
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:PKN1-LDLR (FusionGDB2 ID:65773) |
Fusion Gene Summary for PKN1-LDLR |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: PKN1-LDLR | Fusion gene ID: 65773 | Hgene | Tgene | Gene symbol | PKN1 | LDLR | Gene ID | 5585 | 3949 |
| Gene name | protein kinase N1 | low density lipoprotein receptor | |
| Synonyms | DBK|PAK-1|PAK1|PKN|PKN-ALPHA|PRK1|PRKCL1 | FH|FHC|FHCL1|LDLCQ2 | |
| Cytomap | 19p13.12 | 19p13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | serine/threonine-protein kinase N1protease-activated kinase 1protein kinase C-like 1protein kinase C-like PKNprotein kinase C-related kinase 1protein kinase PKN-alphaserine-threonine kinase Nserine/threonine protein kinase N | low-density lipoprotein receptorLDL receptorlow-density lipoprotein receptor class A domain-containing protein 3 | |
| Modification date | 20200329 | 20200322 | |
| UniProtAcc | . | Q5SW96 | |
| Ensembl transtripts involved in fusion gene | ENST00000242783, ENST00000342216, ENST00000587429, | ENST00000560628, ENST00000455727, ENST00000535915, ENST00000545707, ENST00000557933, ENST00000558013, ENST00000558518, | |
| Fusion gene scores | * DoF score | 10 X 7 X 8=560 | 4 X 6 X 4=96 |
| # samples | 12 | 5 | |
| ** MAII score | log2(12/560*10)=-2.22239242133645 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/96*10)=-0.941106310946431 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: PKN1 [Title/Abstract] AND LDLR [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | PKN1(14557373)-LDLR(11227535), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | PKN1-LDLR seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. PKN1-LDLR seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. PKN1-LDLR seems lost the major protein functional domain in Hgene partner, which is a kinase due to the frame-shifted ORF. PKN1-LDLR seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. PKN1-LDLR seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | PKN1 | GO:0006357 | regulation of transcription by RNA polymerase II | 12514133 |
| Hgene | PKN1 | GO:0006468 | protein phosphorylation | 17332740 |
| Hgene | PKN1 | GO:0035407 | histone H3-T11 phosphorylation | 18066052 |
 Fusion gene breakpoints across PKN1 (5'-gene) Fusion gene breakpoints across PKN1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
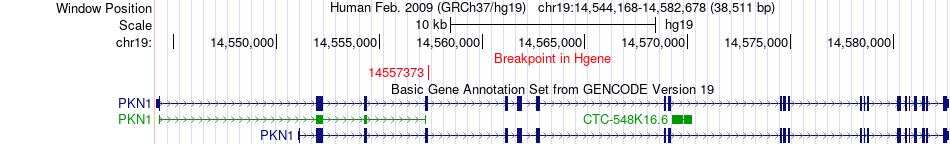 |
 Fusion gene breakpoints across LDLR (3'-gene) Fusion gene breakpoints across LDLR (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
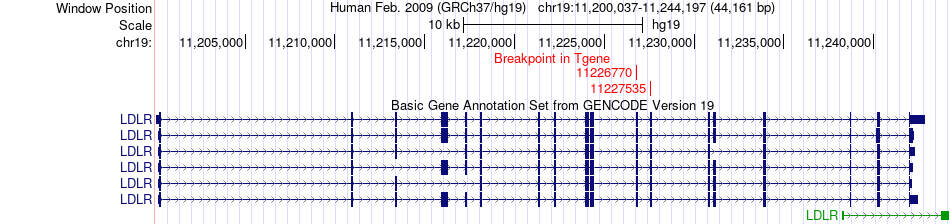 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-C8-A26W-01A | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| ChimerDB4 | BRCA | TCGA-C8-A26W-01A | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
Top |
Fusion Gene ORF analysis for PKN1-LDLR |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000242783 | ENST00000560628 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| 5CDS-intron | ENST00000242783 | ENST00000560628 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| 5CDS-intron | ENST00000342216 | ENST00000560628 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| 5CDS-intron | ENST00000342216 | ENST00000560628 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| Frame-shift | ENST00000242783 | ENST00000455727 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| Frame-shift | ENST00000242783 | ENST00000535915 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| Frame-shift | ENST00000242783 | ENST00000545707 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| Frame-shift | ENST00000242783 | ENST00000557933 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| Frame-shift | ENST00000242783 | ENST00000558013 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| Frame-shift | ENST00000242783 | ENST00000558518 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| Frame-shift | ENST00000342216 | ENST00000455727 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| Frame-shift | ENST00000342216 | ENST00000535915 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| Frame-shift | ENST00000342216 | ENST00000545707 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| Frame-shift | ENST00000342216 | ENST00000557933 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| Frame-shift | ENST00000342216 | ENST00000558013 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| Frame-shift | ENST00000342216 | ENST00000558518 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| In-frame | ENST00000242783 | ENST00000455727 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| In-frame | ENST00000242783 | ENST00000535915 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| In-frame | ENST00000242783 | ENST00000545707 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| In-frame | ENST00000242783 | ENST00000557933 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| In-frame | ENST00000242783 | ENST00000558013 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| In-frame | ENST00000242783 | ENST00000558518 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| In-frame | ENST00000342216 | ENST00000455727 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| In-frame | ENST00000342216 | ENST00000535915 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| In-frame | ENST00000342216 | ENST00000545707 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| In-frame | ENST00000342216 | ENST00000557933 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| In-frame | ENST00000342216 | ENST00000558013 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| In-frame | ENST00000342216 | ENST00000558518 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| intron-3CDS | ENST00000587429 | ENST00000455727 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| intron-3CDS | ENST00000587429 | ENST00000455727 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| intron-3CDS | ENST00000587429 | ENST00000535915 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| intron-3CDS | ENST00000587429 | ENST00000535915 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| intron-3CDS | ENST00000587429 | ENST00000545707 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| intron-3CDS | ENST00000587429 | ENST00000545707 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| intron-3CDS | ENST00000587429 | ENST00000557933 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| intron-3CDS | ENST00000587429 | ENST00000557933 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| intron-3CDS | ENST00000587429 | ENST00000558013 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| intron-3CDS | ENST00000587429 | ENST00000558013 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| intron-3CDS | ENST00000587429 | ENST00000558518 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| intron-3CDS | ENST00000587429 | ENST00000558518 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
