTTGGGGAAGGAGAGGAAGTTTGGGGCTCCAGGTAGCTCCCTGTTGTGGGACTGCTCTGTCCCCTGCC CCTACTGCAGAGATAGCACTGCCGAGTTCCCTTCAGGCCTGGCAGACGGGCAGTGAGGAGGGGCCTCAGTTAGCTCTCAAGGGTGCCTTC CCCTCCTCCCAACCCAGACATACCCTCTGCCAAACTGGGAACCAGCAGTGCTAGTAACTACCTCACAGAGCCCCAGAGGGCCTGCTTGAG CCTTCTTGCTCCACAGGAGAAGCTGGTGCCTCTAGGCAACCCCTTCCTCCCACCTCTCATCAGGGGTGGGGGTTCTCCTTTCTTTCCCCT GAAGTGTTTATGGGGAGATCCTAGTGGCTTTGCCATTCAAACCACTCGACTGTTTGCCTGTTTCTTGAAAACCAGTAGAAGGGAAACAGC ACAGCCTGTCACAGTAATTGCAGGAAGATTGAAGAAAAATCCTCATCAATGCCAGGGGACATAAAAGCCATTTCCCTTCCAAATACTCGA CAATTTAGATGCAGAACATTTCTCTGTATTCAGACTTAGAGTAACACCAGCTGAAAACTGCAGTTTCTTTCCTTTGGATACATAAGGCTT CTCTATCGGGGTACGGGACAGGGAGGAGGCCTCATGTCTGAAGGGGGATTTAGGGGCGAGAGCCCCAGCCCTGACCCTCGGTCCTGTGCA CCGCTTTGGGGCACAGTCTGATGGCGCCTTTGCTGGCGCCTTAGTATGGTTGACTCCGGATGGACAAAAGAAAAAAAATTTTTTTTCTTG AATGAAATAGCAGGAAGCTCCTCGGGAGCATGTGTTTTGATTAACCGCAGGTGATGGATGCTACGAGTATAAATGGATTAACTACCTCAA TCCTTACAGTAAGATTGGAACTAAGGGCAGGGACTCATGCATAAGGGTATGAATCCCAGCCAGGACAAGTGAGTTGAGGCTTGTGCCACA AAAGGTTTGTCCTTGGGGAACAGGCAGGCCTGCCAGGATCCCCCCCATATCGATTGGGCTGGGAGGGCTGGCCATGAGGTCCCCACTTTC TGCTTTCCTTGCCCATGTGTCACCCCTTTGGCCTCCAGCTTGTCCCTCTCTCACTTTCTATAGCTTTGTTGGACCAGATGGTGAGGAAAG GAATGGCCTCTTCCCTTCTAGAGGGGGCTGGCTGGAGTGAGACCTGGGGCTTGGCCTGGAACCCACCACACAGCCCCAAAGTCAGGAAGC CTGGGGAAACCAGAGCTGAGACCTCTTCAACAGGGTTTCTTTGAGATCCTACACCTCCATTGGGCCCTTTTTCAGTCTTCAATGGGGGCC CAGTTGGCTCTAGAAGGAGAAGAGGTGAAGCAGGATCCTTTGCCCTGGGGGAGTCTGAGGGCGCGGTCCTTGGACTCATTCAGGCCGTCT TTGTAGTTGGGGGAGTTCCACTGGGCGATCCCAGCCCCTCCCCACCCACCCTCTAATGGACCTCCTCATAGAAGCCCCATTTCACTTTTG TTTTATCTACCTCTTAGCAAAACAATAGATAAATTAGGTAGTGGCAGCTCCACTTGCTTAGGTTAGGGGGGGAAAAAGATTTCTTTTTCC AAAGGAAAAAAATATTACCTTGAGAATACTTTCCAAAAAATAAAATTAAAAAAAAAAAAACCAAAAAAAAAAATTTTTTTTTAAAAGGGA GACATTTTCCAGTGACCACTGGATTGTTTTAATTTCCCAAGCTTTTTTTTCCCCCATAAATAAGTTTCACTCTTTGGCGATTTTCTTCAC TTGTTTAAGATAACGTGCTAGCTATTCCAACAGGTAACAGCTTTCACAGTCTGCCCCTGGCCTGTCTCACCCCATCCCCCACCCTATTCC TGCCAGTGAGTCCTTCCTGTGCTTCTCTCCCTTCTCCCCTCCCAGCCAGCTGACTTCAGTCACCCCTGTCCCCCCTCCCCTGCCAATAAG CTCCCCCAGGAATAAAGGCTTTGTTTTGGGGATGCTTAAATCTTGACTGGCACTTCCCGGCTGTGGGGGCTGGGGAGCCACTTGTAACAT TTCTGTGCAGATTTTATGTTAGCCACTGCTATGTAAAAGCACGTTCAAAATGAATTTCAGCAGATTATGTGTTACCATAATGAATAAACG >64271_64271_1_PEMT-IGF2BP1_PEMT_chr17_17480234_ENST00000395783_IGF2BP1_chr17_47118740_ENST00000290341_length(amino acids)=321AA_BP=11 