
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:PEPD-GRAMD1A (FusionGDB2 ID:64286) |
Fusion Gene Summary for PEPD-GRAMD1A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: PEPD-GRAMD1A | Fusion gene ID: 64286 | Hgene | Tgene | Gene symbol | PEPD | GRAMD1A | Gene ID | 5184 | 57655 |
| Gene name | peptidase D | GRAM domain containing 1A | |
| Synonyms | PROLIDASE | KIAA1533 | |
| Cytomap | 19q13.11 | 19q13.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | xaa-Pro dipeptidaseX-Pro dipeptidaseaminoacyl-L-proline hydrolaseimidodipeptidaseproline dipeptidasetesticular tissue protein Li 138 | protein Aster-AGRAM domain-containing protein 1A | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q96CP6 | |
| Ensembl transtripts involved in fusion gene | ENST00000244137, ENST00000436370, ENST00000397032, ENST00000591968, | ENST00000598073, ENST00000411896, ENST00000424536, ENST00000504615, ENST00000317991, ENST00000599564, | |
| Fusion gene scores | * DoF score | 11 X 8 X 8=704 | 5 X 5 X 5=125 |
| # samples | 12 | 5 | |
| ** MAII score | log2(12/704*10)=-2.55254102302878 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/125*10)=-1.32192809488736 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: PEPD [Title/Abstract] AND GRAMD1A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | PEPD(33953901)-GRAMD1A(35500023), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across PEPD (5'-gene) Fusion gene breakpoints across PEPD (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
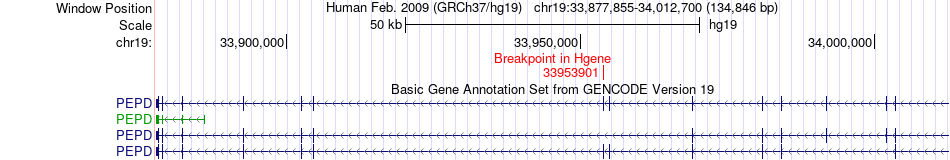 |
 Fusion gene breakpoints across GRAMD1A (3'-gene) Fusion gene breakpoints across GRAMD1A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
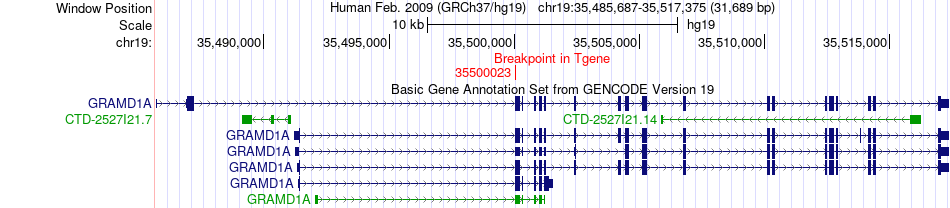 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SKCM | TCGA-D3-A3CC-06A | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
Top |
Fusion Gene ORF analysis for PEPD-GRAMD1A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000244137 | ENST00000598073 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| 5CDS-3UTR | ENST00000436370 | ENST00000598073 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| 5CDS-5UTR | ENST00000244137 | ENST00000411896 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| 5CDS-5UTR | ENST00000244137 | ENST00000424536 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| 5CDS-5UTR | ENST00000244137 | ENST00000504615 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| 5CDS-5UTR | ENST00000436370 | ENST00000411896 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| 5CDS-5UTR | ENST00000436370 | ENST00000424536 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| 5CDS-5UTR | ENST00000436370 | ENST00000504615 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| In-frame | ENST00000244137 | ENST00000317991 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| In-frame | ENST00000244137 | ENST00000599564 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| In-frame | ENST00000436370 | ENST00000317991 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| In-frame | ENST00000436370 | ENST00000599564 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| intron-3CDS | ENST00000397032 | ENST00000317991 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| intron-3CDS | ENST00000397032 | ENST00000599564 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| intron-3CDS | ENST00000591968 | ENST00000317991 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| intron-3CDS | ENST00000591968 | ENST00000599564 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| intron-3UTR | ENST00000397032 | ENST00000598073 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| intron-3UTR | ENST00000591968 | ENST00000598073 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| intron-5UTR | ENST00000397032 | ENST00000411896 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| intron-5UTR | ENST00000397032 | ENST00000424536 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| intron-5UTR | ENST00000397032 | ENST00000504615 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| intron-5UTR | ENST00000591968 | ENST00000411896 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| intron-5UTR | ENST00000591968 | ENST00000424536 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
| intron-5UTR | ENST00000591968 | ENST00000504615 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000244137 | PEPD | chr19 | 33953901 | - | ENST00000599564 | GRAMD1A | chr19 | 35500023 | + | 3190 | 705 | 34 | 2859 | 941 |
| ENST00000244137 | PEPD | chr19 | 33953901 | - | ENST00000317991 | GRAMD1A | chr19 | 35500023 | + | 3200 | 705 | 34 | 2871 | 945 |
| ENST00000436370 | PEPD | chr19 | 33953901 | - | ENST00000599564 | GRAMD1A | chr19 | 35500023 | + | 2992 | 507 | 28 | 2661 | 877 |
| ENST00000436370 | PEPD | chr19 | 33953901 | - | ENST00000317991 | GRAMD1A | chr19 | 35500023 | + | 3002 | 507 | 28 | 2673 | 881 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000244137 | ENST00000599564 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + | 0.004699641 | 0.9953003 |
| ENST00000244137 | ENST00000317991 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + | 0.0046464 | 0.9953536 |
| ENST00000436370 | ENST00000599564 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + | 0.009382281 | 0.9906177 |
| ENST00000436370 | ENST00000317991 | PEPD | chr19 | 33953901 | - | GRAMD1A | chr19 | 35500023 | + | 0.008747364 | 0.9912526 |
Top |
Fusion Genomic Features for PEPD-GRAMD1A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| PEPD | chr19 | 33953900 | - | GRAMD1A | chr19 | 35500022 | + | 5.44E-06 | 0.9999945 |
| PEPD | chr19 | 33953900 | - | GRAMD1A | chr19 | 35500022 | + | 5.44E-06 | 0.9999945 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
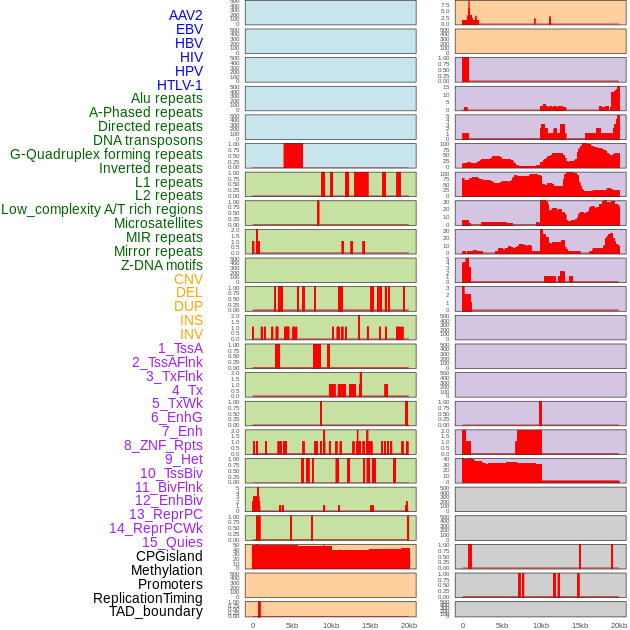 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
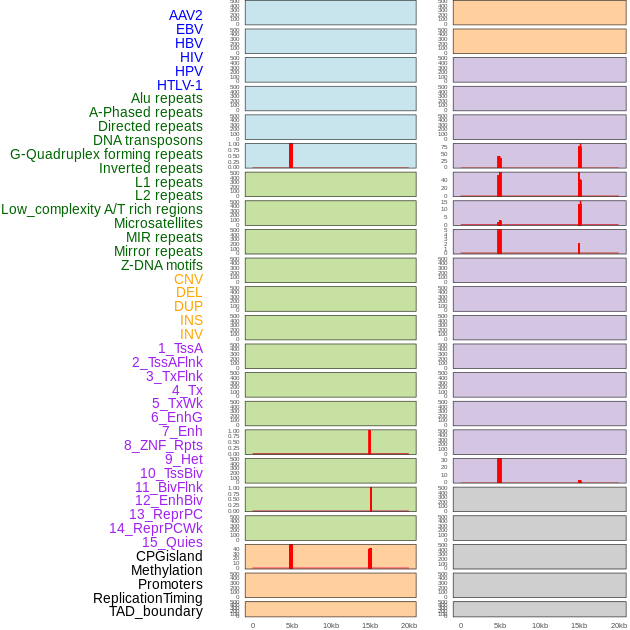 |
Top |
Fusion Protein Features for PEPD-GRAMD1A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:33953901/chr19:35500023) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | GRAMD1A |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Cholesterol transporter that mediates non-vesicular transport of cholesterol from the plasma membrane (PM) to the endoplasmic reticulum (ER) (By similarity). Contains unique domains for binding cholesterol and the PM, thereby serving as a molecular