| intron-intron | ENST00000587429 | ENST00000560628 | PKN1 | chr19 | 14557373 | - | LDLR | chr19 | 11226770 | + |
| intron-intron | ENST00000587429 | ENST00000560628 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000242783 | PKN1 | chr19 | 14557373 | + | ENST00000558518 | LDLR | chr19 | 11227535 | + | 2500 | 775 | 165 | 1652 | 495 |
| ENST00000242783 | PKN1 | chr19 | 14557373 | + | ENST00000557933 | LDLR | chr19 | 11227535 | + | 1924 | 775 | 165 | 1916 | 583 |
| ENST00000242783 | PKN1 | chr19 | 14557373 | + | ENST00000455727 | LDLR | chr19 | 11227535 | + | 1821 | 775 | 165 | 1652 | 495 |
| ENST00000242783 | PKN1 | chr19 | 14557373 | + | ENST00000535915 | LDLR | chr19 | 11227535 | + | 1875 | 775 | 165 | 1652 | 495 |
| ENST00000242783 | PKN1 | chr19 | 14557373 | + | ENST00000545707 | LDLR | chr19 | 11227535 | + | 1794 | 775 | 165 | 1499 | 444 |
| ENST00000242783 | PKN1 | chr19 | 14557373 | + | ENST00000558013 | LDLR | chr19 | 11227535 | + | 2143 | 775 | 165 | 1646 | 493 |
| ENST00000342216 | PKN1 | chr19 | 14557373 | + | ENST00000558518 | LDLR | chr19 | 11227535 | + | 2384 | 659 | 31 | 1536 | 501 |
| ENST00000342216 | PKN1 | chr19 | 14557373 | + | ENST00000557933 | LDLR | chr19 | 11227535 | + | 1808 | 659 | 31 | 1800 | 589 |
| ENST00000342216 | PKN1 | chr19 | 14557373 | + | ENST00000455727 | LDLR | chr19 | 11227535 | + | 1705 | 659 | 31 | 1536 | 501 |
| ENST00000342216 | PKN1 | chr19 | 14557373 | + | ENST00000535915 | LDLR | chr19 | 11227535 | + | 1759 | 659 | 31 | 1536 | 501 |
| ENST00000342216 | PKN1 | chr19 | 14557373 | + | ENST00000545707 | LDLR | chr19 | 11227535 | + | 1678 | 659 | 31 | 1383 | 450 |
| ENST00000342216 | PKN1 | chr19 | 14557373 | + | ENST00000558013 | LDLR | chr19 | 11227535 | + | 2027 | 659 | 31 | 1530 | 499 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000242783 | ENST00000558518 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + | 0.022305634 | 0.9776944 |
| ENST00000242783 | ENST00000557933 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + | 0.10431041 | 0.8956896 |
| ENST00000242783 | ENST00000455727 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + | 0.025160855 | 0.97483915 |
| ENST00000242783 | ENST00000535915 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + | 0.026398083 | 0.97360194 |
| ENST00000242783 | ENST00000545707 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + | 0.012469866 | 0.9875301 |
| ENST00000242783 | ENST00000558013 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + | 0.022939015 | 0.977061 |
| ENST00000342216 | ENST00000558518 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + | 0.04011812 | 0.95988184 |
| ENST00000342216 | ENST00000557933 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + | 0.11569983 | 0.88430023 |
| ENST00000342216 | ENST00000455727 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + | 0.038374744 | 0.9616252 |
| ENST00000342216 | ENST00000535915 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + | 0.04130113 | 0.9586988 |
| ENST00000342216 | ENST00000545707 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + | 0.021241404 | 0.9787586 |
| ENST00000342216 | ENST00000558013 | PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227535 | + | 0.033702962 | 0.96629703 |
Top |
Fusion Genomic Features for PKN1-LDLR |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227534 | + | 1.98E-13 | 1 |
| PKN1 | chr19 | 14557373 | + | LDLR | chr19 | 11227534 | + | 1.98E-13 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
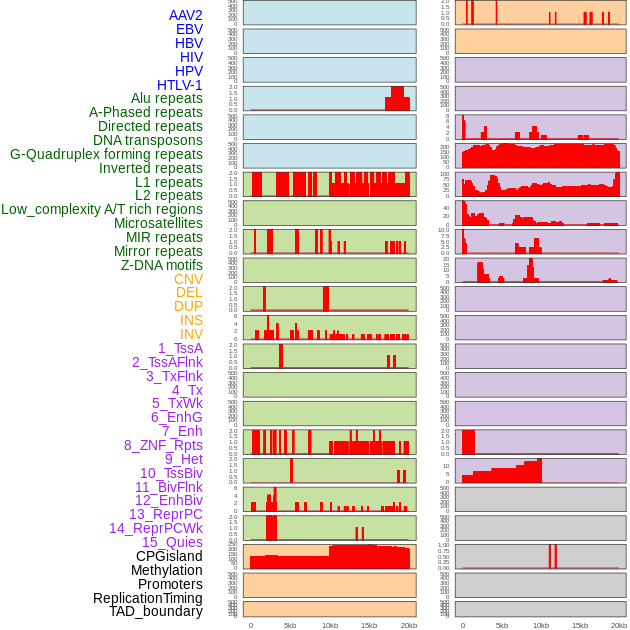 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
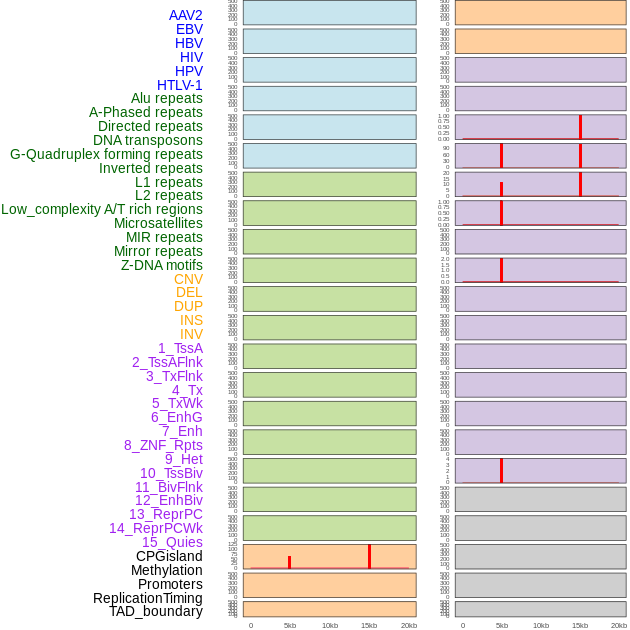 |
Top |
Fusion Protein Features for PKN1-LDLR |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:14557373/chr19:11227535) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | LDLR |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Adapter protein (clathrin-associated sorting protein (CLASP)) required for efficient endocytosis of the LDL receptor (LDLR) in polarized cells such as hepatocytes and lymphocytes, but not in non-polarized cells (fibroblasts). May be required for LDL binding and internalization but not for receptor clustering in coated pits. May facilitate the endocytocis of LDLR and LDLR-LDL complexes from coated pits by stabilizing the interaction between the receptor and the structural components of the pits. May also be involved in the internalization of other LDLR family members. Binds to phosphoinositides, which regulate clathrin bud assembly at the cell surface. Required for trafficking of LRP2 to the endocytic recycling compartment which is necessary for LRP2 proteolysis, releasing a tail fragment which translocates to the nucleus and mediates transcriptional repression (By similarity). {ECO:0000250|UniProtKB:D3ZAR1, ECO:0000269|PubMed:15728179}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PKN1 | chr19:14557373 | chr19:11227535 | ENST00000242783 | + | 4 | 22 | 112_193 | 203 | 943.0 | Domain | REM-1 2 |
| Hgene | PKN1 | chr19:14557373 | chr19:11227535 | ENST00000242783 | + | 4 | 22 | 25_100 | 203 | 943.0 | Domain | REM-1 1 |
| Hgene | PKN1 | chr19:14557373 | chr19:11227535 | ENST00000342216 | + | 4 | 22 | 112_193 | 209 | 949.0 | Domain | REM-1 2 |
| Hgene | PKN1 | chr19:14557373 | chr19:11227535 | ENST00000342216 | + | 4 | 22 | 25_100 | 209 | 949.0 | Domain | REM-1 1 |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 663_712 | 400 | 693.0 | Domain | EGF-like 3 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 663_712 | 527 | 820.0 | Domain | EGF-like 3 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 663_712 | 441 | 683.0 | Domain | EGF-like 3 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 663_712 | 568 | 859.0 | Domain | EGF-like 3 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 663_712 | 568 | 861.0 | Domain | EGF-like 3 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 823_828 | 400 | 693.0 | Motif | NPXY motif | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 823_828 | 527 | 820.0 | Motif | NPXY motif | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 823_828 | 441 | 683.0 | Motif | NPXY motif | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 823_828 | 568 | 859.0 | Motif | NPXY motif | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 823_828 | 568 | 861.0 | Motif | NPXY motif | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 721_768 | 400 | 693.0 | Region | Note=Clustered O-linked oligosaccharides | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 811_860 | 400 | 693.0 | Region | Required for MYLIP-triggered