
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ARID5B-NRBF2 (FusionGDB2 ID:6508) |
Fusion Gene Summary for ARID5B-NRBF2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ARID5B-NRBF2 | Fusion gene ID: 6508 | Hgene | Tgene | Gene symbol | ARID5B | NRBF2 | Gene ID | 84159 | 29982 |
| Gene name | AT-rich interaction domain 5B | nuclear receptor binding factor 2 | |
| Synonyms | DESRT|MRF-2|MRF2 | COPR|COPR1|COPR2|NRBF-2 | |
| Cytomap | 10q21.2 | 10q21.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | AT-rich interactive domain-containing protein 5BARID domain-containing protein 5BAT-rich interactive domain 5B (MRF1-like)MRF1-like proteinmodulator recognition factor 2 (MRF2) | nuclear receptor-binding factor 2comodulator of PPAR and RXR 1comodulator of PPAR and RXR 2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q14865 | Q96F24 | |
| Ensembl transtripts involved in fusion gene | ENST00000279873, ENST00000309334, | ENST00000435510, ENST00000277746, | |
| Fusion gene scores | * DoF score | 18 X 15 X 7=1890 | 4 X 5 X 3=60 |
| # samples | 18 | 5 | |
| ** MAII score | log2(18/1890*10)=-3.39231742277876 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/60*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ARID5B [Title/Abstract] AND NRBF2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ARID5B(63810759)-NRBF2(64905968), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ARID5B | GO:0000122 | negative regulation of transcription by RNA polymerase II | 8649988 |
| Hgene | ARID5B | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 21532585 |
 Fusion gene breakpoints across ARID5B (5'-gene) Fusion gene breakpoints across ARID5B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
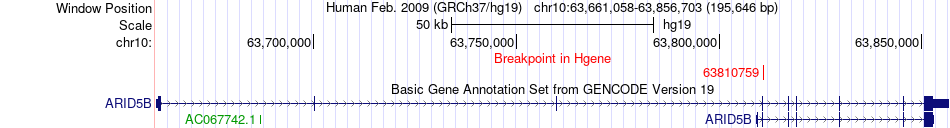 |
 Fusion gene breakpoints across NRBF2 (3'-gene) Fusion gene breakpoints across NRBF2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
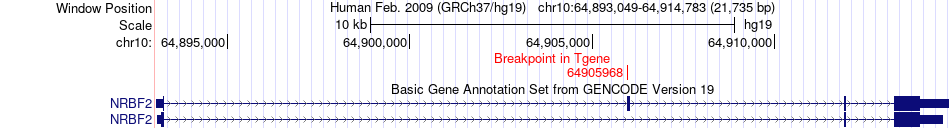 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-FP-A4BE | ARID5B | chr10 | 63810759 | + | NRBF2 | chr10 | 64905968 | + |
Top |
