
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:PI4K2B-INSR (FusionGDB2 ID:65056) |
Fusion Gene Summary for PI4K2B-INSR |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: PI4K2B-INSR | Fusion gene ID: 65056 | Hgene | Tgene | Gene symbol | PI4K2B | INSR | Gene ID | 55300 | 3643 |
| Gene name | phosphatidylinositol 4-kinase type 2 beta | insulin receptor | |
| Synonyms | PI4KIIB|PIK42B | CD220|HHF5 | |
| Cytomap | 4p15.2 | 19p13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | phosphatidylinositol 4-kinase type 2-betaPI4KII-BETAphosphatidylinositol 4-kinase type-II beta | insulin receptorIR | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | P06213 | |
| Ensembl transtripts involved in fusion gene | ENST00000264864, ENST00000507794, ENST00000512921, | ENST00000302850, ENST00000341500, | |
| Fusion gene scores | * DoF score | 5 X 5 X 3=75 | 20 X 13 X 8=2080 |
| # samples | 5 | 20 | |
| ** MAII score | log2(5/75*10)=-0.584962500721156 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(20/2080*10)=-3.37851162325373 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: PI4K2B [Title/Abstract] AND INSR [Title/Abstract] AND fusion [Title/Abstract] | ||
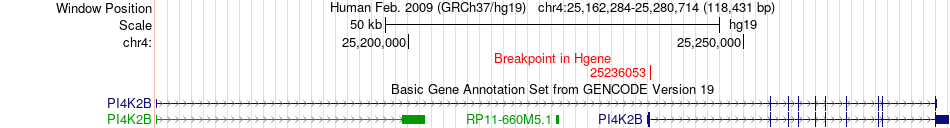
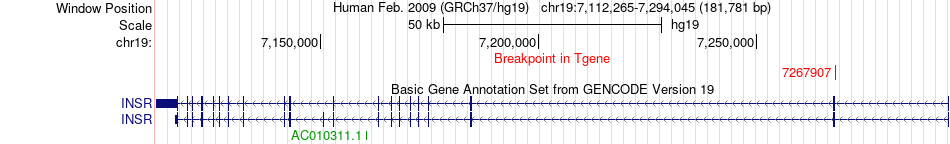
| Most frequent breakpoint | PI4K2B(25236053)-INSR(7267907), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | INSR | GO:0001934 | positive regulation of protein phosphorylation | 7556070 |
| Tgene | INSR | GO:0002092 | positive regulation of receptor internalization | 25401701 |
| Tgene | INSR | GO:0007186 | G protein-coupled receptor signaling pathway | 9092559 |
| Tgene | INSR | GO:0008284 | positive regulation of cell proliferation | 17925406 |
| Tgene | INSR | GO:0008286 | insulin receptor signaling pathway | 6849137|8440175|20455999 |
| Tgene | INSR | GO:0018108 | peptidyl-tyrosine phosphorylation | 8496180 |
| Tgene | INSR | GO:0032148 | activation of protein kinase B activity | 7556070 |
| Tgene | INSR | GO:0032869 | cellular response to insulin stimulus | 8440175 |
| Tgene | INSR | GO:0043410 | positive regulation of MAPK cascade | 20455999 |
| Tgene | INSR | GO:0045725 | positive regulation of glycogen biosynthetic process | 17925406 |
| Tgene | INSR | GO:0046326 | positive regulation of glucose import | 3518947 |
| Tgene | INSR | GO:0046777 | protein autophosphorylation | 6849137|8496180 |
| Tgene | INSR | GO:0060267 | positive regulation of respiratory burst | 9092559 |
 Fusion gene breakpoints across PI4K2B (5'-gene) Fusion gene breakpoints across PI4K2B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across INSR (3'-gene) Fusion gene breakpoints across INSR (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-DX-A1KZ-01A | PI4K2B | chr4 | 25236053 | - | INSR | chr19 | 7267907 | - |
| ChimerDB4 | SARC | TCGA-DX-A1KZ-01A | PI4K2B | chr4 | 25236053 | + | INSR | chr19 | 7267907 | - |
Top |
Fusion Gene ORF analysis for PI4K2B-INSR |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000264864 | ENST00000302850 | PI4K2B | chr4 | 25236053 | + | INSR | chr19 | 7267907 | - |
| In-frame | ENST00000264864 | ENST00000341500 | PI4K2B | chr4 | 25236053 | + | INSR | chr19 | 7267907 | - |
| intron-3CDS | ENST00000507794 | ENST00000302850 | PI4K2B | chr4 | 25236053 | + | INSR | chr19 | 7267907 | - |
| intron-3CDS | ENST00000507794 | ENST00000341500 | PI4K2B | chr4 | 25236053 | + | INSR | chr19 | 7267907 | - |
| intron-3CDS | ENST00000512921 | ENST00000302850 | PI4K2B | chr4 | 25236053 | + | INSR | chr19 | 7267907 | - |
| intron-3CDS | ENST00000512921 | ENST00000341500 | PI4K2B | chr4 | 25236053 | + | INSR | chr19 | 7267907 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000264864 | PI4K2B | chr4 | 25236053 | + | ENST00000341500 | INSR | chr19 | 7267907 | - | 9271 | 457 | 45 | 4469 | 1474 |