down-regulation of LDLR | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 721_768 | 527 | 820.0 | Region | Note=Clustered O-linked oligosaccharides | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 811_860 | 527 | 820.0 | Region | Required for MYLIP-triggered down-regulation of LDLR | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 721_768 | 441 | 683.0 | Region | Note=Clustered O-linked oligosaccharides | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 811_860 | 441 | 683.0 | Region | Required for MYLIP-triggered down-regulation of LDLR | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 721_768 | 568 | 859.0 | Region | Note=Clustered O-linked oligosaccharides | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 811_860 | 568 | 859.0 | Region | Required for MYLIP-triggered down-regulation of LDLR | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 721_768 | 568 | 861.0 | Region | Note=Clustered O-linked oligosaccharides | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 811_860 | 568 | 861.0 | Region | Required for MYLIP-triggered down-regulation of LDLR | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 439_485 | 400 | 693.0 | Repeat | Note=LDL-receptor class B 2 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 486_528 | 400 | 693.0 | Repeat | Note=LDL-receptor class B 3 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 529_572 | 400 | 693.0 | Repeat | Note=LDL-receptor class B 4 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 573_615 | 400 | 693.0 | Repeat | Note=LDL-receptor class B 5 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 616_658 | 400 | 693.0 | Repeat | Note=LDL-receptor class B 6 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 529_572 | 527 | 820.0 | Repeat | Note=LDL-receptor class B 4 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 573_615 | 527 | 820.0 | Repeat | Note=LDL-receptor class B 5 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 616_658 | 527 | 820.0 | Repeat | Note=LDL-receptor class B 6 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 439_485 | 441 | 683.0 | Repeat | Note=LDL-receptor class B 2 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 486_528 | 441 | 683.0 | Repeat | Note=LDL-receptor class B 3 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 529_572 | 441 | 683.0 | Repeat | Note=LDL-receptor class B 4 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 573_615 | 441 | 683.0 | Repeat | Note=LDL-receptor class B 5 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 616_658 | 441 | 683.0 | Repeat | Note=LDL-receptor class B 6 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 573_615 | 568 | 859.0 | Repeat | Note=LDL-receptor class B 5 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 616_658 | 568 | 859.0 | Repeat | Note=LDL-receptor class B 6 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 573_615 | 568 | 861.0 | Repeat | Note=LDL-receptor class B 5 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 616_658 | 568 | 861.0 | Repeat | Note=LDL-receptor class B 6 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 789_810 | 400 | 693.0 | Transmembrane | Helical | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 789_810 | 527 | 820.0 | Transmembrane | Helical | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 789_810 | 441 | 683.0 | Transmembrane | Helical | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 789_810 | 568 | 859.0 | Transmembrane | Helical | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 789_810 | 568 | 861.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PKN1 | chr19:14557373 | chr19:11227535 | ENST00000242783 | + | 4 | 22 | 201_280 | 203 | 943.0 | Domain | REM-1 3 |
| Hgene | PKN1 | chr19:14557373 | chr19:11227535 | ENST00000242783 | + | 4 | 22 | 307_470 | 203 | 943.0 | Domain | C2 |
| Hgene | PKN1 | chr19:14557373 | chr19:11227535 | ENST00000242783 | + | 4 | 22 | 615_874 | 203 | 943.0 | Domain | Protein kinase |
| Hgene | PKN1 | chr19:14557373 | chr19:11227535 | ENST00000242783 | + | 4 | 22 | 875_942 | 203 | 943.0 | Domain | AGC-kinase C-terminal |
| Hgene | PKN1 | chr19:14557373 | chr19:11227535 | ENST00000342216 | + | 4 | 22 | 201_280 | 209 | 949.0 | Domain | REM-1 3 |
| Hgene | PKN1 | chr19:14557373 | chr19:11227535 | ENST00000342216 | + | 4 | 22 | 307_470 | 209 | 949.0 | Domain | C2 |
| Hgene | PKN1 | chr19:14557373 | chr19:11227535 | ENST00000342216 | + | 4 | 22 | 615_874 | 209 | 949.0 | Domain | Protein kinase |
| Hgene | PKN1 | chr19:14557373 | chr19:11227535 | ENST00000342216 | + | 4 | 22 | 875_942 | 209 | 949.0 | Domain | AGC-kinase C-terminal |
| Hgene | PKN1 | chr19:14557373 | chr19:11227535 | ENST00000242783 | + | 4 | 22 | 621_629 | 203 | 943.0 | Nucleotide binding | ATP |
| Hgene | PKN1 | chr19:14557373 | chr19:11227535 | ENST00000342216 | + | 4 | 22 | 621_629 | 209 | 949.0 | Nucleotide binding | ATP |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 107_145 | 400 | 693.0 | Domain | LDL-receptor class A 3 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 146_186 | 400 | 693.0 | Domain | LDL-receptor class A 4 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 195_233 | 400 | 693.0 | Domain | LDL-receptor class A 5 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 234_272 | 400 | 693.0 | Domain | LDL-receptor class A 6 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 25_65 | 400 | 693.0 | Domain | LDL-receptor class A 1 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 274_313 | 400 | 693.0 | Domain | LDL-receptor class A 7 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 314_353 | 400 | 693.0 | Domain | EGF-like 1 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 354_393 | 400 | 693.0 | Domain | EGF-like 2%3B calcium-binding | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 66_106 | 400 | 693.0 | Domain | LDL-receptor class A 2 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 107_145 | 527 | 820.0 | Domain | LDL-receptor class A 3 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 146_186 | 527 | 820.0 | Domain | LDL-receptor class A 4 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 195_233 | 527 | 820.0 | Domain | LDL-receptor class A 5 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 234_272 | 527 | 820.0 | Domain | LDL-receptor class A 6 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 25_65 | 527 | 820.0 | Domain | LDL-receptor class A 1 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 274_313 | 527 | 820.0 | Domain | LDL-receptor class A 7 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 314_353 | 527 | 820.0 | Domain | EGF-like 1 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 354_393 | 527 | 820.0 | Domain | EGF-like 2%3B calcium-binding | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 