bridge for the transfer of cholesterol from the PM to the ER (By similarity). Plays a crucial role in cholesterol homeostasis and has the unique ability to localize to the PM based on the level of membrane cholesterol (By similarity). In lipid-poor conditions localizes to the ER membrane and in response to excess cholesterol in the PM is recruited to the endoplasmic reticulum-plasma membrane contact sites (EPCS) which is mediated by the GRAM domain (By similarity). At the EPCS, the sterol-binding VASt/ASTER domain binds to the cholesterol in the PM and facilitates its transfer from the PM to ER (By similarity). May play a role in tumor progression (By similarity). Plays a role in autophagy regulation and is required for biogenesis of the autophagosome (PubMed:31222192). This function in autophagy requires its cholesterol-transfer activity (PubMed:31222192). {ECO:0000250|UniProtKB:Q8VEF1, ECO:0000269|PubMed:31222192}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | GRAMD1A | chr19:33953901 | chr19:35500023 | ENST00000317991 | 0 | 20 | 367_538 | 2 | 725.0 | Domain | VASt | |
| Tgene | GRAMD1A | chr19:33953901 | chr19:35500023 | ENST00000317991 | 0 | 20 | 91_158 | 2 | 725.0 | Domain | GRAM | |
| Tgene | GRAMD1A | chr19:33953901 | chr19:35500023 | ENST00000411896 | 0 | 18 | 367_538 | 2 | 714.0 | Domain | VASt | |
| Tgene | GRAMD1A | chr19:33953901 | chr19:35500023 | ENST00000411896 | 0 | 18 | 91_158 | 2 | 714.0 | Domain | GRAM | |
| Tgene | GRAMD1A | chr19:33953901 | chr19:35500023 | ENST00000317991 | 0 | 20 | 610_630 | 2 | 725.0 | Transmembrane | Helical | |
| Tgene | GRAMD1A | chr19:33953901 | chr19:35500023 | ENST00000411896 | 0 | 18 | 610_630 | 2 | 714.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Gene Sequence for PEPD-GRAMD1A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >64286_64286_1_PEPD-GRAMD1A_PEPD_chr19_33953901_ENST00000244137_GRAMD1A_chr19_35500023_ENST00000317991_length(transcript)=3200nt_BP=705nt GCCGCACTTCACGTGACGCCGGTGCCGGGCGAACATGGCGGCGGCCACCGGACCCTCGTTTTGGCTGGGGAATGAAACCCTGAAGGTGCC GCTGGCGCTCTTTGCCTTGAACCGGCAGCGCCTGTGTGAGCGGCTGCGGAAGAACCCTGCTGTGCAGGCCGGCTCCATCGTGGTCCTGCA GGGCGGGGAGGAGACTCAGCGCTACTGCACCGACACCGGGGTCCTCTTCCGCCAGGAGTCCTTCTTTCACTGGGCGTTCGGTGTCACTGA GCCAGGCTGCTATGGTGTCATCGATGTTGACACTGGGAAGTCGACCCTGTTTGTGCCCAGGCTTCCTGCCAGCCATGCCACCTGGATGGG AAAGATCCATTCCAAGGAGCACTTCAAGGAGAAGTATGCCGTGGACGACGTCCAGTACGTAGATGAGATTGCCAGCGTCCTGACGTCACA GAAGCCCTCTGTCCTCCTCACTTTGCGTGGCGTCAACACGGACAGCGGCAGTGTCTGCAGGGAGGCCTCCTTTGACGGCATCAGCAAGTT CGAAGTCAACAATACCATTCTTCACCCAGAGATCGTTGAGTGCCGAGTGTTTAAGACGGATATGGAGCTGGAGGTTCTGCGCTATACCAA TAAAATCTCCAGCGAGGCCCACCGTGAGGTAATGAAGGCTGTAAAAGTGGGAATGAAAGAATATGAGTTGGAAAGCACCACACCCCACTC TGGCCGGAGCACGCCAAGCAGCTCCCCATCGCTCCGGAAACGGCTGCAGCTCCTGCCCCCAAGCCGGCCCCCACCTGAGCCAGAACCAGG CACCATGGTGGAGAAGGGATCAGATAGCTCCTCAGAGAAGGGTGGGGTGCCTGGGACCCCCAGCACCCAGAGCCTAGGCAGCCGGAACTT CATCCGCAACAGCAAGAAGATGCAGAGCTGGTACAGTATGCTGAGCCCCACTTATAAGCAGCGTAATGAGGACTTCCGGAAACTGTTCAG CAAACTCCCCGAAGCAGAACGCCTCATTGTGGATTACTCCTGCGCCCTGCAGCGTGAGATCCTGCTCCAGGGCCGCCTCTACCTCTCTGA GAACTGGATCTGCTTCTACAGCAACATCTTCCGCTGGGAGACCACGATCTCCATCCAGCTGAAGGAAGTGACATGTCTGAAGAAGGAAAA GACGGCCAAGCTGATCCCCAACGCCATCCAGATCTGCACGGAGAGCGAGAAGCATTTCTTCACTTCCTTTGGGGCCCGTGACCGCTGCTT