| ENST00000264864 | PI4K2B | chr4 | 25236053 | + | ENST00000302850 | INSR | chr19 | 7267907 | - | 4935 | 457 | 45 | 4505 | 1486 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000264864 | ENST00000341500 | PI4K2B | chr4 | 25236053 | + | INSR | chr19 | 7267907 | - | 0.000197072 | 0.999803 |
| ENST00000264864 | ENST00000302850 | PI4K2B | chr4 | 25236053 | + | INSR | chr19 | 7267907 | - | 0.001532939 | 0.998467 |
Top |
Fusion Genomic Features for PI4K2B-INSR |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
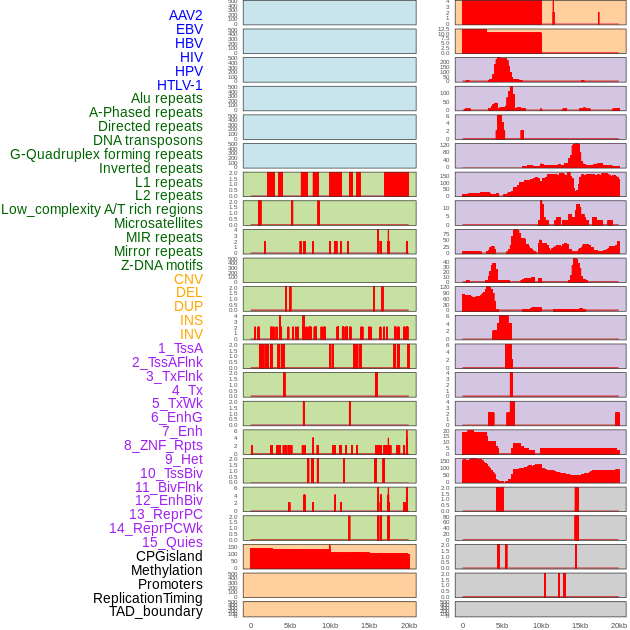
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for PI4K2B-INSR |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr4:25236053/chr19:7267907) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | INSR |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Receptor tyrosine kinase which mediates the pleiotropic actions of insulin. Binding of insulin leads to phosphorylation of several intracellular substrates, including, insulin receptor substrates (IRS1, 2, 3, 4), SHC, GAB1, CBL and other signaling intermediates. Each of these phosphorylated proteins serve as docking proteins for other signaling proteins that contain Src-homology-2 domains (SH2 domain) that specifically recognize different phosphotyrosine residues, including the p85 regulatory subunit of PI3K and SHP2. Phosphorylation of IRSs proteins lead to the activation of two main signaling pathways: the PI3K-AKT/PKB pathway, which is responsible for most of the metabolic actions of insulin, and the Ras-MAPK pathway, which regulates expression of some genes and cooperates with the PI3K pathway to control cell growth and differentiation. Binding of the SH2 domains of PI3K to phosphotyrosines on IRS1 leads to the activation of PI3K and the generation of phosphatidylinositol-(3, 4, 5)-triphosphate (PIP3), a lipid second messenger, which activates several PIP3-dependent serine/threonine kinases, such as PDPK1 and subsequently AKT/PKB. The net effect of this pathway is to produce a translocation of the glucose transporter SLC2A4/GLUT4 from cytoplasmic vesicles to the cell membrane to facilitate glucose transport. Moreover, upon insulin stimulation, activated AKT/PKB is responsible for: anti-apoptotic effect of insulin by inducing phosphorylation of BAD; regulates the expression of gluconeogenic and lipogenic enzymes by controlling the activity of the winged helix or forkhead (FOX) class of transcription factors. Another pathway regulated by PI3K-AKT/PKB activation is mTORC1 signaling pathway which regulates cell growth and metabolism and integrates signals from insulin. AKT mediates insulin-stimulated protein synthesis by phosphorylating TSC2 thereby activating mTORC1 pathway. The Ras/RAF/MAP2K/MAPK pathway is mainly involved in mediating cell growth, survival and cellular differentiation of insulin. Phosphorylated IRS1 recruits GRB2/SOS complex, which triggers the activation of the Ras/RAF/MAP2K/MAPK pathway. In addition to binding insulin, the insulin receptor can bind insulin-like growth factors (IGFI and IGFII). Isoform Short has a higher affinity for IGFII binding. When present in a hybrid receptor with IGF1R, binds IGF1. PubMed:12138094 shows that hybrid receptors composed of IGF1R and INSR isoform Long are activated with a high affinity by IGF1, with low affinity by IGF2 and not significantly activated by insulin, and that hybrid receptors composed of IGF1R and INSR isoform Short are activated by IGF1, IGF2 and insulin. In contrast, PubMed:16831875 shows that hybrid receptors composed of IGF1R and INSR isoform Long and hybrid receptors composed of IGF1R and INSR isoform Short have similar binding characteristics, both bind IGF1 and have a low affinity for insulin. In adipocytes, inhibits lipolysis (By similarity). {ECO:0000250|UniProtKB:P15208, ECO:0000269|PubMed:12138094, ECO:0000269|PubMed:16314505, ECO:0000269|PubMed:16831875, ECO:0000269|PubMed:8257688, ECO:0000269|PubMed:8276809, ECO:0000269|PubMed:8452530, ECO:0000269|PubMed:9428692}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PI4K2B | chr4:25236053 | chr19:7267907 | ENST00000264864 | + | 1 | 10 | 19_22 | 89 | 482.0 | Compositional bias | Note=Poly-Glu |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000302850 | 0 | 22 | 182_339 | 33 | 1383.0 | Compositional bias | Note=Cys-rich | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000341500 | 0 | 21 | 182_339 | 33 | 1371.0 | Compositional bias | Note=Cys-rich | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000302850 | 0 | 22 | 1023_1298 | 33 | 1383.0 | Domain | Protein kinase | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000302850 | 0 | 22 | 624_726 | 33 | 1383.0 | Domain | Fibronectin type-III 1 | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000302850 | 0 | 22 | 757_842 | 33 | 1383.0 | Domain | Fibronectin type-III 2 | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000302850 | 0 | 22 | 853_947 | 33 | 1383.0 | Domain | Fibronectin type-III 3 | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000341500 | 0 | 21 | 1023_1298 | 33 | 1371.0 | Domain | Protein kinase | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000341500 | 0 | 21 | 624_726 | 33 | 1371.0 | Domain | Fibronectin type-III 1 | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000341500 | 0 | 21 | 757_842 | 33 | 1371.0 | Domain | Fibronectin type-III 2 | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000341500 | 0 | 21 | 853_947 | 33 | 1371.0 | Domain | Fibronectin type-III 3 | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000302850 | 0 | 22 | 1104_1110 | 33 | 1383.0 | Nucleotide binding | ATP | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000302850 | 0 | 22 | 1163_1164 | 33 | 1383.0 | Nucleotide binding | ATP | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000341500 | 0 | 21 | 1104_1110 | 33 | 1371.0 | Nucleotide binding | ATP | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000341500 | 0 | 21 | 1163_1164 | 33 | 1371.0 | Nucleotide binding | ATP | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000302850 | 0 | 22 | 1361_1364 | 33 | 1383.0 | Region | Note=PIK3R1-binding | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000302850 | 0 | 22 | 733_741 | 33 | 1383.0 | Region | Note=Insulin-binding | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000341500 | 0 | 21 | 1361_1364 | 33 | 1371.0 | Region | Note=PIK3R1-binding | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000341500 | 0 | 21 | 733_741 | 33 | 1371.0 | Region | Note=Insulin-binding | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000302850 | 0 | 22 | 763_956 | 33 | 1383.0 | Topological domain | Extracellular | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000302850 | 0 | 22 | 980_1382 | 33 | 1383.0 | Topological domain | Cytoplasmic | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000341500 | 0 | 21 | 763_956 | 33 | 1371.0 | Topological domain | Extracellular | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000341500 | 0 | 21 | 980_1382 | 33 | 1371.0 | Topological domain | Cytoplasmic | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000302850 | 0 | 22 | 957_979 | 33 | 1383.0 | Transmembrane | Helical | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000341500 | 0 | 21 | 957_979 | 33 | 1371.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PI4K2B | chr4:25236053 | chr19:7267907 | ENST00000264864 | + | 1 | 10 | 129_436 | 89 | 482.0 | Domain | Note=PI3K/PI4K |
| Hgene | PI4K2B | chr4:25236053 | chr19:7267907 | ENST00000264864 | + | 1 | 10 | 127_133 | 89 | 482.0 | Nucleotide binding | ATP |