66_106 | 527 | 820.0 | Domain | LDL-receptor class A 2 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 107_145 | 441 | 683.0 | Domain | LDL-receptor class A 3 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 146_186 | 441 | 683.0 | Domain | LDL-receptor class A 4 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 195_233 | 441 | 683.0 | Domain | LDL-receptor class A 5 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 234_272 | 441 | 683.0 | Domain | LDL-receptor class A 6 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 25_65 | 441 | 683.0 | Domain | LDL-receptor class A 1 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 274_313 | 441 | 683.0 | Domain | LDL-receptor class A 7 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 314_353 | 441 | 683.0 | Domain | EGF-like 1 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 354_393 | 441 | 683.0 | Domain | EGF-like 2%3B calcium-binding | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 66_106 | 441 | 683.0 | Domain | LDL-receptor class A 2 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 107_145 | 568 | 859.0 | Domain | LDL-receptor class A 3 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 146_186 | 568 | 859.0 | Domain | LDL-receptor class A 4 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 195_233 | 568 | 859.0 | Domain | LDL-receptor class A 5 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 234_272 | 568 | 859.0 | Domain | LDL-receptor class A 6 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 25_65 | 568 | 859.0 | Domain | LDL-receptor class A 1 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 274_313 | 568 | 859.0 | Domain | LDL-receptor class A 7 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 314_353 | 568 | 859.0 | Domain | EGF-like 1 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 354_393 | 568 | 859.0 | Domain | EGF-like 2%3B calcium-binding | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 66_106 | 568 | 859.0 | Domain | LDL-receptor class A 2 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 107_145 | 568 | 861.0 | Domain | LDL-receptor class A 3 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 146_186 | 568 | 861.0 | Domain | LDL-receptor class A 4 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 195_233 | 568 | 861.0 | Domain | LDL-receptor class A 5 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 234_272 | 568 | 861.0 | Domain | LDL-receptor class A 6 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 25_65 | 568 | 861.0 | Domain | LDL-receptor class A 1 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 274_313 | 568 | 861.0 | Domain | LDL-receptor class A 7 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 314_353 | 568 | 861.0 | Domain | EGF-like 1 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 354_393 | 568 | 861.0 | Domain | EGF-like 2%3B calcium-binding | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 66_106 | 568 | 861.0 | Domain | LDL-receptor class A 2 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 397_438 | 400 | 693.0 | Repeat | Note=LDL-receptor class B 1 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 397_438 | 527 | 820.0 | Repeat | Note=LDL-receptor class B 1 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 439_485 | 527 | 820.0 | Repeat | Note=LDL-receptor class B 2 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 486_528 | 527 | 820.0 | Repeat | Note=LDL-receptor class B 3 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 397_438 | 441 | 683.0 | Repeat | Note=LDL-receptor class B 1 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 397_438 | 568 | 859.0 | Repeat | Note=LDL-receptor class B 1 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 439_485 | 568 | 859.0 | Repeat | Note=LDL-receptor class B 2 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 486_528 | 568 | 859.0 | Repeat | Note=LDL-receptor class B 3 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 529_572 | 568 | 859.0 | Repeat | Note=LDL-receptor class B 4 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 397_438 | 568 | 861.0 | Repeat | Note=LDL-receptor class B 1 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 439_485 | 568 | 861.0 | Repeat | Note=LDL-receptor class B 2 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 486_528 | 568 | 861.0 | Repeat | Note=LDL-receptor class B 3 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 529_572 | 568 | 861.0 | Repeat | Note=LDL-receptor class B 4 | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000455727 | 8 | 16 | 22_788 | 400 | 693.0 | Topological domain | Extracellular | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000535915 | 9 | 17 | 22_788 | 527 | 820.0 | Topological domain | Extracellular | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000545707 | 9 | 16 | 22_788 | 441 | 683.0 | Topological domain | Extracellular | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558013 | 10 | 18 | 22_788 | 568 | 859.0 | Topological domain | Extracellular | |
| Tgene | LDLR | chr19:14557373 | chr19:11227535 | ENST00000558518 | 10 | 18 | 22_788 | 568 | 861.0 | Topological domain | Extracellular |
Top |
Fusion Gene Sequence for PKN1-LDLR |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >65773_65773_1_PKN1-LDLR_PKN1_chr19_14557373_ENST00000242783_LDLR_chr19_11227535_ENST00000455727_length(transcript)=1821nt_BP=775nt CGGCGGCCTGGCCAGGCGAGGCGGGAGCCGAGCAGGCCGGAGCGGGCCGCGCGCCGGGGAGGGTCGCGCCGCGCCCGCCCCCTCCCCGCC ACGCCCGGGGCGCACGCGCGCTCCTCTGGCCGCCCCTCCCTCCGCGCGGGGACCCCTGGCGGGCGGCAGGAGGACATGGCCAGCGACGCC GTGCAGAGTGAGCCTCGCAGCTGGTCCCTGCTAGAGCAGCTGGGCCTGGCCGGGGCAGACCTGGCGGCCCCCGGGGTACAGCAGCAGCTG GAGCTGGAGCGGGAGCGGCTGCGGCGGGAAATCCGCAAGGAGCTGAAGCTGAAGGAGGGTGCTGAGAACCTGCGGCGGGCCACCACTGAC CTGGGCCGCAGCCTGGGCCCCGTAGAGCTGCTGCTGCGGGGCTCCTCGCGCCGCCTCGACCTGCTGCACCAGCAGCTGCAGGAGCTGCAC GCCCACGTGGTGCTTCCCGACCCGGCGGCCACCCACGATGGCCCCCAGTCCCCTGGTGCGGGTGGCCCCACCTGCTCGGCCACCAACCTG AGCCGCGTGGCGGGCCTGGAGAAGCAGTTGGCCATTGAGCTGAAGGTGAAGCAGGGGGCGGAGAACATGATCCAGACCTACAGCAATGGC AGCACCAAGGACCGGAAGCTGCTGCTGACAGCCCAGCAGATGTTGCAGGACAGTAAGACCAAGATTGACATCATCCGCATGCAACTCCGC CGGGCGCTGCAGGCCGGCCAGCTGGAGAACCAGGCAGCCCCGGATGACACCCAAGATCTCCTCAGTGGCCGCCTCTACTGGGTTGACTCC AAACTTCACTCCATCTCAAGCATCGATGTCAACGGGGGCAACCGGAAGACCATCTTGGAGGATGAAAAGAGGCTGGCCCACCCCTTCTCC TTGGCCGTCTTTGAGGACAAAGTATTTTGGACAGATATCATCAACGAAGCCATTTTCAGTGCCAACCGCCTCACAGGTTCCGATGTCAAC TTGTTGGCTGAAAACCTACTGTCCCCAGAGGATATGGTTCTCTTCCACAACCTCACCCAGCCAAGAGGAGTGAACTGGTGTGAGAGGACC ACCCTGAGCAATGGCGGCTGCCAGTATCTGTGCCTCCCTGCCCCGCAGATCAACCCCCACTCGCCCAAGTTTACCTGCGCCTGCCCGGAC GGCATGCTGCTGGCCAGGGACATGAGGAGCTGCCTCACAGAGGCTGAGGCTGCAGTGGCCACCCAGGAGACATCCACCGTCAGGCTAAAG GTCAGCTCCACAGCCGTAAGGACACAGCACACAACCACCCGACCTGTTCCCGACACCTCCCGGCTGCCTGGGGCCACCCCTGGGCTCACC