CCTCCTCATCTTCCGCCTCTGGCAGAATGCACTGCTTGAAAAGACGCTGAGTCCCCGCGAGCTCTGGCACCTGGTGCATCAGTGCTACGG CTCAGAGCTGGGCCTCACCAGTGAGGATGAGGACTATGTCTCCCCCTTGCAGCTGAACGGTCTGGGGACCCCCAAGGAAGTGGGAGATGT GATCGCCCTGAGCGACATCACCTCCTCGGGGGCAGCTGACCGCAGCCAGGAGCCAAGCCCAGTGGGTTCGCGCCGTGGCCATGTCACGCC CAACCTTTCCCGAGCCAGCAGCGACGCAGACCATGGGGCAGAGGAGGACAAGGAGGAGCAGGTAGACAGCCAGCCAGACGCCTCCTCCAG CCAGACAGTGACCCCGGTGGCTGAACCCCCGAGCACAGAGCCCACCCAGCCTGACGGGCCCACCACCCTGGGCCCCTTGGATCTGCTGCC CAGTGAGGAGCTATTGACAGACACAAGTAACTCCTCTTCATCCACTGGGGAGGAAGCGGACTTGGCTGCCCTGCTTCCCGACCTCTCCGG CCGCCTCCTCATCAACTCTGTCTTCCATGTGGGCGCTGAGCGGCTCCAGCAGATGCTCTTCTCGGACTCGCCCTTCCTCCAGGGCTTCCT ACAGCAGTGCAAGTTCACAGACGTGACGCTGAGCCCCTGGAGTGGGGACAGCAAGTGCCACCAGCGCCGGGTGCTGACGTACACCATCCC CATCAGCAACCCACTGGGCCCCAAGAGCGCCTCCGTGGTGGAGACACAGACGCTGTTCCGGCGCGGCCCCCAGGCCGGCGGGTGTGTGGT GGACTCCGAGGTGCTGACGCAGGGCATCCCCTACCAGGACTACTTCTACACTGCCCACCGCTACTGCATCCTGGGTCTGGCCCGGAACAA GGCGCGGCTCCGAGTGTCTTCTGAGATCCGCTACCGAAAGCAGCCGTGGAGCCTGGTGAAGTCGCTCATTGAGAAGAACTCGTGGAGCGG CATTGAAGACTATTTCCACCATCTGGAGCGAGAGCTCGCCAAGGCTGAGAAGCTGTCTCTGGAGGAAGGCGGGAAGGATGCCCGGGGCTT GCTATCCGGCCTGCGGCGGCGGAAGCGGCCCCTGAGCTGGCGGGCTCACGGGGACGGGCCCCAGCACCCAGATCCTGACCCCTGTGCCCG GGCCGGCATTCACACCTCGGGCTCCCTCAGCTCCCGCTTCTCCGAACCATCTGTGGACCAGGGCCCCGGGGCAGGCATCCCCAGTGCCCT GGTTCTCATCAGCATTGTGATCTGTGTGAGCCTTATCATCCTCATCGCCCTCAACGTCCTGCTCTTCTACCGCCTCTGGTCCCTGGAAAG GACAGCCCACACCTTTGAGTCCTGGCACAGCCTGGCCCTGGCCAAGGGCAAGTTCCCCCAGACGGCCACAGAGTGGGCCGAGATCCTGGC GCTGCAGAAGCAATTCCACAGCGTGGAGGTGCACAAGTGGAGGCAGATCCTGCGGGCCTCCGTGGAGCTCCTGGATGAGATGAAGTTCTC GCTGGAGAAGCTGCACCAAGGCATCACAGTCTCAGACCCTCCCTTTGACACCCAGCCCCGGCCCGATGACAGCTTTTCCTGAGGACCCCG GCCACGCAGCTGTTCCCCCACATGGACAGATGGACACACAGAGCCTCGGCGGCCACTGCTGGCACGGTGTGAGCGCCAGGCATCTCCCAC CCGCCCCTCCCGACGGCCCAACCAGGGGCTGTGCAGACGTGGGGACCACGGAACCGAGATGCACTTTAGACCAGGGAGCTGGCCCGGCCT CTGGCAGGCCCCCCACTAACTTATTTTGCCCGGCTGAGGTTGTGGGGGGCGCCTCCTGGGGTGCACGATTCCCTCAGCTCTGGGTTTAAT >64286_64286_1_PEPD-GRAMD1A_PEPD_chr19_33953901_ENST00000244137_GRAMD1A_chr19_35500023_ENST00000317991_length(amino acids)=945AA_BP=223 MAAATGPSFWLGNETLKVPLALFALNRQRLCERLRKNPAVQAGSIVVLQGGEETQRYCTDTGVLFRQESFFHWAFGVTEPGCYGVIDVDT GKSTLFVPRLPASHATWMGKIHSKEHFKEKYAVDDVQYVDEIASVLTSQKPSVLLTLRGVNTDSGSVCREASFDGISKFEVNNTILHPEI VECRVFKTDMELEVLRYTNKISSEAHREVMKAVKVGMKEYELESTTPHSGRSTPSSSPSLRKRLQLLPPSRPPPEPEPGTMVEKGSDSSS EKGGVPGTPSTQSLGSRNFIRNSKKMQSWYSMLSPTYKQRNEDFRKLFSKLPEAERLIVDYSCALQREILLQGRLYLSENWICFYSNIFR WETTISIQLKEVTCLKKEKTAKLIPNAIQICTESEKHFFTSFGARDRCFLLIFRLWQNALLEKTLSPRELWHLVHQCYGSELGLTSEDED YVSPLQLNGLGTPKEVGDVIALSDITSSGAADRSQEPSPVGSRRGHVTPNLSRASSDADHGAEEDKEEQVDSQPDASSSQTVTPVAEPPS TEPTQPDGPTTLGPLDLLPSEELLTDTSNSSSSTGEEADLAALLPDLSGRLLINSVFHVGAERLQQMLFSDSPFLQGFLQQCKFTDVTLS PWSGDSKCHQRRVLTYTIPISNPLGPKSASVVETQTLFRRGPQAGGCVVDSEVLTQGIPYQDYFYTAHRYCILGLARNKARLRVSSEIRY RKQPWSLVKSLIEKNSWSGIEDYFHHLERELAKAEKLSLEEGGKDARGLLSGLRRRKRPLSWRAHGDGPQHPDPDPCARAGIHTSGSLSS RFSEPSVDQGPGAGIPSALVLISIVICVSLIILIALNVLLFYRLWSLERTAHTFESWHSLALAKGKFPQTATEWAEILALQKQFHSVEVH -------------------------------------------------------------- >64286_64286_2_PEPD-GRAMD1A_PEPD_chr19_33953901_ENST00000244137_GRAMD1A_chr19_35500023_ENST00000599564_length(transcript)=3190nt_BP=705nt