| Hgene | PI4K2B | chr4:25236053 | chr19:7267907 | ENST00000264864 | + | 1 | 10 | 257_260 | 89 | 482.0 | Nucleotide binding | ATP |
| Hgene | PI4K2B | chr4:25236053 | chr19:7267907 | ENST00000264864 | + | 1 | 10 | 153_155 | 89 | 482.0 | Region | Important for substrate binding |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000302850 | 0 | 22 | 28_174 | 33 | 1383.0 | Compositional bias | Note=Leu-rich | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000341500 | 0 | 21 | 28_174 | 33 | 1371.0 | Compositional bias | Note=Leu-rich | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000302850 | 0 | 22 | 28_758 | 33 | 1383.0 | Topological domain | Extracellular | |
| Tgene | INSR | chr4:25236053 | chr19:7267907 | ENST00000341500 | 0 | 21 | 28_758 | 33 | 1371.0 | Topological domain | Extracellular |
Top |
Fusion Gene Sequence for PI4K2B-INSR |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >65056_65056_1_PI4K2B-INSR_PI4K2B_chr4_25236053_ENST00000264864_INSR_chr19_7267907_ENST00000302850_length(transcript)=4935nt_BP=457nt GGAGGCGCGAGGTGGGGCGCGGCCGGCGGCGAGGCGGGACAACCGCTGGGCGGGCGCCAAGCGTGCCCGTGCGCTGGTGAGGTGGCGTCC GTTCTACCCGGTCGCTCCCGTTCCGCGCCATGCAGAGCCCAGTCTCTGGCACCTGGCTGCTCTGATCTGGTCTCAGCGCGGAGGGAGCAG AGGGAGTCCATGGAGGATCCCTCCGAGCCCGACCGGTTGGCGTCCGCGGACGGCGGGAGCCCGGAGGAGGAGGAGGATGGGGAGCGGGAG CCGCTGCTACCGCGGATCGCCTGGGCCCACCCGCGGAGAGGCGCCCCAGGCAGCGCCGTGAGGCTGCTGGACGCTGCCGGGGAGGAGGGC GAGGCCGGCGACGAGGAGCTGCCCCTCCCGCCCGGGGACGTGGGGGTCTCCCGGAGTTCGTCCGCCGAGCTGGACCGGAGCCGCCCCGCG GTTTCAGTGTGTCCCGGCATGGATATCCGGAACAACCTCACTAGGTTGCATGAGCTGGAGAATTGCTCTGTCATCGAAGGACACTTGCAG ATACTCTTGATGTTCAAAACGAGGCCCGAAGATTTCCGAGACCTCAGTTTCCCCAAACTCATCATGATCACTGATTACTTGCTGCTCTTC CGGGTCTATGGGCTCGAGAGCCTGAAGGACCTGTTCCCCAACCTCACGGTCATCCGGGGATCACGACTGTTCTTTAACTACGCGCTGGTC ATCTTCGAGATGGTTCACCTCAAGGAACTCGGCCTCTACAACCTGATGAACATCACCCGGGGTTCTGTCCGCATCGAGAAGAACAATGAG CTCTGTTACTTGGCCACTATCGACTGGTCCCGTATCCTGGATTCCGTGGAGGATAATTACATCGTGTTGAACAAAGATGACAACGAGGAG TGTGGAGACATCTGTCCGGGTACCGCGAAGGGCAAGACCAACTGCCCCGCCACCGTCATCAACGGGCAGTTTGTCGAACGATGTTGGACT CATAGTCACTGCCAGAAAGTTTGCCCGACCATCTGTAAGTCACACGGCTGCACCGCCGAAGGCCTCTGTTGCCACAGCGAGTGCCTGGGC AACTGTTCTCAGCCCGACGACCCCACCAAGTGCGTGGCCTGCCGCAACTTCTACCTGGACGGCAGGTGTGTGGAGACCTGCCCGCCCCCG TACTACCACTTCCAGGACTGGCGCTGTGTGAACTTCAGCTTCTGCCAGGACCTGCACCACAAATGCAAGAACTCGCGGAGGCAGGGCTGC CACCAGTACGTCATTCACAACAACAAGTGCATCCCTGAGTGTCCCTCCGGGTACACGATGAATTCCAGCAACTTGCTGTGCACCCCATGC CTGGGTCCCTGTCCCAAGGTGTGCCACCTCCTAGAAGGCGAGAAGACCATCGACTCGGTGACGTCTGCCCAGGAGCTCCGAGGATGCACC GTCATCAACGGGAGTCTGATCATCAACATTCGAGGAGGCAACAATCTGGCAGCTGAGCTAGAAGCCAACCTCGGCCTCATTGAAGAAATT TCAGGGTATCTAAAAATCCGCCGATCCTACGCTCTGGTGTCACTTTCCTTCTTCCGGAAGTTACGTCTGATTCGAGGAGAGACCTTGGAA ATTGGGAACTACTCCTTCTATGCCTTGGACAACCAGAACCTAAGGCAGCTCTGGGACTGGAGCAAACACAACCTCACCATCACTCAGGGG AAACTCTTCTTCCACTATAACCCCAAACTCTGCTTGTCAGAAATCCACAAGATGGAAGAAGTTTCAGGAACCAAGGGGCGCCAGGAGAGA AACGACATTGCCCTGAAGACCAATGGGGACCAGGCATCCTGTGAAAATGAGTTACTTAAATTTTCTTACATTCGGACATCTTTTGACAAG ATCTTGCTGAGATGGGAGCCGTACTGGCCCCCCGACTTCCGAGACCTCTTGGGGTTCATGCTGTTCTACAAAGAGGCCCCTTATCAGAAT GTGACGGAGTTCGACGGGCAGGATGCGTGTGGTTCCAACAGTTGGACGGTGGTAGACATTGACCCACCCCTGAGGTCCAACGACCCCAAA TCACAGAACCACCCAGGGTGGCTGATGCGGGGTCTCAAGCCCTGGACCCAGTATGCCATCTTTGTGAAGACCCTGGTCACCTTTTCGGAT GAACGCCGGACCTATGGGGCCAAGAGTGACATCATTTATGTCCAGACAGATGCCACCAACCCCTCTGTGCCCCTGGATCCAATCTCAGTG TCTAACTCATCATCCCAGATTATTCTGAAGTGGAAACCACCCTCCGACCCCAATGGCAACATCACCCACTACCTGGTTTTCTGGGAGAGG CAGGCGGAAGACAGTGAGCTGTTCGAGCTGGATTATTGCCTCAAAGGGCTGAAGCTGCCCTCGAGGACCTGGTCTCCACCATTCGAGTCT GAAGATTCTCAGAAGCACAACCAGAGTGAGTATGAGGATTCGGCCGGCGAATGCTGCTCCTGTCCAAAGACAGACTCTCAGATCCTGAAG GAGCTGGAGGAGTCCTCGTTTAGGAAGACGTTTGAGGATTACCTGCACAACGTGGTTTTCGTCCCCAGAAAAACCTCTTCAGGCACTGGT GCCGAGGACCCTAGGCCATCTCGGAAACGCAGGTCCCTTGGCGATGTTGGGAATGTGACGGTGGCCGTGCCCACGGTGGCAGCTTTCCCC