ACGGTGGAGATAGTGACAATGTCTCACCAAGCTCTGGGCGACGTTGCTGGCAGAGGAAATGAGAAGAAGCCCAGTAGCGTGAGGGCTCTG TCCATTGTCCTCCCCATCGTGCTCCTCGTCTTCCTTTGCCTGGGGGTCTTCCTTCTATGGAAGAACTGGCGGCTTAAGAACATCAACAGC ATCAACTTTGACAACCCCGTCTATCAGAAGACCACAGAGGATGAGGTCCACATTTGCCACAACCAGGACGGCTACAGCTACCCCTCGAGA CAGATGGTCAGTCTGGAGGATGACGTGGCGTGAACATCTGCCTGGAGTCCCGTCCCTGCCCAGAACCCTTCCTGAGACCTCGCCGGCCTT GTTTTATTCAAAGACAGAGAAGACCAAAGCATTGCCTGCCAGAGCTTTGTTTTATATATTTATTCATCTGGGAGGCAGAACAGGCTTCGG >65773_65773_1_PKN1-LDLR_PKN1_chr19_14557373_ENST00000242783_LDLR_chr19_11227535_ENST00000455727_length(amino acids)=495AA_BP=203 MASDAVQSEPRSWSLLEQLGLAGADLAAPGVQQQLELERERLRREIRKELKLKEGAENLRRATTDLGRSLGPVELLLRGSSRRLDLLHQQ LQELHAHVVLPDPAATHDGPQSPGAGGPTCSATNLSRVAGLEKQLAIELKVKQGAENMIQTYSNGSTKDRKLLLTAQQMLQDSKTKIDII RMQLRRALQAGQLENQAAPDDTQDLLSGRLYWVDSKLHSISSIDVNGGNRKTILEDEKRLAHPFSLAVFEDKVFWTDIINEAIFSANRLT GSDVNLLAENLLSPEDMVLFHNLTQPRGVNWCERTTLSNGGCQYLCLPAPQINPHSPKFTCACPDGMLLARDMRSCLTEAEAAVATQETS TVRLKVSSTAVRTQHTTTRPVPDTSRLPGATPGLTTVEIVTMSHQALGDVAGRGNEKKPSSVRALSIVLPIVLLVFLCLGVFLLWKNWRL -------------------------------------------------------------- >65773_65773_2_PKN1-LDLR_PKN1_chr19_14557373_ENST00000242783_LDLR_chr19_11227535_ENST00000535915_length(transcript)=1875nt_BP=775nt CGGCGGCCTGGCCAGGCGAGGCGGGAGCCGAGCAGGCCGGAGCGGGCCGCGCGCCGGGGAGGGTCGCGCCGCGCCCGCCCCCTCCCCGCC ACGCCCGGGGCGCACGCGCGCTCCTCTGGCCGCCCCTCCCTCCGCGCGGGGACCCCTGGCGGGCGGCAGGAGGACATGGCCAGCGACGCC GTGCAGAGTGAGCCTCGCAGCTGGTCCCTGCTAGAGCAGCTGGGCCTGGCCGGGGCAGACCTGGCGGCCCCCGGGGTACAGCAGCAGCTG GAGCTGGAGCGGGAGCGGCTGCGGCGGGAAATCCGCAAGGAGCTGAAGCTGAAGGAGGGTGCTGAGAACCTGCGGCGGGCCACCACTGAC CTGGGCCGCAGCCTGGGCCCCGTAGAGCTGCTGCTGCGGGGCTCCTCGCGCCGCCTCGACCTGCTGCACCAGCAGCTGCAGGAGCTGCAC GCCCACGTGGTGCTTCCCGACCCGGCGGCCACCCACGATGGCCCCCAGTCCCCTGGTGCGGGTGGCCCCACCTGCTCGGCCACCAACCTG AGCCGCGTGGCGGGCCTGGAGAAGCAGTTGGCCATTGAGCTGAAGGTGAAGCAGGGGGCGGAGAACATGATCCAGACCTACAGCAATGGC AGCACCAAGGACCGGAAGCTGCTGCTGACAGCCCAGCAGATGTTGCAGGACAGTAAGACCAAGATTGACATCATCCGCATGCAACTCCGC CGGGCGCTGCAGGCCGGCCAGCTGGAGAACCAGGCAGCCCCGGATGACACCCAAGATCTCCTCAGTGGCCGCCTCTACTGGGTTGACTCC AAACTTCACTCCATCTCAAGCATCGATGTCAACGGGGGCAACCGGAAGACCATCTTGGAGGATGAAAAGAGGCTGGCCCACCCCTTCTCC TTGGCCGTCTTTGAGGACAAAGTATTTTGGACAGATATCATCAACGAAGCCATTTTCAGTGCCAACCGCCTCACAGGTTCCGATGTCAAC TTGTTGGCTGAAAACCTACTGTCCCCAGAGGATATGGTTCTCTTCCACAACCTCACCCAGCCAAGAGGAGTGAACTGGTGTGAGAGGACC ACCCTGAGCAATGGCGGCTGCCAGTATCTGTGCCTCCCTGCCCCGCAGATCAACCCCCACTCGCCCAAGTTTACCTGCGCCTGCCCGGAC GGCATGCTGCTGGCCAGGGACATGAGGAGCTGCCTCACAGAGGCTGAGGCTGCAGTGGCCACCCAGGAGACATCCACCGTCAGGCTAAAG GTCAGCTCCACAGCCGTAAGGACACAGCACACAACCACCCGACCTGTTCCCGACACCTCCCGGCTGCCTGGGGCCACCCCTGGGCTCACC ACGGTGGAGATAGTGACAATGTCTCACCAAGCTCTGGGCGACGTTGCTGGCAGAGGAAATGAGAAGAAGCCCAGTAGCGTGAGGGCTCTG TCCATTGTCCTCCCCATCGTGCTCCTCGTCTTCCTTTGCCTGGGGGTCTTCCTTCTATGGAAGAACTGGCGGCTTAAGAACATCAACAGC ATCAACTTTGACAACCCCGTCTATCAGAAGACCACAGAGGATGAGGTCCACATTTGCCACAACCAGGACGGCTACAGCTACCCCTCGAGA CAGATGGTCAGTCTGGAGGATGACGTGGCGTGAACATCTGCCTGGAGTCCCGTCCCTGCCCAGAACCCTTCCTGAGACCTCGCCGGCCTT GTTTTATTCAAAGACAGAGAAGACCAAAGCATTGCCTGCCAGAGCTTTGTTTTATATATTTATTCATCTGGGAGGCAGAACAGGCTTCGG >65773_65773_2_PKN1-LDLR_PKN1_chr19_14557373_ENST00000242783_LDLR_chr19_11227535_ENST00000535915_length(amino acids)=495AA_BP=203 MASDAVQSEPRSWSLLEQLGLAGADLAAPGVQQQLELERERLRREIRKELKLKEGAENLRRATTDLGRSLGPVELLLRGSSRRLDLLHQQ LQELHAHVVLPDPAATHDGPQSPGAGGPTCSATNLSRVAGLEKQLAIELKVKQGAENMIQTYSNGSTKDRKLLLTAQQMLQDSKTKIDII RMQLRRALQAGQLENQAAPDDTQDLLSGRLYWVDSKLHSISSIDVNGGNRKTILEDEKRLAHPFSLAVFEDKVFWTDIINEAIFSANRLT GSDVNLLAENLLSPEDMVLFHNLTQPRGVNWCERTTLSNGGCQYLCLPAPQINPHSPKFTCACPDGMLLARDMRSCLTEAEAAVATQETS TVRLKVSSTAVRTQHTTTRPVPDTSRLPGATPGLTTVEIVTMSHQALGDVAGRGNEKKPSSVRALSIVLPIVLLVFLCLGVFLLWKNWRL -------------------------------------------------------------- >65773_65773_3_PKN1-LDLR_PKN1_chr19_14557373_ENST00000242783_LDLR_chr19_11227535_ENST00000545707_length(transcript)=1794nt_BP=775nt CGGCGGCCTGGCCAGGCGAGGCGGGAGCCGAGCAGGCCGGAGCGGGCCGCGCGCCGGGGAGGGTCGCGCCGCGCCCGCCCCCTCCCCGCC ACGCCCGGGGCGCACGCGCGCTCCTCTGGCCGCCCCTCCCTCCGCGCGGGGACCCCTGGCGGGCGGCAGGAGGACATGGCCAGCGACGCC GTGCAGAGTGAGCCTCGCAGCTGGTCCCTGCTAGAGCAGCTGGGCCTGGCCGGGGCAGACCTGGCGGCCCCCGGGGTACAGCAGCAGCTG GAGCTGGAGCGGGAGCGGCTGCGGCGGGAAATCCGCAAGGAGCTGAAGCTGAAGGAGGGTGCTGAGAACCTGCGGCGGGCCACCACTGAC CTGGGCCGCAGCCTGGGCCCCGTAGAGCTGCTGCTGCGGGGCTCCTCGCGCCGCCTCGACCTGCTGCACCAGCAGCTGCAGGAGCTGCAC GCCCACGTGGTGCTTCCCGACCCGGCGGCCACCCACGATGGCCCCCAGTCCCCTGGTGCGGGTGGCCCCACCTGCTCGGCCACCAACCTG AGCCGCGTGGCGGGCCTGGAGAAGCAGTTGGCCATTGAGCTGAAGGTGAAGCAGGGGGCGGAGAACATGATCCAGACCTACAGCAATGGC AGCACCAAGGACCGGAAGCTGCTGCTGACAGCCCAGCAGATGTTGCAGGACAGTAAGACCAAGATTGACATCATCCGCATGCAACTCCGC CGGGCGCTGCAGGCCGGCCAGCTGGAGAACCAGGCAGCCCCGGATGACACCCAAGATCTCCTCAGTGGCCGCCTCTACTGGGTTGACTCC AAACTTCACTCCATCTCAAGCATCGATGTCAACGGGGGCAACCGGAAGACCATCTTGGAGGATGAAAAGAGGCTGGCCCACCCCTTCTCC TTGGCCGTCTTTGAGGACAAAGTATTTTGGACAGATATCATCAACGAAGCCATTTTCAGTGCCAACCGCCTCACAGGTTCCGATGTCAAC TTGTTGGCTGAAAACCTACTGTCCCCAGAGGATATGGTTCTCTTCCACAACCTCACCCAGCCAAGAGAGGCTGAGGCTGCAGTGGCCACC CAGGAGACATCCACCGTCAGGCTAAAGGTCAGCTCCACAGCCGTAAGGACACAGCACACAACCACCCGACCTGTTCCCGACACCTCCCGG CTGCCTGGGGCCACCCCTGGGCTCACCACGGTGGAGATAGTGACAATGTCTCACCAAGCTCTGGGCGACGTTGCTGGCAGAGGAAATGAG AAGAAGCCCAGTAGCGTGAGGGCTCTGTCCATTGTCCTCCCCATCGTGCTCCTCGTCTTCCTTTGCCTGGGGGTCTTCCTTCTATGGAAG AACTGGCGGCTTAAGAACATCAACAGCATCAACTTTGACAACCCCGTCTATCAGAAGACCACAGAGGATGAGGTCCACATTTGCCACAAC CAGGACGGCTACAGCTACCCCTCGAGACAGATGGTCAGTCTGGAGGATGACGTGGCGTGAACATCTGCCTGGAGTCCCGTCCCTGCCCAG AACCCTTCCTGAGACCTCGCCGGCCTTGTTTTATTCAAAGACAGAGAAGACCAAAGCATTGCCTGCCAGAGCTTTGTTTTATATATTTAT TCATCTGGGAGGCAGAACAGGCTTCGGACAGTGCCCATGCAATGGCTTGGGTTGGGATTTTGGTTTCTTCCTTTCCTCGTGAAGGATAAG >65773_65773_3_PKN1-LDLR_PKN1_chr19_14557373_ENST00000242783_LDLR_chr19_11227535_ENST00000545707_length(amino acids)=444AA_BP=203 MASDAVQSEPRSWSLLEQLGLAGADLAAPGVQQQLELERERLRREIRKELKLKEGAENLRRATTDLGRSLGPVELLLRGSSRRLDLLHQQ LQELHAHVVLPDPAATHDGPQSPGAGGPTCSATNLSRVAGLEKQLAIELKVKQGAENMIQTYSNGSTKDRKLLLTAQQMLQDSKTKIDII RMQLRRALQAGQLENQAAPDDTQDLLSGRLYWVDSKLHSISSIDVNGGNRKTILEDEKRLAHPFSLAVFEDKVFWTDIINEAIFSANRLT GSDVNLLAENLLSPEDMVLFHNLTQPREAEAAVATQETSTVRLKVSSTAVRTQHTTTRPVPDTSRLPGATPGLTTVEIVTMSHQALGDVA -------------------------------------------------------------- >65773_65773_4_PKN1-LDLR_PKN1_chr19_14557373_ENST00000242783_LDLR_chr19_11227535_ENST00000557933_length(transcript)=1924nt_BP=775nt CGGCGGCCTGGCCAGGCGAGGCGGGAGCCGAGCAGGCCGGAGCGGGCCGCGCGCCGGGGAGGGTCGCGCCGCGCCCGCCCCCTCCCCGCC ACGCCCGGGGCGCACGCGCGCTCCTCTGGCCGCCCCTCCCTCCGCGCGGGGACCCCTGGCGGGCGGCAGGAGGACATGGCCAGCGACGCC GTGCAGAGTGAGCCTCGCAGCTGGTCCCTGCTAGAGCAGCTGGGCCTGGCCGGGGCAGACCTGGCGGCCCCCGGGGTACAGCAGCAGCTG GAGCTGGAGCGGGAGCGGCTGCGGCGGGAAATCCGCAAGGAGCTGAAGCTGAAGGAGGGTGCTGAGAACCTGCGGCGGGCCACCACTGAC CTGGGCCGCAGCCTGGGCCCCGTAGAGCTGCTGCTGCGGGGCTCCTCGCGCCGCCTCGACCTGCTGCACCAGCAGCTGCAGGAGCTGCAC GCCCACGTGGTGCTTCCCGACCCGGCGGCCACCCACGATGGCCCCCAGTCCCCTGGTGCGGGTGGCCCCACCTGCTCGGCCACCAACCTG