GCCGCACTTCACGTGACGCCGGTGCCGGGCGAACATGGCGGCGGCCACCGGACCCTCGTTTTGGCTGGGGAATGAAACCCTGAAGGTGCC GCTGGCGCTCTTTGCCTTGAACCGGCAGCGCCTGTGTGAGCGGCTGCGGAAGAACCCTGCTGTGCAGGCCGGCTCCATCGTGGTCCTGCA GGGCGGGGAGGAGACTCAGCGCTACTGCACCGACACCGGGGTCCTCTTCCGCCAGGAGTCCTTCTTTCACTGGGCGTTCGGTGTCACTGA GCCAGGCTGCTATGGTGTCATCGATGTTGACACTGGGAAGTCGACCCTGTTTGTGCCCAGGCTTCCTGCCAGCCATGCCACCTGGATGGG AAAGATCCATTCCAAGGAGCACTTCAAGGAGAAGTATGCCGTGGACGACGTCCAGTACGTAGATGAGATTGCCAGCGTCCTGACGTCACA GAAGCCCTCTGTCCTCCTCACTTTGCGTGGCGTCAACACGGACAGCGGCAGTGTCTGCAGGGAGGCCTCCTTTGACGGCATCAGCAAGTT CGAAGTCAACAATACCATTCTTCACCCAGAGATCGTTGAGTGCCGAGTGTTTAAGACGGATATGGAGCTGGAGGTTCTGCGCTATACCAA TAAAATCTCCAGCGAGGCCCACCGTGAGGTAATGAAGGCTGTAAAAGTGGGAATGAAAGAATATGAGTTGGAAAGCACCACACCCCACTC TGGCCGGAGCACGCCAAGCAGCTCCCCATCGCTCCGGAAACGGCTGCAGCTCCTGCCCCCAAGCCGGCCCCCACCTGAGCCAGAACCAGG CACCATGGTGGAGAAGGGATCAGATAGCTCCTCAGAGAAGGGTGGGGTGCCTGGGACCCCCAGCACCCAGAGCCTAGGCAGCCGGAACTT CATCCGCAACAGCAAGAAGATGCAGAGCTGGTACAGTATGCTGAGCCCCACTTATAAGCAGCGTAATGAGGACTTCCGGAAACTGTTCAG CAAACTCCCCGAAGCAGAACGCCTCATTGTGGATTACTCCTGCGCCCTGCAGCGTGAGATCCTGCTCCAGGGCCGCCTCTACCTCTCTGA GAACTGGATCTGCTTCTACAGCAACATCTTCCGCTGGGAGACCACGATCTCCATCCAGCTGAAGGAAGTGACATGTCTGAAGAAGGAAAA GACGGCCAAGCTGATCCCCAACGCCATCCAGATCTGCACGGAGAGCGAGAAGCATTTCTTCACTTCCTTTGGGGCCCGTGACCGCTGCTT CCTCCTCATCTTCCGCCTCTGGCAGAATGCACTGCTTGAAAAGACGCTGAGTCCCCGCGAGCTCTGGCACCTGGTGCATCAGTGCTACGG CTCAGAGCTGGGCCTCACCAGTGAGGATGAGGACTATGTCTCCCCCTTGCAGCTGAACGGTCTGGGGACCCCCAAGGAAGTGGGAGATGT GATCGCCCTGAGCGACATCACCTCCTCGGGGGCAGCTGACCGCAGCCAGGAGCCAAGCCCAGTGGGTTCGCGCCGTGGCCATGTCACGCC CAACCTTTCCCGAGCCAGCAGCGACGCAGACCATGGGGCAGAGGAGGACAAGGAGGAGCAGGTAGACAGCCAGCCAGACGCCTCCTCCAG CCAGACAGTGACCCCGGTGGCTGAACCCCCGAGCACAGAGCCCACCCAGCCTGACGGGCCCACCACCCTGGGCCCCTTGGATCTGCTGCC CAGTGAGGAGCTATTGACAGACACAAGTAACTCCTCTTCATCCACTGGGGAGGAAGCGGACTTGGCTGCCCTGCTTCCCGACCTCTCCGG CCGCCTCCTCATCAACTCTGTCTTCCATGTGGGCGCTGAGCGGCTCCAGCAGATGCTCTTCTCGGACTCGCCCTTCCTCCAGGGCTTCCT ACAGCAGTGCAAGTTCACAGACGTGACGCTGAGCCCCTGGAGTGGGGACAGCAAGTGCCACCAGCGCCGGGTGCTGACGTACACCATCCC CATCAGCAACCCACTGGGCCCCAAGAGCGCCTCCGTGGTGGAGACACAGACGCTGTTCCGGCGCGGCCCCCAGGCCGGCGGGTGTGTGGT GGACTCCGAGGTGCTGACGCAGGGCATCCCCTACCAGGACTACTTCTACACTGCCCACCGCTACTGCATCCTGGGTCTGGCCCGGAACAA GGCGCGGCTCCGAGTGTCTTCTGAGATCCGCTACCGAAAGCAGCCGTGGAGCCTGGTGAAGTCGCTCATTGAGAAGAACTCGTGGAGCGG CATTGAAGACTATTTCCACCATCTGGAGCGAGAGCTCGCCAAGGCTGAGAAGCTGTCTCTGGAGGAAGGCGGGAAGGATGCCCGGGGCTT GCTATCCGGCCTGCGGCGGCGGAAGCGGCCCCTGAGCTGGCGGGCTCACGGGGACGGGCCCCAGCACCCAGATCCTGACCCCTGTGCCCG GGCCGGCATTCACACCTCGGGCTCCCTCAGCTCCCGCTTCTCCGAACCATCTGTGGACCAGGGCCCCGGGGCAGGCATCCCCAGTGCCCT GGTTCTCATCAGCATTGTCCTTATCATCCTCATCGCCCTCAACGTCCTGCTCTTCTACCGCCTCTGGTCCCTGGAAAGGACAGCCCACAC CTTTGAGTCCTGGCACAGCCTGGCCCTGGCCAAGGGCAAGTTCCCCCAGACGGCCACAGAGTGGGCCGAGATCCTGGCGCTGCAGAAGCA ATTCCACAGCGTGGAGGTGCACAAGTGGAGGCAGATCCTGCGGGCCTCCGTGGAGCTCCTGGATGAGATGAAGTTCTCGCTGGAGAAGCT GCACCAAGGCATCACAGTCTCAGACCCTCCCTTTGACACCCAGCCCCGGCCCGATGACAGCTTTTCCTGAGGACCCCGGCCACGCAGCTG TTCCCCCACATGGACAGATGGACACACAGAGCCTCGGCGGCCACTGCTGGCACGGTGTGAGCGCCAGGCATCTCCCACCCGCCCCTCCCG ACGGCCCAACCAGGGGCTGTGCAGACGTGGGGACCACGGAACCGAGATGCACTTTAGACCAGGGAGCTGGCCCGGCCTCTGGCAGGCCCC CCACTAACTTATTTTGCCCGGCTGAGGTTGTGGGGGGCGCCTCCTGGGGTGCACGATTCCCTCAGCTCTGGGTTTAATGTATTATATTTA >64286_64286_2_PEPD-GRAMD1A_PEPD_chr19_33953901_ENST00000244137_GRAMD1A_chr19_35500023_ENST00000599564_length(amino acids)=941AA_BP=223 MAAATGPSFWLGNETLKVPLALFALNRQRLCERLRKNPAVQAGSIVVLQGGEETQRYCTDTGVLFRQESFFHWAFGVTEPGCYGVIDVDT GKSTLFVPRLPASHATWMGKIHSKEHFKEKYAVDDVQYVDEIASVLTSQKPSVLLTLRGVNTDSGSVCREASFDGISKFEVNNTILHPEI VECRVFKTDMELEVLRYTNKISSEAHREVMKAVKVGMKEYELESTTPHSGRSTPSSSPSLRKRLQLLPPSRPPPEPEPGTMVEKGSDSSS EKGGVPGTPSTQSLGSRNFIRNSKKMQSWYSMLSPTYKQRNEDFRKLFSKLPEAERLIVDYSCALQREILLQGRLYLSENWICFYSNIFR WETTISIQLKEVTCLKKEKTAKLIPNAIQICTESEKHFFTSFGARDRCFLLIFRLWQNALLEKTLSPRELWHLVHQCYGSELGLTSEDED YVSPLQLNGLGTPKEVGDVIALSDITSSGAADRSQEPSPVGSRRGHVTPNLSRASSDADHGAEEDKEEQVDSQPDASSSQTVTPVAEPPS TEPTQPDGPTTLGPLDLLPSEELLTDTSNSSSSTGEEADLAALLPDLSGRLLINSVFHVGAERLQQMLFSDSPFLQGFLQQCKFTDVTLS PWSGDSKCHQRRVLTYTIPISNPLGPKSASVVETQTLFRRGPQAGGCVVDSEVLTQGIPYQDYFYTAHRYCILGLARNKARLRVSSEIRY RKQPWSLVKSLIEKNSWSGIEDYFHHLERELAKAEKLSLEEGGKDARGLLSGLRRRKRPLSWRAHGDGPQHPDPDPCARAGIHTSGSLSS RFSEPSVDQGPGAGIPSALVLISIVLIILIALNVLLFYRLWSLERTAHTFESWHSLALAKGKFPQTATEWAEILALQKQFHSVEVHKWRQ -------------------------------------------------------------- >64286_64286_3_PEPD-GRAMD1A_PEPD_chr19_33953901_ENST00000436370_GRAMD1A_chr19_35500023_ENST00000317991_length(transcript)=3002nt_BP=507nt CTTCACGTGACGCCGGTGCCGGGCGAACATGGCGGCGGCCACCGGACCCTCGTTTTGGCTGGGGAATGAAACCCTGAAGGTGCCGCTGGC GCTCTTTGCCTTGAACCGGCAGCGCCTGTGTGAGCGGCTGCGGAAGAACCCTGCTGTGCAGGCCGGCTCCATCGTGGTCCTGCAGGGCGG GGAGGAGACTCAGCGCTACTGCACCGACACCGGGGTCCTCTTCCGCCAGATTGCCAGCGTCCTGACGTCACAGAAGCCCTCTGTCCTCCT CACTTTGCGTGGCGTCAACACGGACAGCGGCAGTGTCTGCAGGGAGGCCTCCTTTGACGGCATCAGCAAGTTCGAAGTCAACAATACCAT TCTTCACCCAGAGATCGTTGAGTGCCGAGTGTTTAAGACGGATATGGAGCTGGAGGTTCTGCGCTATACCAATAAAATCTCCAGCGAGGC CCACCGTGAGGTAATGAAGGCTGTAAAAGTGGGAATGAAAGAATATGAGTTGGAAAGCACCACACCCCACTCTGGCCGGAGCACGCCAAG CAGCTCCCCATCGCTCCGGAAACGGCTGCAGCTCCTGCCCCCAAGCCGGCCCCCACCTGAGCCAGAACCAGGCACCATGGTGGAGAAGGG ATCAGATAGCTCCTCAGAGAAGGGTGGGGTGCCTGGGACCCCCAGCACCCAGAGCCTAGGCAGCCGGAACTTCATCCGCAACAGCAAGAA GATGCAGAGCTGGTACAGTATGCTGAGCCCCACTTATAAGCAGCGTAATGAGGACTTCCGGAAACTGTTCAGCAAACTCCCCGAAGCAGA ACGCCTCATTGTGGATTACTCCTGCGCCCTGCAGCGTGAGATCCTGCTCCAGGGCCGCCTCTACCTCTCTGAGAACTGGATCTGCTTCTA CAGCAACATCTTCCGCTGGGAGACCACGATCTCCATCCAGCTGAAGGAAGTGACATGTCTGAAGAAGGAAAAGACGGCCAAGCTGATCCC CAACGCCATCCAGATCTGCACGGAGAGCGAGAAGCATTTCTTCACTTCCTTTGGGGCCCGTGACCGCTGCTTCCTCCTCATCTTCCGCCT CTGGCAGAATGCACTGCTTGAAAAGACGCTGAGTCCCCGCGAGCTCTGGCACCTGGTGCATCAGTGCTACGGCTCAGAGCTGGGCCTCAC CAGTGAGGATGAGGACTATGTCTCCCCCTTGCAGCTGAACGGTCTGGGGACCCCCAAGGAAGTGGGAGATGTGATCGCCCTGAGCGACAT CACCTCCTCGGGGGCAGCTGACCGCAGCCAGGAGCCAAGCCCAGTGGGTTCGCGCCGTGGCCATGTCACGCCCAACCTTTCCCGAGCCAG CAGCGACGCAGACCATGGGGCAGAGGAGGACAAGGAGGAGCAGGTAGACAGCCAGCCAGACGCCTCCTCCAGCCAGACAGTGACCCCGGT GGCTGAACCCCCGAGCACAGAGCCCACCCAGCCTGACGGGCCCACCACCCTGGGCCCCTTGGATCTGCTGCCCAGTGAGGAGCTATTGAC AGACACAAGTAACTCCTCTTCATCCACTGGGGAGGAAGCGGACTTGGCTGCCCTGCTTCCCGACCTCTCCGGCCGCCTCCTCATCAACTC TGTCTTCCATGTGGGCGCTGAGCGGCTCCAGCAGATGCTCTTCTCGGACTCGCCCTTCCTCCAGGGCTTCCTACAGCAGTGCAAGTTCAC AGACGTGACGCTGAGCCCCTGGAGTGGGGACAGCAAGTGCCACCAGCGCCGGGTGCTGACGTACACCATCCCCATCAGCAACCCACTGGG CCCCAAGAGCGCCTCCGTGGTGGAGACACAGACGCTGTTCCGGCGCGGCCCCCAGGCCGGCGGGTGTGTGGTGGACTCCGAGGTGCTGAC GCAGGGCATCCCCTACCAGGACTACTTCTACACTGCCCACCGCTACTGCATCCTGGGTCTGGCCCGGAACAAGGCGCGGCTCCGAGTGTC