AACACTTCCTCGACCAGCGTGCCCACGAGTCCGGAGGAGCACAGGCCTTTTGAGAAGGTGGTGAACAAGGAGTCGCTGGTCATCTCCGGC TTGCGACACTTCACGGGCTATCGCATCGAGCTGCAGGCTTGCAACCAGGACACCCCTGAGGAACGGTGCAGTGTGGCAGCCTACGTCAGT GCGAGGACCATGCCTGAAGCCAAGGCTGATGACATTGTTGGCCCTGTGACGCATGAAATCTTTGAGAACAACGTCGTCCACTTGATGTGG CAGGAGCCGAAGGAGCCCAATGGTCTGATCGTGCTGTATGAAGTGAGTTATCGGCGATATGGTGATGAGGAGCTGCATCTCTGCGTCTCC CGCAAGCACTTCGCTCTGGAACGGGGCTGCAGGCTGCGTGGGCTGTCACCGGGGAACTACAGCGTGCGAATCCGGGCCACCTCCCTTGCG GGCAACGGCTCTTGGACGGAACCCACCTATTTCTACGTGACAGACTATTTAGACGTCCCGTCAAATATTGCAAAAATTATCATCGGCCCC CTCATCTTTGTCTTTCTCTTCAGTGTTGTGATTGGAAGTATTTATCTATTCCTGAGAAAGAGGCAGCCAGATGGGCCGCTGGGACCGCTT TACGCTTCTTCAAACCCTGAGTATCTCAGTGCCAGTGATGTGTTTCCATGCTCTGTGTACGTGCCGGACGAGTGGGAGGTGTCTCGAGAG AAGATCACCCTCCTTCGAGAGCTGGGGCAGGGCTCCTTCGGCATGGTGTATGAGGGCAATGCCAGGGACATCATCAAGGGTGAGGCAGAG ACCCGCGTGGCGGTGAAGACGGTCAACGAGTCAGCCAGTCTCCGAGAGCGGATTGAGTTCCTCAATGAGGCCTCGGTCATGAAGGGCTTC ACCTGCCATCACGTGGTGCGCCTCCTGGGAGTGGTGTCCAAGGGCCAGCCCACGCTGGTGGTGATGGAGCTGATGGCTCACGGAGACCTG AAGAGCTACCTCCGTTCTCTGCGGCCAGAGGCTGAGAATAATCCTGGCCGCCCTCCCCCTACCCTTCAAGAGATGATTCAGATGGCGGCA GAGATTGCTGACGGGATGGCCTACCTGAACGCCAAGAAGTTTGTGCATCGGGACCTGGCAGCGAGAAACTGCATGGTCGCCCATGATTTT ACTGTCAAAATTGGAGACTTTGGAATGACCAGAGACATCTATGAAACGGATTACTACCGGAAAGGGGGCAAGGGTCTGCTCCCTGTACGG TGGATGGCACCGGAGTCCCTGAAGGATGGGGTCTTCACCACTTCTTCTGACATGTGGTCCTTTGGCGTGGTCCTTTGGGAAATCACCAGC TTGGCAGAACAGCCTTACCAAGGCCTGTCTAATGAACAGGTGTTGAAATTTGTCATGGATGGAGGGTATCTGGATCAACCCGACAACTGT CCAGAGAGAGTCACTGACCTCATGCGCATGTGCTGGCAATTCAACCCCAAGATGAGGCCAACCTTCCTGGAGATTGTCAACCTGCTCAAG GACGACCTGCACCCCAGCTTTCCAGAGGTGTCGTTCTTCCACAGCGAGGAGAACAAGGCTCCCGAGAGTGAGGAGCTGGAGATGGAGTTT GAGGACATGGAGAATGTGCCCCTGGACCGTTCCTCGCACTGTCAGAGGGAGGAGGCGGGGGGCCGGGATGGAGGGTCCTCGCTGGGTTTC AAGCGGAGCTACGAGGAACACATCCCTTACACACACATGAACGGAGGCAAGAAAAACGGGCGGATTCTGACCTTGCCTCGGTCCAATCCT TCCTAACAGTGCCTACCGTGGCGGGGGCGGGCAGGGGTTCCCATTTTCGCTTTCCTCTGGTTTGAAAGCCTCTGGAAAACTCAGGATTCT CACGACTCTACCATGTCCAATGGAGTTCAGAGATCGTTCCTATACATTTCTGTTCATCTTAAGGTGGACTCGTTTGGTTACCAATTTAAC TAGTCCTGCAGAGGATTTAACTGTGAACCTGGAGGGCAAGGGGTTTCCACAGTTGCTGCTCCTTTGGGGCAACGACGGTTTCAAACCAGG ATTTTGTGTTTTTTCGTTCCCCCCACCCGCCCCCAGCAGATGGAAAGAAAGCACCTGTTTTTACAAATTCTTTTTTTTTTTTTTTTTTTT >65056_65056_1_PI4K2B-INSR_PI4K2B_chr4_25236053_ENST00000264864_INSR_chr19_7267907_ENST00000302850_length(amino acids)=1486AA_BP=137 MGGRQACPCAGEVASVLPGRSRSAPCRAQSLAPGCSDLVSARREQRESMEDPSEPDRLASADGGSPEEEEDGEREPLLPRIAWAHPRRGA PGSAVRLLDAAGEEGEAGDEELPLPPGDVGVSRSSSAELDRSRPAVSVCPGMDIRNNLTRLHELENCSVIEGHLQILLMFKTRPEDFRDL SFPKLIMITDYLLLFRVYGLESLKDLFPNLTVIRGSRLFFNYALVIFEMVHLKELGLYNLMNITRGSVRIEKNNELCYLATIDWSRILDS VEDNYIVLNKDDNEECGDICPGTAKGKTNCPATVINGQFVERCWTHSHCQKVCPTICKSHGCTAEGLCCHSECLGNCSQPDDPTKCVACR NFYLDGRCVETCPPPYYHFQDWRCVNFSFCQDLHHKCKNSRRQGCHQYVIHNNKCIPECPSGYTMNSSNLLCTPCLGPCPKVCHLLEGEK TIDSVTSAQELRGCTVINGSLIINIRGGNNLAAELEANLGLIEEISGYLKIRRSYALVSLSFFRKLRLIRGETLEIGNYSFYALDNQNLR QLWDWSKHNLTITQGKLFFHYNPKLCLSEIHKMEEVSGTKGRQERNDIALKTNGDQASCENELLKFSYIRTSFDKILLRWEPYWPPDFRD LLGFMLFYKEAPYQNVTEFDGQDACGSNSWTVVDIDPPLRSNDPKSQNHPGWLMRGLKPWTQYAIFVKTLVTFSDERRTYGAKSDIIYVQ TDATNPSVPLDPISVSNSSSQIILKWKPPSDPNGNITHYLVFWERQAEDSELFELDYCLKGLKLPSRTWSPPFESEDSQKHNQSEYEDSA GECCSCPKTDSQILKELEESSFRKTFEDYLHNVVFVPRKTSSGTGAEDPRPSRKRRSLGDVGNVTVAVPTVAAFPNTSSTSVPTSPEEHR PFEKVVNKESLVISGLRHFTGYRIELQACNQDTPEERCSVAAYVSARTMPEAKADDIVGPVTHEIFENNVVHLMWQEPKEPNGLIVLYEV SYRRYGDEELHLCVSRKHFALERGCRLRGLSPGNYSVRIRATSLAGNGSWTEPTYFYVTDYLDVPSNIAKIIIGPLIFVFLFSVVIGSIY LFLRKRQPDGPLGPLYASSNPEYLSASDVFPCSVYVPDEWEVSREKITLLRELGQGSFGMVYEGNARDIIKGEAETRVAVKTVNESASLR ERIEFLNEASVMKGFTCHHVVRLLGVVSKGQPTLVVMELMAHGDLKSYLRSLRPEAENNPGRPPPTLQEMIQMAAEIADGMAYLNAKKFV HRDLAARNCMVAHDFTVKIGDFGMTRDIYETDYYRKGGKGLLPVRWMAPESLKDGVFTTSSDMWSFGVVLWEITSLAEQPYQGLSNEQVL KFVMDGGYLDQPDNCPERVTDLMRMCWQFNPKMRPTFLEIVNLLKDDLHPSFPEVSFFHSEENKAPESEELEMEFEDMENVPLDRSSHCQ -------------------------------------------------------------- >65056_65056_2_PI4K2B-INSR_PI4K2B_chr4_25236053_ENST00000264864_INSR_chr19_7267907_ENST00000341500_length(transcript)=9271nt_BP=457nt GGAGGCGCGAGGTGGGGCGCGGCCGGCGGCGAGGCGGGACAACCGCTGGGCGGGCGCCAAGCGTGCCCGTGCGCTGGTGAGGTGGCGTCC GTTCTACCCGGTCGCTCCCGTTCCGCGCCATGCAGAGCCCAGTCTCTGGCACCTGGCTGCTCTGATCTGGTCTCAGCGCGGAGGGAGCAG AGGGAGTCCATGGAGGATCCCTCCGAGCCCGACCGGTTGGCGTCCGCGGACGGCGGGAGCCCGGAGGAGGAGGAGGATGGGGAGCGGGAG CCGCTGCTACCGCGGATCGCCTGGGCCCACCCGCGGAGAGGCGCCCCAGGCAGCGCCGTGAGGCTGCTGGACGCTGCCGGGGAGGAGGGC GAGGCCGGCGACGAGGAGCTGCCCCTCCCGCCCGGGGACGTGGGGGTCTCCCGGAGTTCGTCCGCCGAGCTGGACCGGAGCCGCCCCGCG GTTTCAGTGTGTCCCGGCATGGATATCCGGAACAACCTCACTAGGTTGCATGAGCTGGAGAATTGCTCTGTCATCGAAGGACACTTGCAG ATACTCTTGATGTTCAAAACGAGGCCCGAAGATTTCCGAGACCTCAGTTTCCCCAAACTCATCATGATCACTGATTACTTGCTGCTCTTC CGGGTCTATGGGCTCGAGAGCCTGAAGGACCTGTTCCCCAACCTCACGGTCATCCGGGGATCACGACTGTTCTTTAACTACGCGCTGGTC ATCTTCGAGATGGTTCACCTCAAGGAACTCGGCCTCTACAACCTGATGAACATCACCCGGGGTTCTGTCCGCATCGAGAAGAACAATGAG CTCTGTTACTTGGCCACTATCGACTGGTCCCGTATCCTGGATTCCGTGGAGGATAATTACATCGTGTTGAACAAAGATGACAACGAGGAG TGTGGAGACATCTGTCCGGGTACCGCGAAGGGCAAGACCAACTGCCCCGCCACCGTCATCAACGGGCAGTTTGTCGAACGATGTTGGACT CATAGTCACTGCCAGAAAGTTTGCCCGACCATCTGTAAGTCACACGGCTGCACCGCCGAAGGCCTCTGTTGCCACAGCGAGTGCCTGGGC AACTGTTCTCAGCCCGACGACCCCACCAAGTGCGTGGCCTGCCGCAACTTCTACCTGGACGGCAGGTGTGTGGAGACCTGCCCGCCCCCG TACTACCACTTCCAGGACTGGCGCTGTGTGAACTTCAGCTTCTGCCAGGACCTGCACCACAAATGCAAGAACTCGCGGAGGCAGGGCTGC CACCAGTACGTCATTCACAACAACAAGTGCATCCCTGAGTGTCCCTCCGGGTACACGATGAATTCCAGCAACTTGCTGTGCACCCCATGC CTGGGTCCCTGTCCCAAGGTGTGCCACCTCCTAGAAGGCGAGAAGACCATCGACTCGGTGACGTCTGCCCAGGAGCTCCGAGGATGCACC GTCATCAACGGGAGTCTGATCATCAACATTCGAGGAGGCAACAATCTGGCAGCTGAGCTAGAAGCCAACCTCGGCCTCATTGAAGAAATT TCAGGGTATCTAAAAATCCGCCGATCCTACGCTCTGGTGTCACTTTCCTTCTTCCGGAAGTTACGTCTGATTCGAGGAGAGACCTTGGAA ATTGGGAACTACTCCTTCTATGCCTTGGACAACCAGAACCTAAGGCAGCTCTGGGACTGGAGCAAACACAACCTCACCATCACTCAGGGG AAACTCTTCTTCCACTATAACCCCAAACTCTGCTTGTCAGAAATCCACAAGATGGAAGAAGTTTCAGGAACCAAGGGGCGCCAGGAGAGA AACGACATTGCCCTGAAGACCAATGGGGACCAGGCATCCTGTGAAAATGAGTTACTTAAATTTTCTTACATTCGGACATCTTTTGACAAG ATCTTGCTGAGATGGGAGCCGTACTGGCCCCCCGACTTCCGAGACCTCTTGGGGTTCATGCTGTTCTACAAAGAGGCCCCTTATCAGAAT GTGACGGAGTTCGACGGGCAGGATGCGTGTGGTTCCAACAGTTGGACGGTGGTAGACATTGACCCACCCCTGAGGTCCAACGACCCCAAA TCACAGAACCACCCAGGGTGGCTGATGCGGGGTCTCAAGCCCTGGACCCAGTATGCCATCTTTGTGAAGACCCTGGTCACCTTTTCGGAT GAACGCCGGACCTATGGGGCCAAGAGTGACATCATTTATGTCCAGACAGATGCCACCAACCCCTCTGTGCCCCTGGATCCAATCTCAGTG TCTAACTCATCATCCCAGATTATTCTGAAGTGGAAACCACCCTCCGACCCCAATGGCAACATCACCCACTACCTGGTTTTCTGGGAGAGG CAGGCGGAAGACAGTGAGCTGTTCGAGCTGGATTATTGCCTCAAAGGGCTGAAGCTGCCCTCGAGGACCTGGTCTCCACCATTCGAGTCT GAAGATTCTCAGAAGCACAACCAGAGTGAGTATGAGGATTCGGCCGGCGAATGCTGCTCCTGTCCAAAGACAGACTCTCAGATCCTGAAG GAGCTGGAGGAGTCCTCGTTTAGGAAGACGTTTGAGGATTACCTGCACAACGTGGTTTTCGTCCCCAGGCCATCTCGGAAACGCAGGTCC CTTGGCGATGTTGGGAATGTGACGGTGGCCGTGCCCACGGTGGCAGCTTTCCCCAACACTTCCTCGACCAGCGTGCCCACGAGTCCGGAG GAGCACAGGCCTTTTGAGAAGGTGGTGAACAAGGAGTCGCTGGTCATCTCCGGCTTGCGACACTTCACGGGCTATCGCATCGAGCTGCAG GCTTGCAACCAGGACACCCCTGAGGAACGGTGCAGTGTGGCAGCCTACGTCAGTGCGAGGACCATGCCTGAAGCCAAGGCTGATGACATT GTTGGCCCTGTGACGCATGAAATCTTTGAGAACAACGTCGTCCACTTGATGTGGCAGGAGCCGAAGGAGCCCAATGGTCTGATCGTGCTG TATGAAGTGAGTTATCGGCGATATGGTGATGAGGAGCTGCATCTCTGCGTCTCCCGCAAGCACTTCGCTCTGGAACGGGGCTGCAGGCTG CGTGGGCTGTCACCGGGGAACTACAGCGTGCGAATCCGGGCCACCTCCCTTGCGGGCAACGGCTCTTGGACGGAACCCACCTATTTCTAC GTGACAGACTATTTAGACGTCCCGTCAAATATTGCAAAAATTATCATCGGCCCCCTCATCTTTGTCTTTCTCTTCAGTGTTGTGATTGGA AGTATTTATCTATTCCTGAGAAAGAGGCAGCCAGATGGGCCGCTGGGACCGCTTTACGCTTCTTCAAACCCTGAGTATCTCAGTGCCAGT GATGTGTTTCCATGCTCTGTGTACGTGCCGGACGAGTGGGAGGTGTCTCGAGAGAAGATCACCCTCCTTCGAGAGCTGGGGCAGGGCTCC TTCGGCATGGTGTATGAGGGCAATGCCAGGGACATCATCAAGGGTGAGGCAGAGACCCGCGTGGCGGTGAAGACGGTCAACGAGTCAGCC AGTCTCCGAGAGCGGATTGAGTTCCTCAATGAGGCCTCGGTCATGAAGGGCTTCACCTGCCATCACGTGGTGCGCCTCCTGGGAGTGGTG