Fusion Gene ORF analysis for ARID5B-NRBF2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000279873 | ENST00000435510 | ARID5B | chr10 | 63810759 | + | NRBF2 | chr10 | 64905968 | + |
| 5CDS-intron | ENST00000309334 | ENST00000435510 | ARID5B | chr10 | 63810759 | + | NRBF2 | chr10 | 64905968 | + |
| In-frame | ENST00000279873 | ENST00000277746 | ARID5B | chr10 | 63810759 | + | NRBF2 | chr10 | 64905968 | + |
| In-frame | ENST00000309334 | ENST00000277746 | ARID5B | chr10 | 63810759 | + | NRBF2 | chr10 | 64905968 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000279873 | ARID5B | chr10 | 63810759 | + | ENST00000277746 | NRBF2 | chr10 | 64905968 | + | 2895 | 1256 | 410 | 2089 | 559 |
| ENST00000309334 | ARID5B | chr10 | 63810759 | + | ENST00000277746 | NRBF2 | chr10 | 64905968 | + | 2025 | 386 | 161 | 1219 | 352 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000279873 | ENST00000277746 | ARID5B | chr10 | 63810759 | + | NRBF2 | chr10 | 64905968 | + | 0.002422288 | 0.9975777 |
| ENST00000309334 | ENST00000277746 | ARID5B | chr10 | 63810759 | + | NRBF2 | chr10 | 64905968 | + | 0.001147095 | 0.99885285 |
Top |
Fusion Genomic Features for ARID5B-NRBF2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ARID5B | chr10 | 63810759 | + | NRBF2 | chr10 | 64905968 | + | 0.043421455 | 0.9565785 |
| ARID5B | chr10 | 63810759 | + | NRBF2 | chr10 | 64905968 | + | 0.043421455 | 0.9565785 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
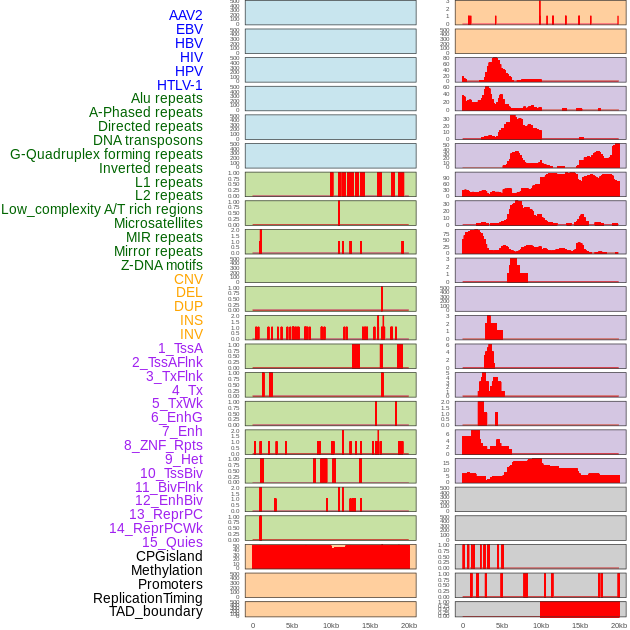 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
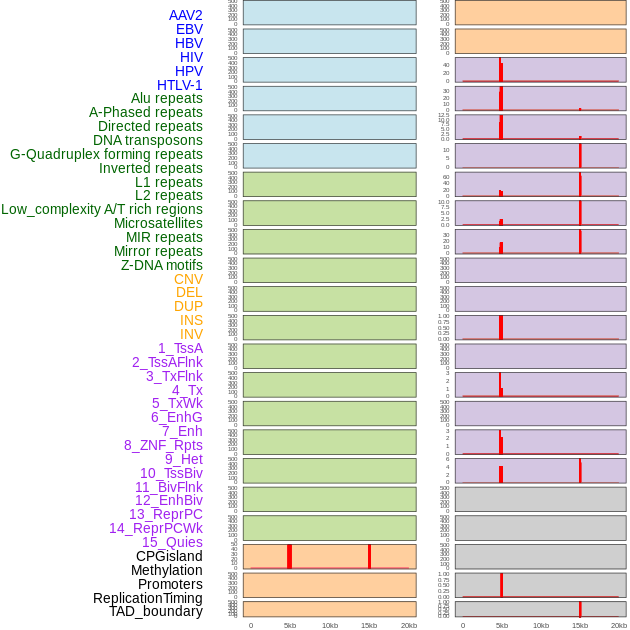 |
Top |
Fusion Protein Features for ARID5B-NRBF2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:63810759/chr10:64905968) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ARID5B | NRBF2 |
| FUNCTION: Transcription coactivator that binds to the 5'-AATA[CT]-3' core sequence and plays a key role in adipogenesis and liver development. Acts by forming a complex with phosphorylated PHF2, which mediates demethylation at Lys-336, leading to target the PHF2-ARID5B complex to target promoters, where PHF2 mediates demethylation of dimethylated 'Lys-9' of histone H3 (H3K9me2), followed by transcription activation of target genes. The PHF2-ARID5B complex acts as a coactivator of HNF4A in liver. Required for adipogenesis: regulates triglyceride metabolism in adipocytes by regulating expression of adipogenic genes. Overexpression leads to induction of smooth muscle marker genes, suggesting that it may also act as a regulator of smooth muscle cell differentiation and proliferation. Represses the cytomegalovirus enhancer. {ECO:0000269|PubMed:21532585}. | FUNCTION: May modulate transcriptional activation by target nuclear receptors. Can act as transcriptional activator (in vitro). {ECO:0000269|PubMed:15610520}.; FUNCTION: Involved in starvation-induced autophagy probably by its association with PI3K complex I (PI3KC3-C1). However, effects has been described variably. Involved in the induction of starvation-induced autophagy (PubMed:24785657). Stabilzes PI3KC3-C1 assembly and enhances ATG14-linked lipid kinase activity of PIK3C3 (By similarity). Proposed to negatively regulate basal and starvation-induced autophagy and to inhibit PIK3C3 activity by modulating interactions in PI3KC3-C1 (PubMed:25086043). May be involved in autophagosome biogenesis (PubMed:25086043). May play a role in neural progenitor cell survival during differentiation (By similarity). {ECO:0000250|UniProtKB:Q8VCQ3, ECO:0000269|PubMed:24785657, ECO:0000269|PubMed:25086043}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | NRBF2 | chr10:63810759 | chr10:64905968 | ENST00000277746 | 0 | 4 | 168_209 | 10 | 288.0 | Coiled coil | Ontology_term=ECO:0000255 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ARID5B | chr10:63810759 | chr10:64905968 | ENST00000279873 | + | 5 | 10 | 318_410 | 282 | 1189.0 | Domain | ARID |
| Hgene | ARID5B | chr10:63810759 | chr10:64905968 | ENST00000309334 | + | 2 | 7 | 318_410 | 39 | 946.0 | Domain | ARID |
Top |