TTCTGAGATCCGCTACCGAAAGCAGCCGTGGAGCCTGGTGAAGTCGCTCATTGAGAAGAACTCGTGGAGCGGCATTGAAGACTATTTCCA CCATCTGGAGCGAGAGCTCGCCAAGGCTGAGAAGCTGTCTCTGGAGGAAGGCGGGAAGGATGCCCGGGGCTTGCTATCCGGCCTGCGGCG GCGGAAGCGGCCCCTGAGCTGGCGGGCTCACGGGGACGGGCCCCAGCACCCAGATCCTGACCCCTGTGCCCGGGCCGGCATTCACACCTC GGGCTCCCTCAGCTCCCGCTTCTCCGAACCATCTGTGGACCAGGGCCCCGGGGCAGGCATCCCCAGTGCCCTGGTTCTCATCAGCATTGT GATCTGTGTGAGCCTTATCATCCTCATCGCCCTCAACGTCCTGCTCTTCTACCGCCTCTGGTCCCTGGAAAGGACAGCCCACACCTTTGA GTCCTGGCACAGCCTGGCCCTGGCCAAGGGCAAGTTCCCCCAGACGGCCACAGAGTGGGCCGAGATCCTGGCGCTGCAGAAGCAATTCCA CAGCGTGGAGGTGCACAAGTGGAGGCAGATCCTGCGGGCCTCCGTGGAGCTCCTGGATGAGATGAAGTTCTCGCTGGAGAAGCTGCACCA AGGCATCACAGTCTCAGACCCTCCCTTTGACACCCAGCCCCGGCCCGATGACAGCTTTTCCTGAGGACCCCGGCCACGCAGCTGTTCCCC CACATGGACAGATGGACACACAGAGCCTCGGCGGCCACTGCTGGCACGGTGTGAGCGCCAGGCATCTCCCACCCGCCCCTCCCGACGGCC CAACCAGGGGCTGTGCAGACGTGGGGACCACGGAACCGAGATGCACTTTAGACCAGGGAGCTGGCCCGGCCTCTGGCAGGCCCCCCACTA ACTTATTTTGCCCGGCTGAGGTTGTGGGGGGCGCCTCCTGGGGTGCACGATTCCCTCAGCTCTGGGTTTAATGTATTATATTTATTTGGG >64286_64286_3_PEPD-GRAMD1A_PEPD_chr19_33953901_ENST00000436370_GRAMD1A_chr19_35500023_ENST00000317991_length(amino acids)=881AA_BP=159 MAAATGPSFWLGNETLKVPLALFALNRQRLCERLRKNPAVQAGSIVVLQGGEETQRYCTDTGVLFRQIASVLTSQKPSVLLTLRGVNTDS GSVCREASFDGISKFEVNNTILHPEIVECRVFKTDMELEVLRYTNKISSEAHREVMKAVKVGMKEYELESTTPHSGRSTPSSSPSLRKRL QLLPPSRPPPEPEPGTMVEKGSDSSSEKGGVPGTPSTQSLGSRNFIRNSKKMQSWYSMLSPTYKQRNEDFRKLFSKLPEAERLIVDYSCA LQREILLQGRLYLSENWICFYSNIFRWETTISIQLKEVTCLKKEKTAKLIPNAIQICTESEKHFFTSFGARDRCFLLIFRLWQNALLEKT LSPRELWHLVHQCYGSELGLTSEDEDYVSPLQLNGLGTPKEVGDVIALSDITSSGAADRSQEPSPVGSRRGHVTPNLSRASSDADHGAEE DKEEQVDSQPDASSSQTVTPVAEPPSTEPTQPDGPTTLGPLDLLPSEELLTDTSNSSSSTGEEADLAALLPDLSGRLLINSVFHVGAERL QQMLFSDSPFLQGFLQQCKFTDVTLSPWSGDSKCHQRRVLTYTIPISNPLGPKSASVVETQTLFRRGPQAGGCVVDSEVLTQGIPYQDYF YTAHRYCILGLARNKARLRVSSEIRYRKQPWSLVKSLIEKNSWSGIEDYFHHLERELAKAEKLSLEEGGKDARGLLSGLRRRKRPLSWRA HGDGPQHPDPDPCARAGIHTSGSLSSRFSEPSVDQGPGAGIPSALVLISIVICVSLIILIALNVLLFYRLWSLERTAHTFESWHSLALAK -------------------------------------------------------------- >64286_64286_4_PEPD-GRAMD1A_PEPD_chr19_33953901_ENST00000436370_GRAMD1A_chr19_35500023_ENST00000599564_length(transcript)=2992nt_BP=507nt CTTCACGTGACGCCGGTGCCGGGCGAACATGGCGGCGGCCACCGGACCCTCGTTTTGGCTGGGGAATGAAACCCTGAAGGTGCCGCTGGC GCTCTTTGCCTTGAACCGGCAGCGCCTGTGTGAGCGGCTGCGGAAGAACCCTGCTGTGCAGGCCGGCTCCATCGTGGTCCTGCAGGGCGG GGAGGAGACTCAGCGCTACTGCACCGACACCGGGGTCCTCTTCCGCCAGATTGCCAGCGTCCTGACGTCACAGAAGCCCTCTGTCCTCCT CACTTTGCGTGGCGTCAACACGGACAGCGGCAGTGTCTGCAGGGAGGCCTCCTTTGACGGCATCAGCAAGTTCGAAGTCAACAATACCAT TCTTCACCCAGAGATCGTTGAGTGCCGAGTGTTTAAGACGGATATGGAGCTGGAGGTTCTGCGCTATACCAATAAAATCTCCAGCGAGGC CCACCGTGAGGTAATGAAGGCTGTAAAAGTGGGAATGAAAGAATATGAGTTGGAAAGCACCACACCCCACTCTGGCCGGAGCACGCCAAG CAGCTCCCCATCGCTCCGGAAACGGCTGCAGCTCCTGCCCCCAAGCCGGCCCCCACCTGAGCCAGAACCAGGCACCATGGTGGAGAAGGG ATCAGATAGCTCCTCAGAGAAGGGTGGGGTGCCTGGGACCCCCAGCACCCAGAGCCTAGGCAGCCGGAACTTCATCCGCAACAGCAAGAA GATGCAGAGCTGGTACAGTATGCTGAGCCCCACTTATAAGCAGCGTAATGAGGACTTCCGGAAACTGTTCAGCAAACTCCCCGAAGCAGA ACGCCTCATTGTGGATTACTCCTGCGCCCTGCAGCGTGAGATCCTGCTCCAGGGCCGCCTCTACCTCTCTGAGAACTGGATCTGCTTCTA CAGCAACATCTTCCGCTGGGAGACCACGATCTCCATCCAGCTGAAGGAAGTGACATGTCTGAAGAAGGAAAAGACGGCCAAGCTGATCCC