AGCCGCGTGGCGGGCCTGGAGAAGCAGTTGGCCATTGAGCTGAAGGTGAAGCAGGGGGCGGAGAACATGATCCAGACCTACAGCAATGGC AGCACCAAGGACCGGAAGCTGCTGCTGACAGCCCAGCAGATGTTGCAGGACAGTAAGACCAAGATTGACATCATCCGCATGCAACTCCGC CGGGCGCTGCAGGCCGGCCAGCTGGAGAACCAGGCAGCCCCGGATGACACCCAAGATCTCCTCAGTGGCCGCCTCTACTGGGTTGACTCC AAACTTCACTCCATCTCAAGCATCGATGTCAACGGGGGCAACCGGAAGACCATCTTGGAGGATGAAAAGAGGCTGGCCCACCCCTTCTCC TTGGCCGTCTTTGAGGACAAAGTATTTTGGACAGATATCATCAACGAAGCCATTTTCAGTGCCAACCGCCTCACAGGTTCCGATGTCAAC TTGTTGGCTGAAAACCTACTGTCCCCAGAGGATATGGTTCTCTTCCACAACCTCACCCAGCCAAGAGGAGTGAACTGGTGTGAGAGGACC ACCCTGAGCAATGGCGGCTGCCAGTATCTGTGCCTCCCTGCCCCGCAGATCAACCCCCACTCGCCCAAGTTTACCTGCGCCTGCCCGGAC GGCATGCTGCTGGCCAGGGACATGAGGAGCTGCCTCACAGAGGCTGAGGCTGCAGTGGCCACCCAGGAGACATCCACCGTCAGGCTAAAG GTCAGCTCCACAGCCGTAAGGACACAGCACACAACCACCCGACCTGTTCCCGACACCTCCCGGCTGCCTGGGGCCACCCCTGGGCTCACC ACGGTGGAGATAGTGACAATGTCTCACCAAGCTCTGGGCGACGTTGCTGGCAGAGGAAATGAGAAGAAGCCCAGTAGCGTGAGGGCTCTG TCCATTGTCCTCCCCATCGCCACGGAGCTGGGTCTCTGGTCTCGGGGGCAGCTGTGTGACAGAGCGTGCCTCTCCCTACAGTGCTCCTCG TCTTCCTTTGCCTGGGGGTCTTCCTTCTATGGAAGAACTGGCGGCTTAAGAACATCAACAGCATCAACTTTGACAACCCCGTCTATCAGA AGACCACAGAGGATGAGGTCCACATTTGCCACAACCAGGACGGCTACAGCTACCCCTCGAGACAGATGGTCAGTCTGGAGGATGACGTGG CGTGAACATCTGCCTGGAGTCCCGTCCCTGCCCAGAACCCTTCCTGAGACCTCGCCGGCCTTGTTTTATTCAAAGACAGAGAAGACCAAA GCATTGCCTGCCAGAGCTTTGTTTTATATATTTATTCATCTGGGAGGCAGAACAGGCTTCGGACAGTGCCCATGCAATGGCTTGGGTTGG >65773_65773_4_PKN1-LDLR_PKN1_chr19_14557373_ENST00000242783_LDLR_chr19_11227535_ENST00000557933_length(amino acids)=583AA_BP=203 MASDAVQSEPRSWSLLEQLGLAGADLAAPGVQQQLELERERLRREIRKELKLKEGAENLRRATTDLGRSLGPVELLLRGSSRRLDLLHQQ LQELHAHVVLPDPAATHDGPQSPGAGGPTCSATNLSRVAGLEKQLAIELKVKQGAENMIQTYSNGSTKDRKLLLTAQQMLQDSKTKIDII RMQLRRALQAGQLENQAAPDDTQDLLSGRLYWVDSKLHSISSIDVNGGNRKTILEDEKRLAHPFSLAVFEDKVFWTDIINEAIFSANRLT GSDVNLLAENLLSPEDMVLFHNLTQPRGVNWCERTTLSNGGCQYLCLPAPQINPHSPKFTCACPDGMLLARDMRSCLTEAEAAVATQETS TVRLKVSSTAVRTQHTTTRPVPDTSRLPGATPGLTTVEIVTMSHQALGDVAGRGNEKKPSSVRALSIVLPIATELGLWSRGQLCDRACLS LQCSSSSFAWGSSFYGRTGGLRTSTASTLTTPSIRRPQRMRSTFATTRTATATPRDRWSVWRMTWREHLPGVPSLPRTLPETSPALFYSK -------------------------------------------------------------- >65773_65773_5_PKN1-LDLR_PKN1_chr19_14557373_ENST00000242783_LDLR_chr19_11227535_ENST00000558013_length(transcript)=2143nt_BP=775nt CGGCGGCCTGGCCAGGCGAGGCGGGAGCCGAGCAGGCCGGAGCGGGCCGCGCGCCGGGGAGGGTCGCGCCGCGCCCGCCCCCTCCCCGCC ACGCCCGGGGCGCACGCGCGCTCCTCTGGCCGCCCCTCCCTCCGCGCGGGGACCCCTGGCGGGCGGCAGGAGGACATGGCCAGCGACGCC GTGCAGAGTGAGCCTCGCAGCTGGTCCCTGCTAGAGCAGCTGGGCCTGGCCGGGGCAGACCTGGCGGCCCCCGGGGTACAGCAGCAGCTG GAGCTGGAGCGGGAGCGGCTGCGGCGGGAAATCCGCAAGGAGCTGAAGCTGAAGGAGGGTGCTGAGAACCTGCGGCGGGCCACCACTGAC CTGGGCCGCAGCCTGGGCCCCGTAGAGCTGCTGCTGCGGGGCTCCTCGCGCCGCCTCGACCTGCTGCACCAGCAGCTGCAGGAGCTGCAC GCCCACGTGGTGCTTCCCGACCCGGCGGCCACCCACGATGGCCCCCAGTCCCCTGGTGCGGGTGGCCCCACCTGCTCGGCCACCAACCTG AGCCGCGTGGCGGGCCTGGAGAAGCAGTTGGCCATTGAGCTGAAGGTGAAGCAGGGGGCGGAGAACATGATCCAGACCTACAGCAATGGC AGCACCAAGGACCGGAAGCTGCTGCTGACAGCCCAGCAGATGTTGCAGGACAGTAAGACCAAGATTGACATCATCCGCATGCAACTCCGC CGGGCGCTGCAGGCCGGCCAGCTGGAGAACCAGGCAGCCCCGGATGACACCCAAGATCTCCTCAGTGGCCGCCTCTACTGGGTTGACTCC AAACTTCACTCCATCTCAAGCATCGATGTCAACGGGGGCAACCGGAAGACCATCTTGGAGGATGAAAAGAGGCTGGCCCACCCCTTCTCC TTGGCCGTCTTTGAGGACAAAGTATTTTGGACAGATATCATCAACGAAGCCATTTTCAGTGCCAACCGCCTCACAGGTTCCGATGTCAAC TTGTTGGCTGAAAACCTACTGTCCCCAGAGGATATGGTTCTCTTCCACAACCTCACCCAGCCAAGAGGAGTGAACTGGTGTGAGAGGACC ACCCTGAGCAATGGCGGCTGCCAGTATCTGTGCCTCCCTGCCCCGCAGATCAACCCCCACTCGCCCAAGTTTACCTGCGCCTGCCCGGAC GGCATGCTGCTGGCCAGGGACATGAGGAGCTGCCTCACAGAGGCTGAGGCTGCAGTGGCCACCCAGGAGACATCCACCGTCAGGCTAAAG GTCAGCTCCACAGCCGTAAGGACACAGCACACAACCACCCGACCTGTTCCCGACACCTCCCGGCTGCCTGGGGCCACCCCTGGGCTCACC ACGGTGGAGATAGTGACAATGTCTCACCAAGCTCTGGGCGACGTTGCTGGCAGAGGAAATGAGAAGAAGCCCAGTAGCGTGAGGGCTCTG TCCATTGTCCTCCCCATCGTGCTCCTCGTCTTCCTTTGCCTGGGGGTCTTCCTTCTATGGAAGAACTGGCGGCTTAAGAACATCAACAGC ATCAACTTTGACAACCCCGTCTATCAGAAGACCACAGAGGATGAGGTCCACATTTGCCACAACCAGGACGGCTACAGCTACCCCTCGATG GTCAGTCTGGAGGATGACGTGGCGTGAACATCTGCCTGGAGTCCCGTCCCTGCCCAGAACCCTTCCTGAGACCTCGCCGGCCTTGTTTTA TTCAAAGACAGAGAAGACCAAAGCATTGCCTGCCAGAGCTTTGTTTTATATATTTATTCATCTGGGAGGCAGAACAGGCTTCGGACAGTG CCCATGCAATGGCTTGGGTTGGGATTTTGGTTTCTTCCTTTCCTCGTGAAGGATAAGAGAAACAGGCCCGGGGGGACCAGGATGACACCT CCATTTCTCTCCAGGAAGTTTTGAGTTTCTCTCCACCGTGACACAATCCTCAAACATGGAAGATGAAAGGGGAGGGGATGTCAGGCCCAG AGAAGCAAGTGGCTTTCAACACACAACAGCAGATGGCACCAACGGGACCCCCTGGCCCTGCCTCATCCACCAATCTCTAAGCCAAACCCC >65773_65773_5_PKN1-LDLR_PKN1_chr19_14557373_ENST00000242783_LDLR_chr19_11227535_ENST00000558013_length(amino acids)=493AA_BP=203 MASDAVQSEPRSWSLLEQLGLAGADLAAPGVQQQLELERERLRREIRKELKLKEGAENLRRATTDLGRSLGPVELLLRGSSRRLDLLHQQ LQELHAHVVLPDPAATHDGPQSPGAGGPTCSATNLSRVAGLEKQLAIELKVKQGAENMIQTYSNGSTKDRKLLLTAQQMLQDSKTKIDII RMQLRRALQAGQLENQAAPDDTQDLLSGRLYWVDSKLHSISSIDVNGGNRKTILEDEKRLAHPFSLAVFEDKVFWTDIINEAIFSANRLT GSDVNLLAENLLSPEDMVLFHNLTQPRGVNWCERTTLSNGGCQYLCLPAPQINPHSPKFTCACPDGMLLARDMRSCLTEAEAAVATQETS TVRLKVSSTAVRTQHTTTRPVPDTSRLPGATPGLTTVEIVTMSHQALGDVAGRGNEKKPSSVRALSIVLPIVLLVFLCLGVFLLWKNWRL -------------------------------------------------------------- >65773_65773_6_PKN1-LDLR_PKN1_chr19_14557373_ENST00000242783_LDLR_chr19_11227535_ENST00000558518_length(transcript)=2500nt_BP=775nt CGGCGGCCTGGCCAGGCGAGGCGGGAGCCGAGCAGGCCGGAGCGGGCCGCGCGCCGGGGAGGGTCGCGCCGCGCCCGCCCCCTCCCCGCC ACGCCCGGGGCGCACGCGCGCTCCTCTGGCCGCCCCTCCCTCCGCGCGGGGACCCCTGGCGGGCGGCAGGAGGACATGGCCAGCGACGCC GTGCAGAGTGAGCCTCGCAGCTGGTCCCTGCTAGAGCAGCTGGGCCTGGCCGGGGCAGACCTGGCGGCCCCCGGGGTACAGCAGCAGCTG GAGCTGGAGCGGGAGCGGCTGCGGCGGGAAATCCGCAAGGAGCTGAAGCTGAAGGAGGGTGCTGAGAACCTGCGGCGGGCCACCACTGAC CTGGGCCGCAGCCTGGGCCCCGTAGAGCTGCTGCTGCGGGGCTCCTCGCGCCGCCTCGACCTGCTGCACCAGCAGCTGCAGGAGCTGCAC GCCCACGTGGTGCTTCCCGACCCGGCGGCCACCCACGATGGCCCCCAGTCCCCTGGTGCGGGTGGCCCCACCTGCTCGGCCACCAACCTG AGCCGCGTGGCGGGCCTGGAGAAGCAGTTGGCCATTGAGCTGAAGGTGAAGCAGGGGGCGGAGAACATGATCCAGACCTACAGCAATGGC AGCACCAAGGACCGGAAGCTGCTGCTGACAGCCCAGCAGATGTTGCAGGACAGTAAGACCAAGATTGACATCATCCGCATGCAACTCCGC CGGGCGCTGCAGGCCGGCCAGCTGGAGAACCAGGCAGCCCCGGATGACACCCAAGATCTCCTCAGTGGCCGCCTCTACTGGGTTGACTCC AAACTTCACTCCATCTCAAGCATCGATGTCAACGGGGGCAACCGGAAGACCATCTTGGAGGATGAAAAGAGGCTGGCCCACCCCTTCTCC TTGGCCGTCTTTGAGGACAAAGTATTTTGGACAGATATCATCAACGAAGCCATTTTCAGTGCCAACCGCCTCACAGGTTCCGATGTCAAC TTGTTGGCTGAAAACCTACTGTCCCCAGAGGATATGGTTCTCTTCCACAACCTCACCCAGCCAAGAGGAGTGAACTGGTGTGAGAGGACC ACCCTGAGCAATGGCGGCTGCCAGTATCTGTGCCTCCCTGCCCCGCAGATCAACCCCCACTCGCCCAAGTTTACCTGCGCCTGCCCGGAC GGCATGCTGCTGGCCAGGGACATGAGGAGCTGCCTCACAGAGGCTGAGGCTGCAGTGGCCACCCAGGAGACATCCACCGTCAGGCTAAAG GTCAGCTCCACAGCCGTAAGGACACAGCACACAACCACCCGACCTGTTCCCGACACCTCCCGGCTGCCTGGGGCCACCCCTGGGCTCACC ACGGTGGAGATAGTGACAATGTCTCACCAAGCTCTGGGCGACGTTGCTGGCAGAGGAAATGAGAAGAAGCCCAGTAGCGTGAGGGCTCTG TCCATTGTCCTCCCCATCGTGCTCCTCGTCTTCCTTTGCCTGGGGGTCTTCCTTCTATGGAAGAACTGGCGGCTTAAGAACATCAACAGC ATCAACTTTGACAACCCCGTCTATCAGAAGACCACAGAGGATGAGGTCCACATTTGCCACAACCAGGACGGCTACAGCTACCCCTCGAGA