Fusion Gene Sequence for ARID5B-NRBF2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >6508_6508_1_ARID5B-NRBF2_ARID5B_chr10_63810759_ENST00000279873_NRBF2_chr10_64905968_ENST00000277746_length(transcript)=2895nt_BP=1256nt ATCAGACGATATCCCAGACAGGAGCGGTTAGAGAGAGAGGAATCACATCTCCACACAGTTTTAGGGTGCTTTTTATTTTTACAAATCTTC TTGTGTGTTTTTTGCCTTGATCCATCCTCTTCCCGCCGAGATCGTATGGCGCCTTTCTCTCGATTATGAATTTGATCAATCCATCTTTGG AAGAAAACCCACATAGTTTTTTCAGGAGCTGAAAATTGAGTCGTTATAGAAATATTAGGACATATTTTCAATCATTTCGGTGCCCGAAGG GAGGCAAGAGCTCAGTTTTATATTGAGACATTACGCCGGCTGAAGGCAGAGAATGCGTTTCCCTGCCAGGACCTGATGCAATCCATTCAA GCCAACAAGTTTGGAGAGAATGTTGAGTTCAATCAATTCAGAACGTCGAGATGGAGCCCAACTCACTCCAGTGGGTCGGCTCACCGTGTG GCTTGCACGGACCTTACATTTTCTACAAGGCTTTTCAATTCCACCTTGAAGGCAAACCAAGAATTTTGTCCCTTGGCGACTTTTTCTTTG TAAGATGTACGCCAAAGGATCCGATTTGCATAGCGGAGCTCCAGCTGTTGTGGGAAGAGAGGACCAGCCGGCAACTTTTATCCAGCTCTA AACTTTATTTCCTCCCAGAAGACACTCCCCAGGGCAGAAATAGCGACCATGGCGAGGATGAAGTCATTGCTGTTTCCGAAAAGGTGATTG TGAAGCTTGAAGACCTGGTCAAGTGGGTACATTCTGATTTCTCCAAGTGGAGATGTGGCTTCCACGCTGGACCAGTGAAAACTGAGGCCT TGGGAAGGAATGGACAGAAGGAAGCTCTGCTGAAGTACAGGCAGTCAACCCTAAACAGTGGACTCAACTTCAAAGACGTTCTCAAGGAGA AGGCAGACCTGGGGGAGGACGAGGAAGAAACGAACGTGATAGTTCTCAGCTACCCCCAGTACTGCCGGTACCGCTCGATGCTGAAACGCA TCCAGGATAAGCCATCTTCCATTCTAACGGACCAGTTTGCATTGGCCCTGGGGGGCATTGCAGTGGTCAGCAGGAACCCTCAGATCCTGT ACTGTCGGGACACCTTTGACCACCCGACTCTCATAGAAAACGAGAGTATATGCGATGAGTTTGCGCCAAATCTTAAAGGCAGACCACGCA AAAAGAAACCATGCCCACAAAGAAGAGATTCATTCAGTGGTGTTAAGGATTCCAACAACAATTCCGATGGCAAAGCCGTTGCCAAGGCTC ATCAACAGAGCAGACGAGCAGACCGTTTATTAGCTGCAGGCAAATACGAAGAGGCTATTTCTTGTCACAAAAAGGCTGCAGCATATCTTT CTGAAGCCATGAAGCTGACACAGTCAGAGCAGGCTCATCTTTCACTGGAATTGCAAAGGGATAGCCATATGAAACAGCTCCTCCTCATCC AAGAGAGATGGAAAAGGGCCCAGCGTGAAGAAAGATTGAAAGCCCAGCAGAACACAGACAAGGATGCAGCTGCCCATCTTCAGACATCTC ACAAACCCTCTGCAGAGGATGCAGAGGGCCAGAGTCCCCTTTCTCAGAAGTACAGCCCTTCCACAGAGAAATGCCTGCCTGAGATTCAGG GGATCTTTGACAGGGATCCAGACACACTACTTTATTTACTTCAGCAAAAGAGTGAGCCAGCAGAGCCATGTATTGGAAGCAAAGCCCCAA AAGATGATAAAACAATTATAGAGGAGCAGGCAACCAAAATTGCAGATTTGAAGAGGCATGTGGAATTCCTTGTGGCTGAGAATGAAAGAT TAAGGAAAGAAAATAAACAACTAAAGGCTGAAAAGGCCAGACTTCTAAAAGGTCCAATAGAAAAGGAGCTGGATGTAGATGCTGATTTTG TAGAAACGTCAGAGTTATGGAGCTTGCCACCACATGCAGAAACTGCTACAGCCTCCTCAACCTGGCAGAAGTTCGCAGCAAATACTGGGA AAGCCAAGGACATTCCAATCCCCAATCTTCCTCCCTTGGATTTTCCATCTCCAGAACTTCCTCTTATGGAGCTCTCTGAGGATATTCTGA AAGGATTTATGAATAATTAAAATGGAAGGCCACAGAAAAGGGGAAAAGAGGAAATAATACAGTAATCGTTAATCCAGCAAAAAGAAATGA