TCCAAGGGCCAGCCCACGCTGGTGGTGATGGAGCTGATGGCTCACGGAGACCTGAAGAGCTACCTCCGTTCTCTGCGGCCAGAGGCTGAG AATAATCCTGGCCGCCCTCCCCCTACCCTTCAAGAGATGATTCAGATGGCGGCAGAGATTGCTGACGGGATGGCCTACCTGAACGCCAAG AAGTTTGTGCATCGGGACCTGGCAGCGAGAAACTGCATGGTCGCCCATGATTTTACTGTCAAAATTGGAGACTTTGGAATGACCAGAGAC ATCTATGAAACGGATTACTACCGGAAAGGGGGCAAGGGTCTGCTCCCTGTACGGTGGATGGCACCGGAGTCCCTGAAGGATGGGGTCTTC ACCACTTCTTCTGACATGTGGTCCTTTGGCGTGGTCCTTTGGGAAATCACCAGCTTGGCAGAACAGCCTTACCAAGGCCTGTCTAATGAA CAGGTGTTGAAATTTGTCATGGATGGAGGGTATCTGGATCAACCCGACAACTGTCCAGAGAGAGTCACTGACCTCATGCGCATGTGCTGG CAATTCAACCCCAAGATGAGGCCAACCTTCCTGGAGATTGTCAACCTGCTCAAGGACGACCTGCACCCCAGCTTTCCAGAGGTGTCGTTC TTCCACAGCGAGGAGAACAAGGCTCCCGAGAGTGAGGAGCTGGAGATGGAGTTTGAGGACATGGAGAATGTGCCCCTGGACCGTTCCTCG CACTGTCAGAGGGAGGAGGCGGGGGGCCGGGATGGAGGGTCCTCGCTGGGTTTCAAGCGGAGCTACGAGGAACACATCCCTTACACACAC ATGAACGGAGGCAAGAAAAACGGGCGGATTCTGACCTTGCCTCGGTCCAATCCTTCCTAACAGTGCCTACCGTGGCGGGGGCGGGCAGGG GTTCCCATTTTCGCTTTCCTCTGGTTTGAAAGCCTCTGGAAAACTCAGGATTCTCACGACTCTACCATGTCCAATGGAGTTCAGAGATCG TTCCTATACATTTCTGTTCATCTTAAGGTGGACTCGTTTGGTTACCAATTTAACTAGTCCTGCAGAGGATTTAACTGTGAACCTGGAGGG CAAGGGGTTTCCACAGTTGCTGCTCCTTTGGGGCAACGACGGTTTCAAACCAGGATTTTGTGTTTTTTCGTTCCCCCCACCCGCCCCCAG CAGATGGAAAGAAAGCACCTGTTTTTACAAATTCTTTTTTTTTTTTTTTTTTTTTGCTGGTGTCTGAGCTTCAGTATAAAAGACAAAACT TCCTGTTTGTGGAACAAAAGTTCGAAAGAAAAAACAAAACAAAAACACCCAGCCCTGTTCCAGGAGAATTTCAAGTTTTACAGGTTGAGC TTCAAGATGGTTTTTTTGGTTTTTTTTTTTTCTCTCATCCAGGCTGAAGGATTTTTTTTTTCTTTACAAAATGAGTTCCTCAAATTGACC AATAGCTGCTGCTTTCATATTTTGGATAAGGGTCTGTGGTCCCGGCGTGTGCTCACGTGTGTATGCACGTGTGTGTGTCCATTAGACACG GCTGATGTGTGTGCAAAGTATCCATGCGGAGTTGATGCTTTGGGAATTGGCTCATGAAGGTTCTTCTCAAGGGTGCGAGCTCATCCCCCT CTCTCCTTCCTTCTTATTGACTGGGAGACTGTGCTCTCGACAGATTCTTCTTGTGTCAGAAGTCTAGCCTCAGGTTTCTACCCTCCCTTC ACATTGGTGGCCAAGGGAGGAGCATTTCATTTGGAGTGATTATGAATCTTTTCAAGACCAAACCAAGCTAGGACATTAAAAAAAAAAAAA GAAAAAGAAAGAAAAAACAAAATGGAAAAAGGAAAAAAAAAAAGAACTGAGATGACAGAGTTTTGAGAATATATTTGTACCATATTTAAT TTTTAAAGTCTCTGGTATTAGCCTCATAAGTTATTGACTATTCCCCGGGGTTGGCGGGGAGTGGGGACATGAGTTGGTCTGCCTGTTGTG GGGCCGGGAAGGGGAGGGAGTCAGGCACAAGTGGCCTCTTTGTTTGGTCTTAAAGGCATCCATTTCTGGGAATGAAGCCATGTTCGCTGC TAACACTTTTGGATGTTGTGAGGCCACGTGGAGTGTGTGAGAGACTAGGTTTTATGGATGGTCTGGTTCAGGTACCAGGTCTGCTGGAAG GTTCCTGTTCGGATAAGCTGGTAGCTACCTAGCTCTGAGCCTGCCTTCAAGAACACCTGTGTTCATCCTCTGATTCTCTGTGTGTACCTC TTGTGGCGTTTCCTCTCCCGGGTGTGAACATCCTAACCGTTATTGTGCAAACCCAAGAACGTCAGATCCCAAAGCACAACAACCTGGATG GACTTTGGGAACATCTAAGCAATGTAAGAGAGAGGTGCACTGAGAGTACGTCTTGGTCCCCTCCACCCTGAGAGCATCTGACGGTCCTCA GTACTGAACTCCCGGAAGCTGCTCTGAGCCCGGTGACCTCATCTGGGCCAGGTGTGGTGCCTGAGCTGAATGCTCAGGTGCTTACAGTGT TGCAATCCCTAAGAGAGTAGAGTCTGGAGGAGAAACCGTGAAAAAGACCTTACACACCACCAAGAACTTCCGAATGGGCGTGAATCCACC GTTTCTTCTCTTTGCAAAAAGAACCACCACAGCTGCTCAAAGAACACAGTGAACTCATCACTTTGGTTCATCAAAAAATCATCGCCCATG CGTTATTCCTGAGTGCATTTTCTTACAACTTTTTGACTGCTTCCTTTTCTTCTTCTCTTAAGAGTTGTGGGCTTAAGAATGGGATAGAGT CATAATGGCAACCTCCAAGCCCTCTCAATTCTTGATTAAGAACACAGGTAGACATGAATCCCAATTGTCTATTGCTATCTTATTTATATG ATTCGGGAAAATACAGCATGTAAAAATATTGCTGAGGAGCCTCAGTGATTGGGTACAAGAAGCAAGAGTACAGAAATTATTTTTGCCAAA TTTATTTTGTAAATATGAGGGTCTGTACCTAAATTTAAAAAAAAAACACGTAGAACTAGGTATTTTGTTCTCTTCTTAGTAAATTTGTAG TGGTTGTATACTACACTAGCTGCAATTTTCACATTTTTCTAATTCAGAAAGGTTTTTCTTATATTAGGGGAAAAAGTATTTATTTTAATA TATAAAATCACTCTGAAAATCACTCTCATAAAAAATGGAGCGCATGTAAATTTTTATCAAAGAAAAATAAACAGGTGAATGGGGGATAGT GATTTTCTTTTTTCAGCACAGTCTACCTCAGTGTATTGTTAAGATGTGATTCAATCATGGACATCTTTGAGATTTCAGAATTCTACCTGG AACCGGTCTGAATCAGGGAACGTGTGTATCAGCTGATTCGAATGCCAGGGACCAGTAAGAATTTTGAGGGAGGGAGTTGGGATGGAGAAG GTATGGCCTTTATGCGAGCATAGATCCTTTTCTTCCTGGCTGGTAATATTCTTCTCTGAATTTAATCTTCCTTTAAAAAAAAATCCTCCA TCTATTGTCACTATGTTCCCCAAACATAAACTAAGTTCCAGGCTGTCATGATGTATCTGATATATGGGGTAACCCAGCAAGGTGTACCTT CCTTTGGTGAGAGATGGCTGCCGGGGCAAAGACGGGCTTTGATTCAGAGCAAGCATTCCCACCTGTTCCATGGAATCCCCCTGAAGTGAG CACAAAGGTGCCCTGGGCTCCCTGATGGTTTATGCCCACTCCTTTCAGGCTGGTGATGCACCTTACACACAAACACCTAATGCAATGTCT