CAGATGGTCAGTCTGGAGGATGACGTGGCGTGAACATCTGCCTGGAGTCCCGTCCCTGCCCAGAACCCTTCCTGAGACCTCGCCGGCCTT GTTTTATTCAAAGACAGAGAAGACCAAAGCATTGCCTGCCAGAGCTTTGTTTTATATATTTATTCATCTGGGAGGCAGAACAGGCTTCGG ACAGTGCCCATGCAATGGCTTGGGTTGGGATTTTGGTTTCTTCCTTTCCTCGTGAAGGATAAGAGAAACAGGCCCGGGGGGACCAGGATG ACACCTCCATTTCTCTCCAGGAAGTTTTGAGTTTCTCTCCACCGTGACACAATCCTCAAACATGGAAGATGAAAGGGGAGGGGATGTCAG GCCCAGAGAAGCAAGTGGCTTTCAACACACAACAGCAGATGGCACCAACGGGACCCCCTGGCCCTGCCTCATCCACCAATCTCTAAGCCA AACCCCTAAACTCAGGAGTCAACGTGTTTACCTCTTCTATGCAAGCCTTGCTAGACAGCCAGGTTAGCCTTTGCCCTGTCACCCCCGAAT CATGACCCACCCAGTGTCTTTCGAGGTGGGTTTGTACCTTCCTTAAGCCAGGAAAGGGATTCATGGCGTCGGAAATGATCTGGCTGAATC CGTGGTGGCACCGAGACCAAACTCATTCACCAAATGATGCCACTTCCCAGAGGCAGAGCCTGAGTCACTGGTCACCCTTAATATTTATTA AGTGCCTGAGACACCCGGTTACCTTGGCCGTGAGGACACGTGGCCTGCACCCAGGTGTGGCTGTCAGGACACCAGCCTGGTGCCCATCCT >65773_65773_6_PKN1-LDLR_PKN1_chr19_14557373_ENST00000242783_LDLR_chr19_11227535_ENST00000558518_length(amino acids)=495AA_BP=203 MASDAVQSEPRSWSLLEQLGLAGADLAAPGVQQQLELERERLRREIRKELKLKEGAENLRRATTDLGRSLGPVELLLRGSSRRLDLLHQQ LQELHAHVVLPDPAATHDGPQSPGAGGPTCSATNLSRVAGLEKQLAIELKVKQGAENMIQTYSNGSTKDRKLLLTAQQMLQDSKTKIDII RMQLRRALQAGQLENQAAPDDTQDLLSGRLYWVDSKLHSISSIDVNGGNRKTILEDEKRLAHPFSLAVFEDKVFWTDIINEAIFSANRLT GSDVNLLAENLLSPEDMVLFHNLTQPRGVNWCERTTLSNGGCQYLCLPAPQINPHSPKFTCACPDGMLLARDMRSCLTEAEAAVATQETS TVRLKVSSTAVRTQHTTTRPVPDTSRLPGATPGLTTVEIVTMSHQALGDVAGRGNEKKPSSVRALSIVLPIVLLVFLCLGVFLLWKNWRL -------------------------------------------------------------- >65773_65773_7_PKN1-LDLR_PKN1_chr19_14557373_ENST00000342216_LDLR_chr19_11227535_ENST00000455727_length(transcript)=1705nt_BP=659nt AAGAGGGCGGGGCCCTCAGCCCGGACCCAGGATGGCGGAGGCCAATAACCCCTCGGAGCAGGAGCTGGAGAGTGAGCCTCGCAGCTGGTC CCTGCTAGAGCAGCTGGGCCTGGCCGGGGCAGACCTGGCGGCCCCCGGGGTACAGCAGCAGCTGGAGCTGGAGCGGGAGCGGCTGCGGCG GGAAATCCGCAAGGAGCTGAAGCTGAAGGAGGGTGCTGAGAACCTGCGGCGGGCCACCACTGACCTGGGCCGCAGCCTGGGCCCCGTAGA GCTGCTGCTGCGGGGCTCCTCGCGCCGCCTCGACCTGCTGCACCAGCAGCTGCAGGAGCTGCACGCCCACGTGGTGCTTCCCGACCCGGC GGCCACCCACGATGGCCCCCAGTCCCCTGGTGCGGGTGGCCCCACCTGCTCGGCCACCAACCTGAGCCGCGTGGCGGGCCTGGAGAAGCA GTTGGCCATTGAGCTGAAGGTGAAGCAGGGGGCGGAGAACATGATCCAGACCTACAGCAATGGCAGCACCAAGGACCGGAAGCTGCTGCT GACAGCCCAGCAGATGTTGCAGGACAGTAAGACCAAGATTGACATCATCCGCATGCAACTCCGCCGGGCGCTGCAGGCCGGCCAGCTGGA GAACCAGGCAGCCCCGGATGACACCCAAGATCTCCTCAGTGGCCGCCTCTACTGGGTTGACTCCAAACTTCACTCCATCTCAAGCATCGA TGTCAACGGGGGCAACCGGAAGACCATCTTGGAGGATGAAAAGAGGCTGGCCCACCCCTTCTCCTTGGCCGTCTTTGAGGACAAAGTATT TTGGACAGATATCATCAACGAAGCCATTTTCAGTGCCAACCGCCTCACAGGTTCCGATGTCAACTTGTTGGCTGAAAACCTACTGTCCCC AGAGGATATGGTTCTCTTCCACAACCTCACCCAGCCAAGAGGAGTGAACTGGTGTGAGAGGACCACCCTGAGCAATGGCGGCTGCCAGTA TCTGTGCCTCCCTGCCCCGCAGATCAACCCCCACTCGCCCAAGTTTACCTGCGCCTGCCCGGACGGCATGCTGCTGGCCAGGGACATGAG GAGCTGCCTCACAGAGGCTGAGGCTGCAGTGGCCACCCAGGAGACATCCACCGTCAGGCTAAAGGTCAGCTCCACAGCCGTAAGGACACA GCACACAACCACCCGACCTGTTCCCGACACCTCCCGGCTGCCTGGGGCCACCCCTGGGCTCACCACGGTGGAGATAGTGACAATGTCTCA CCAAGCTCTGGGCGACGTTGCTGGCAGAGGAAATGAGAAGAAGCCCAGTAGCGTGAGGGCTCTGTCCATTGTCCTCCCCATCGTGCTCCT CGTCTTCCTTTGCCTGGGGGTCTTCCTTCTATGGAAGAACTGGCGGCTTAAGAACATCAACAGCATCAACTTTGACAACCCCGTCTATCA GAAGACCACAGAGGATGAGGTCCACATTTGCCACAACCAGGACGGCTACAGCTACCCCTCGAGACAGATGGTCAGTCTGGAGGATGACGT GGCGTGAACATCTGCCTGGAGTCCCGTCCCTGCCCAGAACCCTTCCTGAGACCTCGCCGGCCTTGTTTTATTCAAAGACAGAGAAGACCA >65773_65773_7_PKN1-LDLR_PKN1_chr19_14557373_ENST00000342216_LDLR_chr19_11227535_ENST00000455727_length(amino acids)=501AA_BP=209 MAEANNPSEQELESEPRSWSLLEQLGLAGADLAAPGVQQQLELERERLRREIRKELKLKEGAENLRRATTDLGRSLGPVELLLRGSSRRL DLLHQQLQELHAHVVLPDPAATHDGPQSPGAGGPTCSATNLSRVAGLEKQLAIELKVKQGAENMIQTYSNGSTKDRKLLLTAQQMLQDSK TKIDIIRMQLRRALQAGQLENQAAPDDTQDLLSGRLYWVDSKLHSISSIDVNGGNRKTILEDEKRLAHPFSLAVFEDKVFWTDIINEAIF SANRLTGSDVNLLAENLLSPEDMVLFHNLTQPRGVNWCERTTLSNGGCQYLCLPAPQINPHSPKFTCACPDGMLLARDMRSCLTEAEAAV ATQETSTVRLKVSSTAVRTQHTTTRPVPDTSRLPGATPGLTTVEIVTMSHQALGDVAGRGNEKKPSSVRALSIVLPIVLLVFLCLGVFLL -------------------------------------------------------------- >65773_65773_8_PKN1-LDLR_PKN1_chr19_14557373_ENST00000342216_LDLR_chr19_11227535_ENST00000535915_length(transcript)=1759nt_BP=659nt AAGAGGGCGGGGCCCTCAGCCCGGACCCAGGATGGCGGAGGCCAATAACCCCTCGGAGCAGGAGCTGGAGAGTGAGCCTCGCAGCTGGTC CCTGCTAGAGCAGCTGGGCCTGGCCGGGGCAGACCTGGCGGCCCCCGGGGTACAGCAGCAGCTGGAGCTGGAGCGGGAGCGGCTGCGGCG GGAAATCCGCAAGGAGCTGAAGCTGAAGGAGGGTGCTGAGAACCTGCGGCGGGCCACCACTGACCTGGGCCGCAGCCTGGGCCCCGTAGA GCTGCTGCTGCGGGGCTCCTCGCGCCGCCTCGACCTGCTGCACCAGCAGCTGCAGGAGCTGCACGCCCACGTGGTGCTTCCCGACCCGGC GGCCACCCACGATGGCCCCCAGTCCCCTGGTGCGGGTGGCCCCACCTGCTCGGCCACCAACCTGAGCCGCGTGGCGGGCCTGGAGAAGCA GTTGGCCATTGAGCTGAAGGTGAAGCAGGGGGCGGAGAACATGATCCAGACCTACAGCAATGGCAGCACCAAGGACCGGAAGCTGCTGCT GACAGCCCAGCAGATGTTGCAGGACAGTAAGACCAAGATTGACATCATCCGCATGCAACTCCGCCGGGCGCTGCAGGCCGGCCAGCTGGA GAACCAGGCAGCCCCGGATGACACCCAAGATCTCCTCAGTGGCCGCCTCTACTGGGTTGACTCCAAACTTCACTCCATCTCAAGCATCGA TGTCAACGGGGGCAACCGGAAGACCATCTTGGAGGATGAAAAGAGGCTGGCCCACCCCTTCTCCTTGGCCGTCTTTGAGGACAAAGTATT TTGGACAGATATCATCAACGAAGCCATTTTCAGTGCCAACCGCCTCACAGGTTCCGATGTCAACTTGTTGGCTGAAAACCTACTGTCCCC AGAGGATATGGTTCTCTTCCACAACCTCACCCAGCCAAGAGGAGTGAACTGGTGTGAGAGGACCACCCTGAGCAATGGCGGCTGCCAGTA TCTGTGCCTCCCTGCCCCGCAGATCAACCCCCACTCGCCCAAGTTTACCTGCGCCTGCCCGGACGGCATGCTGCTGGCCAGGGACATGAG GAGCTGCCTCACAGAGGCTGAGGCTGCAGTGGCCACCCAGGAGACATCCACCGTCAGGCTAAAGGTCAGCTCCACAGCCGTAAGGACACA GCACACAACCACCCGACCTGTTCCCGACACCTCCCGGCTGCCTGGGGCCACCCCTGGGCTCACCACGGTGGAGATAGTGACAATGTCTCA CCAAGCTCTGGGCGACGTTGCTGGCAGAGGAAATGAGAAGAAGCCCAGTAGCGTGAGGGCTCTGTCCATTGTCCTCCCCATCGTGCTCCT CGTCTTCCTTTGCCTGGGGGTCTTCCTTCTATGGAAGAACTGGCGGCTTAAGAACATCAACAGCATCAACTTTGACAACCCCGTCTATCA GAAGACCACAGAGGATGAGGTCCACATTTGCCACAACCAGGACGGCTACAGCTACCCCTCGAGACAGATGGTCAGTCTGGAGGATGACGT GGCGTGAACATCTGCCTGGAGTCCCGTCCCTGCCCAGAACCCTTCCTGAGACCTCGCCGGCCTTGTTTTATTCAAAGACAGAGAAGACCA AAGCATTGCCTGCCAGAGCTTTGTTTTATATATTTATTCATCTGGGAGGCAGAACAGGCTTCGGACAGTGCCCATGCAATGGCTTGGGTT >65773_65773_8_PKN1-LDLR_PKN1_chr19_14557373_ENST00000342216_LDLR_chr19_11227535_ENST00000535915_length(amino acids)=501AA_BP=209 MAEANNPSEQELESEPRSWSLLEQLGLAGADLAAPGVQQQLELERERLRREIRKELKLKEGAENLRRATTDLGRSLGPVELLLRGSSRRL DLLHQQLQELHAHVVLPDPAATHDGPQSPGAGGPTCSATNLSRVAGLEKQLAIELKVKQGAENMIQTYSNGSTKDRKLLLTAQQMLQDSK TKIDIIRMQLRRALQAGQLENQAAPDDTQDLLSGRLYWVDSKLHSISSIDVNGGNRKTILEDEKRLAHPFSLAVFEDKVFWTDIINEAIF SANRLTGSDVNLLAENLLSPEDMVLFHNLTQPRGVNWCERTTLSNGGCQYLCLPAPQINPHSPKFTCACPDGMLLARDMRSCLTEAEAAV ATQETSTVRLKVSSTAVRTQHTTTRPVPDTSRLPGATPGLTTVEIVTMSHQALGDVAGRGNEKKPSSVRALSIVLPIVLLVFLCLGVFLL -------------------------------------------------------------- >65773_65773_9_PKN1-LDLR_PKN1_chr19_14557373_ENST00000342216_LDLR_chr19_11227535_ENST00000545707_length(transcript)=1678nt_BP=659nt AAGAGGGCGGGGCCCTCAGCCCGGACCCAGGATGGCGGAGGCCAATAACCCCTCGGAGCAGGAGCTGGAGAGTGAGCCTCGCAGCTGGTC CCTGCTAGAGCAGCTGGGCCTGGCCGGGGCAGACCTGGCGGCCCCCGGGGTACAGCAGCAGCTGGAGCTGGAGCGGGAGCGGCTGCGGCG GGAAATCCGCAAGGAGCTGAAGCTGAAGGAGGGTGCTGAGAACCTGCGGCGGGCCACCACTGACCTGGGCCGCAGCCTGGGCCCCGTAGA