AAAGGGAAAACCACATAGAAGGGTAATCCCGGAAATGCTTCATCTGGTGGACTGTGGGAGCAGAGGCATTGCCAGGACTTGGGAAACAGT CACTGTGAAATGCGCTGCGTATCTCATTCACTCACTTCAGCTAATGATTCCGACTTGGCAGACGCTAAACTCATGGAGGTTCGGTTTCTC CTGATACAAACCAAATGGCTACCTGGAAGAATTTCTTTCAAGCAACAGTTATTTTTCTTATCTTCAGGGTTAAAATGTATAAAAGTTATG TGTAATTAATCTATAATGCCATAAATGATAATGCAAAACCTAAATAATATGGTGGCCGGAGGGGCTGCCTTATATTTGAAACATGCTTTC TATCATGCATTGACTGTATGCATTTTGTTAATGCACATTCTGTTTGTTTAAGGTGTGTAAGATACACACCCTTCTAGATGAAACTATATG TGCCACACTTTGCACTACTCATAATGATAACCTCAAGACTATCAGAAGAAATATTTAAATTTCCATTTTATGAAGAAAGGAACCAAATTA TTATGCTTTTTAAAACAAATTACCAGTTTACATAATTAATCAGGGTGCATTTTAAGTTCTAACTTCGTTTATTGTATAATGCATCATTTG AAAATACCAAGGAGGAAATACCCTTTGTTTTTAATGATGCAAGAGTGGACGTAATGCTAGTTGGCAGTATTTTATTGTAAGAAATCAATA >6508_6508_1_ARID5B-NRBF2_ARID5B_chr10_63810759_ENST00000279873_NRBF2_chr10_64905968_ENST00000277746_length(amino acids)=559AA_BP=0 MEPNSLQWVGSPCGLHGPYIFYKAFQFHLEGKPRILSLGDFFFVRCTPKDPICIAELQLLWEERTSRQLLSSSKLYFLPEDTPQGRNSDH GEDEVIAVSEKVIVKLEDLVKWVHSDFSKWRCGFHAGPVKTEALGRNGQKEALLKYRQSTLNSGLNFKDVLKEKADLGEDEEETNVIVLS YPQYCRYRSMLKRIQDKPSSILTDQFALALGGIAVVSRNPQILYCRDTFDHPTLIENESICDEFAPNLKGRPRKKKPCPQRRDSFSGVKD SNNNSDGKAVAKAHQQSRRADRLLAAGKYEEAISCHKKAAAYLSEAMKLTQSEQAHLSLELQRDSHMKQLLLIQERWKRAQREERLKAQQ NTDKDAAAHLQTSHKPSAEDAEGQSPLSQKYSPSTEKCLPEIQGIFDRDPDTLLYLLQQKSEPAEPCIGSKAPKDDKTIIEEQATKIADL KRHVEFLVAENERLRKENKQLKAEKARLLKGPIEKELDVDADFVETSELWSLPPHAETATASSTWQKFAANTGKAKDIPIPNLPPLDFPS -------------------------------------------------------------- >6508_6508_2_ARID5B-NRBF2_ARID5B_chr10_63810759_ENST00000309334_NRBF2_chr10_64905968_ENST00000277746_length(transcript)=2025nt_BP=386nt AGAGCCCTCCCTGCCAGTCTGGCAACTCCAGCAAAGTCCTCAGGCTCATCCCACACACTCGGTGCTAGTCACTGCTGTGTGTTTACATGA GTAATGGAGCTGCCGGGGGAGGGGAGTTGTAAGCAGAGCGCTGAGCCTCGCAGCTCGCATTCGGAGGGAAGCTGACATCCACACCAAGTC GAGACTTCCAGGGATGTGGCCGGGGAGCAGTCACATGCTGTAGCTTTCATGAGCACAGGCATCAGTCAGGCAGATGTTTGTCGACTGGAA TGGCGCCAAATCTTAAAGGCAGACCACGCAAAAAGAAACCATGCCCACAAAGAAGAGATTCATTCAGTGGTGTTAAGGATTCCAACAACA ATTCCGATGGCAAAGCCGTTGCCAAGGCTCATCAACAGAGCAGACGAGCAGACCGTTTATTAGCTGCAGGCAAATACGAAGAGGCTATTT CTTGTCACAAAAAGGCTGCAGCATATCTTTCTGAAGCCATGAAGCTGACACAGTCAGAGCAGGCTCATCTTTCACTGGAATTGCAAAGGG