MWLPSSPSPSIRSTGMWADEVPLKILAHNNFVGRLIGKEGRNLKKVEQDTETKITISSLQDLTLYNPERTITVKGAIENCCRAEQEIMKK VREAYENDVAAMSLQSHLIPGLNLAAVGLFPASSSAVPPPPSSVTGAAPYSSFMQAPEQEMVQVFIPAQAVGAIIGKKGQHIKQLSRFAS ASIKIAPPETPDSKVRMVIITGPPEAQFKAQGRIYGKLKEENFFGPKEEVKLETHIRVPASAAGRVIGKGGKTVNELQNLTAAEVVVPRD -------------------------------------------------------------- >64271_64271_2_PEMT-IGF2BP1_PEMT_chr17_17480234_ENST00000395783_IGF2BP1_chr17_47118740_ENST00000431824_length(transcript)=1189nt_BP=273nt CTGACCACAGAGCGCTGCTCCCGAGAACCCTGCACCCCTCAATGGAGTAAATTACCATAAAGCCTCTTCCTTACCCATGCTTTGGGGTGT TAACAGCTGAGGCTATTCGTCGGTGACCTGTGGGACTCGAGCTATTCCTGCAGCTCAGCAGACCTCCTGGCCGTGGCAGACTTCTGCGTT ATGACCCGGCTGCTGGGCTACGTGGACCCCCTGGATCCCAGCTTTGTGGCTGCCGTCATCACCATCACCTTCAATCCGCTCTACTGGAAT GTGGGCTGACGAGGTTCCCCTGAAGATCCTGGCCCATAATAACTTTGTAGGGCGTCTCATTGGCAAGGAAGGACGGAACCTGAAGAAGGT AGAGCAAGATACCGAGACAAAAATCACCATCTCCTCGTTGCAAGACCTTACCCTTTACAACCCTGAGAGGACCATCACTGTGAAGGGGGC CATCGAGAATTGTTGCAGGGCCGAGCAGGAAATAATGAAGAAAGTTCGGGAGGCCTATGAGAATGATGTGGCTGCCATGAGCCTGCAGTC TCACCTGATCCCTGGCCTGAACCTGGCTGCTGTAGGTCTTTTCCCAGCTTCATCCAGCGCAGTCCCGCCGCCTCCCAGCAGCGTTACTGG GGCTGCTCCCTATAGCTCCTTTATGCAGGCTCCCGAGCAGGAGATGGTGCAGGTGTTTATCCCCGCCCAGGCAGTGGGCGCCATCATCGG CAAGAAGGGGCAGCACATCAAACAGCTCTCCCGGTTTGCCAGCGCCTCCATCAAGATTGCACCACCCGAAACACCTGACTCCAAAGTTCG TATGGTTATCATCACTGGACCGCCAGAGGCCCAATTCAAGGCTCAGGGAAGAATCTATGGCAAACTCAAGGAGGAGAACTTCTTTGGTCC CAAGGAGGAAGTGAAGCTGGAGACCCACATACGTGTGCCAGCATCAGCAGCTGGCCGGGTCATTGGCAAAGGTGGAAAAACGGTGAACGA GTTGCAGAATTTGACGGCAGCTGAGGTGGTAGTACCAAGAGACCAGACCCCTGATGAGAACGACCAGGTCATCGTGAAAATCATCGGACA TTTCTATGCCAGTCAGATGGCTCAACGGAAGATCCGAGACATCCTGGCCCAGGTTAAGCAGCAGCATCAGAAGGGACAGAGTAACCAGGC >64271_64271_2_PEMT-IGF2BP1_PEMT_chr17_17480234_ENST00000395783_IGF2BP1_chr17_47118740_ENST00000431824_length(amino acids)=322AA_BP=11 MWLPSSPSPSIRSTGMWADEVPLKILAHNNFVGRLIGKEGRNLKKVEQDTETKITISSLQDLTLYNPERTITVKGAIENCCRAEQEIMKK VREAYENDVAAMSLQSHLIPGLNLAAVGLFPASSSAVPPPPSSVTGAAPYSSFMQAPEQEMVQVFIPAQAVGAIIGKKGQHIKQLSRFAS ASIKIAPPETPDSKVRMVIITGPPEAQFKAQGRIYGKLKEENFFGPKEEVKLETHIRVPASAAGRVIGKGGKTVNELQNLTAAEVVVPRD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for PEMT-IGF2BP1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for PEMT-IGF2BP1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for PEMT-IGF2BP1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