TTTTAAATTCTCCAAGTGGGATGGGAGCATGTGAGGGAAATTCCAATCCAAAACCCATTAATGTGCTGAACGCTTTTTTTTTTTTTTTTT TTTTTTTTGCAACAACACCTTGGACCTCTGTGTTGGGGTTTGACTGACCTCAAGCTGATATTATTGGACCTTGTGCAGCTTTGATAACCC ATGTGAGAGTCTAGGCAGGACCAGTGGGGCCCAAATCTTGCTGCTCTTGTACTTTTAGGCACTGCCCTTGCAGACTCACCTTTCTCCACC TGCCCTGGAGAAAGGTAGGGTGTGCTGGGCCTGCCCCTTGCAAATGGGATTCACCAGTTTCATTTATTTGACTCTACTGCCACAGTGAAA AGAGCAAACAGCTATTGGGTTGCAAACCTCCTTTGACATTAGGAAATGTTGACTTTGTAACAATAAAACTTTGGTCCTAGAAAGACACGG TTGTCCTGGGAGTTTGTAGTGTTAAGTTGCAACAACAACAACAAAAAGCAACAAAACCAGCTTAGGATAACACTTTTTGTTGCTTGTTCT TAAAGATGTCTCACTATGATTAAAACCCTTTTCATTAATGTAGTGAAAGCCACACAGGAGTTCCTTCTTCCAGGAGGAGAATACCAAGCA CATCACTTTCTCTCTGCATCAGTGATGTCAAATACGCATCAGAAAATGTTCAGGTTTTAGGAGCTGTCCTAGGTGCTGTTTCATCATTGG AAGCAGTGAGAAAGAGAAGCACTGCTGCTTGTCTGGATATAGGCTGAGGATGATTGAGAGAAGCTGTGGGAACTGACACAAGGGTCTGCA TAGGTCATCCTGTGACCCTGGGGACTATGTTACCAACTGACAGACAGATCTTTCACTGTATCCTAGCAGGGCAGGTAGTCCACCAAGAAA TGTGCTTATTGGATTGGGAGGTGTTTATTTGTAGTCTGCTGTAACACGTGTGAAAGAGCAGGAGCGTCATCAGCATATGACTTGCGCTGG TCATCCGGTAAATGGATGTGCTGTAGTCCCAGTGCTAATCATTTCTCTCCTTCACAGTGGGTGGAAGTTTAGGGTTAAATGTCCTTTGAA TGTCACCTGGTGAGTCCTTGACACCTTAGGCTCTTCAGAAACAATGGTTTTGTTGAGGATGGGGAACAGGGAATGCCGATTTTATATACA TGGTACACAGAGAGGGGTGTCACTTCAGAAAATCTTCCAGCATGTTCTTCAGAATATTAATTTATATGCGAGGTGAGGTTGGGAATGAAA AGAACAGGTCAGCACTTTTTTTTTTCCTAGAACATACAAAAGAACATGGTGGACTTTCAGGGAGTGCAATGGAAGGTGAATATTTCCTTA AGGGTCCCCGAGAAATGGGAGTGAGGGGAGGGGACACAATGGCTTTTTGAGCTTACTTTTACCTTCTGATACTAGTCAAGGTCCAGAACC AGCCACCAGCCAAATTTCTATCTGGGTGCGGGCCACTGAAAATCCTTGTTAAAAACCAGATCACAAATCTGGGGCTCTTGGTCCCATTGG AGAAGGAAGGAAGAGCCTCAAAATAAGTGTGCACCCATGCACATATTCAGGAACAGCTTGTTTAGTCTTTACACTTTGCCTGAAAGTTGC TTCTCCTCGTCCCTTTGTGTGCCTGGGTGGCCTCGGCCCTGTGCGTTGGCAACGCAGGATCAAATGTGCTGCAGCTTTTGCAGAAAACAA CTCAGAAACACAAAACCCCCCAACAGCTCAATTATTATTTTTTCAATGTTTTCCTACAAGAGCCAAGTAGCACCATGTACAGAAGACGCC TTTTTTTTTGGAATATTGAAATCGTTCTGCATGTAAAATATGGGATAATGACCTGTTTATATTAAAATTCTGATTAAATTATCTGAGAAT >65056_65056_2_PI4K2B-INSR_PI4K2B_chr4_25236053_ENST00000264864_INSR_chr19_7267907_ENST00000341500_length(amino acids)=1474AA_BP=137 MGGRQACPCAGEVASVLPGRSRSAPCRAQSLAPGCSDLVSARREQRESMEDPSEPDRLASADGGSPEEEEDGEREPLLPRIAWAHPRRGA PGSAVRLLDAAGEEGEAGDEELPLPPGDVGVSRSSSAELDRSRPAVSVCPGMDIRNNLTRLHELENCSVIEGHLQILLMFKTRPEDFRDL SFPKLIMITDYLLLFRVYGLESLKDLFPNLTVIRGSRLFFNYALVIFEMVHLKELGLYNLMNITRGSVRIEKNNELCYLATIDWSRILDS VEDNYIVLNKDDNEECGDICPGTAKGKTNCPATVINGQFVERCWTHSHCQKVCPTICKSHGCTAEGLCCHSECLGNCSQPDDPTKCVACR NFYLDGRCVETCPPPYYHFQDWRCVNFSFCQDLHHKCKNSRRQGCHQYVIHNNKCIPECPSGYTMNSSNLLCTPCLGPCPKVCHLLEGEK TIDSVTSAQELRGCTVINGSLIINIRGGNNLAAELEANLGLIEEISGYLKIRRSYALVSLSFFRKLRLIRGETLEIGNYSFYALDNQNLR QLWDWSKHNLTITQGKLFFHYNPKLCLSEIHKMEEVSGTKGRQERNDIALKTNGDQASCENELLKFSYIRTSFDKILLRWEPYWPPDFRD LLGFMLFYKEAPYQNVTEFDGQDACGSNSWTVVDIDPPLRSNDPKSQNHPGWLMRGLKPWTQYAIFVKTLVTFSDERRTYGAKSDIIYVQ TDATNPSVPLDPISVSNSSSQIILKWKPPSDPNGNITHYLVFWERQAEDSELFELDYCLKGLKLPSRTWSPPFESEDSQKHNQSEYEDSA GECCSCPKTDSQILKELEESSFRKTFEDYLHNVVFVPRPSRKRRSLGDVGNVTVAVPTVAAFPNTSSTSVPTSPEEHRPFEKVVNKESLV ISGLRHFTGYRIELQACNQDTPEERCSVAAYVSARTMPEAKADDIVGPVTHEIFENNVVHLMWQEPKEPNGLIVLYEVSYRRYGDEELHL CVSRKHFALERGCRLRGLSPGNYSVRIRATSLAGNGSWTEPTYFYVTDYLDVPSNIAKIIIGPLIFVFLFSVVIGSIYLFLRKRQPDGPL GPLYASSNPEYLSASDVFPCSVYVPDEWEVSREKITLLRELGQGSFGMVYEGNARDIIKGEAETRVAVKTVNESASLRERIEFLNEASVM KGFTCHHVVRLLGVVSKGQPTLVVMELMAHGDLKSYLRSLRPEAENNPGRPPPTLQEMIQMAAEIADGMAYLNAKKFVHRDLAARNCMVA HDFTVKIGDFGMTRDIYETDYYRKGGKGLLPVRWMAPESLKDGVFTTSSDMWSFGVVLWEITSLAEQPYQGLSNEQVLKFVMDGGYLDQP DNCPERVTDLMRMCWQFNPKMRPTFLEIVNLLKDDLHPSFPEVSFFHSEENKAPESEELEMEFEDMENVPLDRSSHCQREEAGGRDGGSS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for PI4K2B-INSR |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for PI4K2B-INSR |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for PI4K2B-INSR |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