CAACGCCATCCAGATCTGCACGGAGAGCGAGAAGCATTTCTTCACTTCCTTTGGGGCCCGTGACCGCTGCTTCCTCCTCATCTTCCGCCT CTGGCAGAATGCACTGCTTGAAAAGACGCTGAGTCCCCGCGAGCTCTGGCACCTGGTGCATCAGTGCTACGGCTCAGAGCTGGGCCTCAC CAGTGAGGATGAGGACTATGTCTCCCCCTTGCAGCTGAACGGTCTGGGGACCCCCAAGGAAGTGGGAGATGTGATCGCCCTGAGCGACAT CACCTCCTCGGGGGCAGCTGACCGCAGCCAGGAGCCAAGCCCAGTGGGTTCGCGCCGTGGCCATGTCACGCCCAACCTTTCCCGAGCCAG CAGCGACGCAGACCATGGGGCAGAGGAGGACAAGGAGGAGCAGGTAGACAGCCAGCCAGACGCCTCCTCCAGCCAGACAGTGACCCCGGT GGCTGAACCCCCGAGCACAGAGCCCACCCAGCCTGACGGGCCCACCACCCTGGGCCCCTTGGATCTGCTGCCCAGTGAGGAGCTATTGAC AGACACAAGTAACTCCTCTTCATCCACTGGGGAGGAAGCGGACTTGGCTGCCCTGCTTCCCGACCTCTCCGGCCGCCTCCTCATCAACTC TGTCTTCCATGTGGGCGCTGAGCGGCTCCAGCAGATGCTCTTCTCGGACTCGCCCTTCCTCCAGGGCTTCCTACAGCAGTGCAAGTTCAC AGACGTGACGCTGAGCCCCTGGAGTGGGGACAGCAAGTGCCACCAGCGCCGGGTGCTGACGTACACCATCCCCATCAGCAACCCACTGGG CCCCAAGAGCGCCTCCGTGGTGGAGACACAGACGCTGTTCCGGCGCGGCCCCCAGGCCGGCGGGTGTGTGGTGGACTCCGAGGTGCTGAC GCAGGGCATCCCCTACCAGGACTACTTCTACACTGCCCACCGCTACTGCATCCTGGGTCTGGCCCGGAACAAGGCGCGGCTCCGAGTGTC TTCTGAGATCCGCTACCGAAAGCAGCCGTGGAGCCTGGTGAAGTCGCTCATTGAGAAGAACTCGTGGAGCGGCATTGAAGACTATTTCCA CCATCTGGAGCGAGAGCTCGCCAAGGCTGAGAAGCTGTCTCTGGAGGAAGGCGGGAAGGATGCCCGGGGCTTGCTATCCGGCCTGCGGCG GCGGAAGCGGCCCCTGAGCTGGCGGGCTCACGGGGACGGGCCCCAGCACCCAGATCCTGACCCCTGTGCCCGGGCCGGCATTCACACCTC GGGCTCCCTCAGCTCCCGCTTCTCCGAACCATCTGTGGACCAGGGCCCCGGGGCAGGCATCCCCAGTGCCCTGGTTCTCATCAGCATTGT CCTTATCATCCTCATCGCCCTCAACGTCCTGCTCTTCTACCGCCTCTGGTCCCTGGAAAGGACAGCCCACACCTTTGAGTCCTGGCACAG CCTGGCCCTGGCCAAGGGCAAGTTCCCCCAGACGGCCACAGAGTGGGCCGAGATCCTGGCGCTGCAGAAGCAATTCCACAGCGTGGAGGT GCACAAGTGGAGGCAGATCCTGCGGGCCTCCGTGGAGCTCCTGGATGAGATGAAGTTCTCGCTGGAGAAGCTGCACCAAGGCATCACAGT CTCAGACCCTCCCTTTGACACCCAGCCCCGGCCCGATGACAGCTTTTCCTGAGGACCCCGGCCACGCAGCTGTTCCCCCACATGGACAGA TGGACACACAGAGCCTCGGCGGCCACTGCTGGCACGGTGTGAGCGCCAGGCATCTCCCACCCGCCCCTCCCGACGGCCCAACCAGGGGCT GTGCAGACGTGGGGACCACGGAACCGAGATGCACTTTAGACCAGGGAGCTGGCCCGGCCTCTGGCAGGCCCCCCACTAACTTATTTTGCC CGGCTGAGGTTGTGGGGGGCGCCTCCTGGGGTGCACGATTCCCTCAGCTCTGGGTTTAATGTATTATATTTATTTGGGGCCGACAGTGCC >64286_64286_4_PEPD-GRAMD1A_PEPD_chr19_33953901_ENST00000436370_GRAMD1A_chr19_35500023_ENST00000599564_length(amino acids)=877AA_BP=159 MAAATGPSFWLGNETLKVPLALFALNRQRLCERLRKNPAVQAGSIVVLQGGEETQRYCTDTGVLFRQIASVLTSQKPSVLLTLRGVNTDS GSVCREASFDGISKFEVNNTILHPEIVECRVFKTDMELEVLRYTNKISSEAHREVMKAVKVGMKEYELESTTPHSGRSTPSSSPSLRKRL QLLPPSRPPPEPEPGTMVEKGSDSSSEKGGVPGTPSTQSLGSRNFIRNSKKMQSWYSMLSPTYKQRNEDFRKLFSKLPEAERLIVDYSCA LQREILLQGRLYLSENWICFYSNIFRWETTISIQLKEVTCLKKEKTAKLIPNAIQICTESEKHFFTSFGARDRCFLLIFRLWQNALLEKT LSPRELWHLVHQCYGSELGLTSEDEDYVSPLQLNGLGTPKEVGDVIALSDITSSGAADRSQEPSPVGSRRGHVTPNLSRASSDADHGAEE DKEEQVDSQPDASSSQTVTPVAEPPSTEPTQPDGPTTLGPLDLLPSEELLTDTSNSSSSTGEEADLAALLPDLSGRLLINSVFHVGAERL QQMLFSDSPFLQGFLQQCKFTDVTLSPWSGDSKCHQRRVLTYTIPISNPLGPKSASVVETQTLFRRGPQAGGCVVDSEVLTQGIPYQDYF YTAHRYCILGLARNKARLRVSSEIRYRKQPWSLVKSLIEKNSWSGIEDYFHHLERELAKAEKLSLEEGGKDARGLLSGLRRRKRPLSWRA HGDGPQHPDPDPCARAGIHTSGSLSSRFSEPSVDQGPGAGIPSALVLISIVLIILIALNVLLFYRLWSLERTAHTFESWHSLALAKGKFP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for PEPD-GRAMD1A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for PEPD-GRAMD1A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for PEPD-GRAMD1A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