ATAGCCATATGAAACAGCTCCTCCTCATCCAAGAGAGATGGAAAAGGGCCCAGCGTGAAGAAAGATTGAAAGCCCAGCAGAACACAGACA AGGATGCAGCTGCCCATCTTCAGACATCTCACAAACCCTCTGCAGAGGATGCAGAGGGCCAGAGTCCCCTTTCTCAGAAGTACAGCCCTT CCACAGAGAAATGCCTGCCTGAGATTCAGGGGATCTTTGACAGGGATCCAGACACACTACTTTATTTACTTCAGCAAAAGAGTGAGCCAG CAGAGCCATGTATTGGAAGCAAAGCCCCAAAAGATGATAAAACAATTATAGAGGAGCAGGCAACCAAAATTGCAGATTTGAAGAGGCATG TGGAATTCCTTGTGGCTGAGAATGAAAGATTAAGGAAAGAAAATAAACAACTAAAGGCTGAAAAGGCCAGACTTCTAAAAGGTCCAATAG AAAAGGAGCTGGATGTAGATGCTGATTTTGTAGAAACGTCAGAGTTATGGAGCTTGCCACCACATGCAGAAACTGCTACAGCCTCCTCAA CCTGGCAGAAGTTCGCAGCAAATACTGGGAAAGCCAAGGACATTCCAATCCCCAATCTTCCTCCCTTGGATTTTCCATCTCCAGAACTTC CTCTTATGGAGCTCTCTGAGGATATTCTGAAAGGATTTATGAATAATTAAAATGGAAGGCCACAGAAAAGGGGAAAAGAGGAAATAATAC AGTAATCGTTAATCCAGCAAAAAGAAATGAAAAGGGAAAACCACATAGAAGGGTAATCCCGGAAATGCTTCATCTGGTGGACTGTGGGAG CAGAGGCATTGCCAGGACTTGGGAAACAGTCACTGTGAAATGCGCTGCGTATCTCATTCACTCACTTCAGCTAATGATTCCGACTTGGCA GACGCTAAACTCATGGAGGTTCGGTTTCTCCTGATACAAACCAAATGGCTACCTGGAAGAATTTCTTTCAAGCAACAGTTATTTTTCTTA TCTTCAGGGTTAAAATGTATAAAAGTTATGTGTAATTAATCTATAATGCCATAAATGATAATGCAAAACCTAAATAATATGGTGGCCGGA GGGGCTGCCTTATATTTGAAACATGCTTTCTATCATGCATTGACTGTATGCATTTTGTTAATGCACATTCTGTTTGTTTAAGGTGTGTAA GATACACACCCTTCTAGATGAAACTATATGTGCCACACTTTGCACTACTCATAATGATAACCTCAAGACTATCAGAAGAAATATTTAAAT TTCCATTTTATGAAGAAAGGAACCAAATTATTATGCTTTTTAAAACAAATTACCAGTTTACATAATTAATCAGGGTGCATTTTAAGTTCT AACTTCGTTTATTGTATAATGCATCATTTGAAAATACCAAGGAGGAAATACCCTTTGTTTTTAATGATGCAAGAGTGGACGTAATGCTAG >6508_6508_2_ARID5B-NRBF2_ARID5B_chr10_63810759_ENST00000309334_NRBF2_chr10_64905968_ENST00000277746_length(amino acids)=352AA_BP=1 MTSTPSRDFQGCGRGAVTCCSFHEHRHQSGRCLSTGMAPNLKGRPRKKKPCPQRRDSFSGVKDSNNNSDGKAVAKAHQQSRRADRLLAAG KYEEAISCHKKAAAYLSEAMKLTQSEQAHLSLELQRDSHMKQLLLIQERWKRAQREERLKAQQNTDKDAAAHLQTSHKPSAEDAEGQSPL SQKYSPSTEKCLPEIQGIFDRDPDTLLYLLQQKSEPAEPCIGSKAPKDDKTIIEEQATKIADLKRHVEFLVAENERLRKENKQLKAEKAR -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ARID5B-NRBF2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ARID5B-NRBF2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ARID5B-NRBF2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