GCTGCTGCTGCGGGGCTCCTCGCGCCGCCTCGACCTGCTGCACCAGCAGCTGCAGGAGCTGCACGCCCACGTGGTGCTTCCCGACCCGGC GGCCACCCACGATGGCCCCCAGTCCCCTGGTGCGGGTGGCCCCACCTGCTCGGCCACCAACCTGAGCCGCGTGGCGGGCCTGGAGAAGCA GTTGGCCATTGAGCTGAAGGTGAAGCAGGGGGCGGAGAACATGATCCAGACCTACAGCAATGGCAGCACCAAGGACCGGAAGCTGCTGCT GACAGCCCAGCAGATGTTGCAGGACAGTAAGACCAAGATTGACATCATCCGCATGCAACTCCGCCGGGCGCTGCAGGCCGGCCAGCTGGA GAACCAGGCAGCCCCGGATGACACCCAAGATCTCCTCAGTGGCCGCCTCTACTGGGTTGACTCCAAACTTCACTCCATCTCAAGCATCGA TGTCAACGGGGGCAACCGGAAGACCATCTTGGAGGATGAAAAGAGGCTGGCCCACCCCTTCTCCTTGGCCGTCTTTGAGGACAAAGTATT TTGGACAGATATCATCAACGAAGCCATTTTCAGTGCCAACCGCCTCACAGGTTCCGATGTCAACTTGTTGGCTGAAAACCTACTGTCCCC AGAGGATATGGTTCTCTTCCACAACCTCACCCAGCCAAGAGAGGCTGAGGCTGCAGTGGCCACCCAGGAGACATCCACCGTCAGGCTAAA GGTCAGCTCCACAGCCGTAAGGACACAGCACACAACCACCCGACCTGTTCCCGACACCTCCCGGCTGCCTGGGGCCACCCCTGGGCTCAC CACGGTGGAGATAGTGACAATGTCTCACCAAGCTCTGGGCGACGTTGCTGGCAGAGGAAATGAGAAGAAGCCCAGTAGCGTGAGGGCTCT GTCCATTGTCCTCCCCATCGTGCTCCTCGTCTTCCTTTGCCTGGGGGTCTTCCTTCTATGGAAGAACTGGCGGCTTAAGAACATCAACAG CATCAACTTTGACAACCCCGTCTATCAGAAGACCACAGAGGATGAGGTCCACATTTGCCACAACCAGGACGGCTACAGCTACCCCTCGAG ACAGATGGTCAGTCTGGAGGATGACGTGGCGTGAACATCTGCCTGGAGTCCCGTCCCTGCCCAGAACCCTTCCTGAGACCTCGCCGGCCT TGTTTTATTCAAAGACAGAGAAGACCAAAGCATTGCCTGCCAGAGCTTTGTTTTATATATTTATTCATCTGGGAGGCAGAACAGGCTTCG GACAGTGCCCATGCAATGGCTTGGGTTGGGATTTTGGTTTCTTCCTTTCCTCGTGAAGGATAAGAGAAACAGGCCCGGGGGGACCAGGAT >65773_65773_9_PKN1-LDLR_PKN1_chr19_14557373_ENST00000342216_LDLR_chr19_11227535_ENST00000545707_length(amino acids)=450AA_BP=209 MAEANNPSEQELESEPRSWSLLEQLGLAGADLAAPGVQQQLELERERLRREIRKELKLKEGAENLRRATTDLGRSLGPVELLLRGSSRRL DLLHQQLQELHAHVVLPDPAATHDGPQSPGAGGPTCSATNLSRVAGLEKQLAIELKVKQGAENMIQTYSNGSTKDRKLLLTAQQMLQDSK TKIDIIRMQLRRALQAGQLENQAAPDDTQDLLSGRLYWVDSKLHSISSIDVNGGNRKTILEDEKRLAHPFSLAVFEDKVFWTDIINEAIF SANRLTGSDVNLLAENLLSPEDMVLFHNLTQPREAEAAVATQETSTVRLKVSSTAVRTQHTTTRPVPDTSRLPGATPGLTTVEIVTMSHQ ALGDVAGRGNEKKPSSVRALSIVLPIVLLVFLCLGVFLLWKNWRLKNINSINFDNPVYQKTTEDEVHICHNQDGYSYPSRQMVSLEDDVA -------------------------------------------------------------- >65773_65773_10_PKN1-LDLR_PKN1_chr19_14557373_ENST00000342216_LDLR_chr19_11227535_ENST00000557933_length(transcript)=1808nt_BP=659nt AAGAGGGCGGGGCCCTCAGCCCGGACCCAGGATGGCGGAGGCCAATAACCCCTCGGAGCAGGAGCTGGAGAGTGAGCCTCGCAGCTGGTC CCTGCTAGAGCAGCTGGGCCTGGCCGGGGCAGACCTGGCGGCCCCCGGGGTACAGCAGCAGCTGGAGCTGGAGCGGGAGCGGCTGCGGCG GGAAATCCGCAAGGAGCTGAAGCTGAAGGAGGGTGCTGAGAACCTGCGGCGGGCCACCACTGACCTGGGCCGCAGCCTGGGCCCCGTAGA GCTGCTGCTGCGGGGCTCCTCGCGCCGCCTCGACCTGCTGCACCAGCAGCTGCAGGAGCTGCACGCCCACGTGGTGCTTCCCGACCCGGC GGCCACCCACGATGGCCCCCAGTCCCCTGGTGCGGGTGGCCCCACCTGCTCGGCCACCAACCTGAGCCGCGTGGCGGGCCTGGAGAAGCA GTTGGCCATTGAGCTGAAGGTGAAGCAGGGGGCGGAGAACATGATCCAGACCTACAGCAATGGCAGCACCAAGGACCGGAAGCTGCTGCT GACAGCCCAGCAGATGTTGCAGGACAGTAAGACCAAGATTGACATCATCCGCATGCAACTCCGCCGGGCGCTGCAGGCCGGCCAGCTGGA GAACCAGGCAGCCCCGGATGACACCCAAGATCTCCTCAGTGGCCGCCTCTACTGGGTTGACTCCAAACTTCACTCCATCTCAAGCATCGA TGTCAACGGGGGCAACCGGAAGACCATCTTGGAGGATGAAAAGAGGCTGGCCCACCCCTTCTCCTTGGCCGTCTTTGAGGACAAAGTATT TTGGACAGATATCATCAACGAAGCCATTTTCAGTGCCAACCGCCTCACAGGTTCCGATGTCAACTTGTTGGCTGAAAACCTACTGTCCCC AGAGGATATGGTTCTCTTCCACAACCTCACCCAGCCAAGAGGAGTGAACTGGTGTGAGAGGACCACCCTGAGCAATGGCGGCTGCCAGTA TCTGTGCCTCCCTGCCCCGCAGATCAACCCCCACTCGCCCAAGTTTACCTGCGCCTGCCCGGACGGCATGCTGCTGGCCAGGGACATGAG GAGCTGCCTCACAGAGGCTGAGGCTGCAGTGGCCACCCAGGAGACATCCACCGTCAGGCTAAAGGTCAGCTCCACAGCCGTAAGGACACA GCACACAACCACCCGACCTGTTCCCGACACCTCCCGGCTGCCTGGGGCCACCCCTGGGCTCACCACGGTGGAGATAGTGACAATGTCTCA CCAAGCTCTGGGCGACGTTGCTGGCAGAGGAAATGAGAAGAAGCCCAGTAGCGTGAGGGCTCTGTCCATTGTCCTCCCCATCGCCACGGA GCTGGGTCTCTGGTCTCGGGGGCAGCTGTGTGACAGAGCGTGCCTCTCCCTACAGTGCTCCTCGTCTTCCTTTGCCTGGGGGTCTTCCTT CTATGGAAGAACTGGCGGCTTAAGAACATCAACAGCATCAACTTTGACAACCCCGTCTATCAGAAGACCACAGAGGATGAGGTCCACATT TGCCACAACCAGGACGGCTACAGCTACCCCTCGAGACAGATGGTCAGTCTGGAGGATGACGTGGCGTGAACATCTGCCTGGAGTCCCGTC CCTGCCCAGAACCCTTCCTGAGACCTCGCCGGCCTTGTTTTATTCAAAGACAGAGAAGACCAAAGCATTGCCTGCCAGAGCTTTGTTTTA TATATTTATTCATCTGGGAGGCAGAACAGGCTTCGGACAGTGCCCATGCAATGGCTTGGGTTGGGATTTTGGTTTCTTCCTTTCCTCGTG >65773_65773_10_PKN1-LDLR_PKN1_chr19_14557373_ENST00000342216_LDLR_chr19_11227535_ENST00000557933_length(amino acids)=589AA_BP=209 MAEANNPSEQELESEPRSWSLLEQLGLAGADLAAPGVQQQLELERERLRREIRKELKLKEGAENLRRATTDLGRSLGPVELLLRGSSRRL DLLHQQLQELHAHVVLPDPAATHDGPQSPGAGGPTCSATNLSRVAGLEKQLAIELKVKQGAENMIQTYSNGSTKDRKLLLTAQQMLQDSK TKIDIIRMQLRRALQAGQLENQAAPDDTQDLLSGRLYWVDSKLHSISSIDVNGGNRKTILEDEKRLAHPFSLAVFEDKVFWTDIINEAIF SANRLTGSDVNLLAENLLSPEDMVLFHNLTQPRGVNWCERTTLSNGGCQYLCLPAPQINPHSPKFTCACPDGMLLARDMRSCLTEAEAAV ATQETSTVRLKVSSTAVRTQHTTTRPVPDTSRLPGATPGLTTVEIVTMSHQALGDVAGRGNEKKPSSVRALSIVLPIATELGLWSRGQLC DRACLSLQCSSSSFAWGSSFYGRTGGLRTSTASTLTTPSIRRPQRMRSTFATTRTATATPRDRWSVWRMTWREHLPGVPSLPRTLPETSP -------------------------------------------------------------- >65773_65773_11_PKN1-LDLR_PKN1_chr19_14557373_ENST00000342216_LDLR_chr19_11227535_ENST00000558013_length(transcript)=2027nt_BP=659nt AAGAGGGCGGGGCCCTCAGCCCGGACCCAGGATGGCGGAGGCCAATAACCCCTCGGAGCAGGAGCTGGAGAGTGAGCCTCGCAGCTGGTC CCTGCTAGAGCAGCTGGGCCTGGCCGGGGCAGACCTGGCGGCCCCCGGGGTACAGCAGCAGCTGGAGCTGGAGCGGGAGCGGCTGCGGCG GGAAATCCGCAAGGAGCTGAAGCTGAAGGAGGGTGCTGAGAACCTGCGGCGGGCCACCACTGACCTGGGCCGCAGCCTGGGCCCCGTAGA GCTGCTGCTGCGGGGCTCCTCGCGCCGCCTCGACCTGCTGCACCAGCAGCTGCAGGAGCTGCACGCCCACGTGGTGCTTCCCGACCCGGC GGCCACCCACGATGGCCCCCAGTCCCCTGGTGCGGGTGGCCCCACCTGCTCGGCCACCAACCTGAGCCGCGTGGCGGGCCTGGAGAAGCA GTTGGCCATTGAGCTGAAGGTGAAGCAGGGGGCGGAGAACATGATCCAGACCTACAGCAATGGCAGCACCAAGGACCGGAAGCTGCTGCT GACAGCCCAGCAGATGTTGCAGGACAGTAAGACCAAGATTGACATCATCCGCATGCAACTCCGCCGGGCGCTGCAGGCCGGCCAGCTGGA GAACCAGGCAGCCCCGGATGACACCCAAGATCTCCTCAGTGGCCGCCTCTACTGGGTTGACTCCAAACTTCACTCCATCTCAAGCATCGA TGTCAACGGGGGCAACCGGAAGACCATCTTGGAGGATGAAAAGAGGCTGGCCCACCCCTTCTCCTTGGCCGTCTTTGAGGACAAAGTATT TTGGACAGATATCATCAACGAAGCCATTTTCAGTGCCAACCGCCTCACAGGTTCCGATGTCAACTTGTTGGCTGAAAACCTACTGTCCCC AGAGGATATGGTTCTCTTCCACAACCTCACCCAGCCAAGAGGAGTGAACTGGTGTGAGAGGACCACCCTGAGCAATGGCGGCTGCCAGTA TCTGTGCCTCCCTGCCCCGCAGATCAACCCCCACTCGCCCAAGTTTACCTGCGCCTGCCCGGACGGCATGCTGCTGGCCAGGGACATGAG GAGCTGCCTCACAGAGGCTGAGGCTGCAGTGGCCACCCAGGAGACATCCACCGTCAGGCTAAAGGTCAGCTCCACAGCCGTAAGGACACA GCACACAACCACCCGACCTGTTCCCGACACCTCCCGGCTGCCTGGGGCCACCCCTGGGCTCACCACGGTGGAGATAGTGACAATGTCTCA CCAAGCTCTGGGCGACGTTGCTGGCAGAGGAAATGAGAAGAAGCCCAGTAGCGTGAGGGCTCTGTCCATTGTCCTCCCCATCGTGCTCCT CGTCTTCCTTTGCCTGGGGGTCTTCCTTCTATGGAAGAACTGGCGGCTTAAGAACATCAACAGCATCAACTTTGACAACCCCGTCTATCA GAAGACCACAGAGGATGAGGTCCACATTTGCCACAACCAGGACGGCTACAGCTACCCCTCGATGGTCAGTCTGGAGGATGACGTGGCGTG AACATCTGCCTGGAGTCCCGTCCCTGCCCAGAACCCTTCCTGAGACCTCGCCGGCCTTGTTTTATTCAAAGACAGAGAAGACCAAAGCAT TGCCTGCCAGAGCTTTGTTTTATATATTTATTCATCTGGGAGGCAGAACAGGCTTCGGACAGTGCCCATGCAATGGCTTGGGTTGGGATT TTGGTTTCTTCCTTTCCTCGTGAAGGATAAGAGAAACAGGCCCGGGGGGACCAGGATGACACCTCCATTTCTCTCCAGGAAGTTTTGAGT TTCTCTCCACCGTGACACAATCCTCAAACATGGAAGATGAAAGGGGAGGGGATGTCAGGCCCAGAGAAGCAAGTGGCTTTCAACACACAA CAGCAGATGGCACCAACGGGACCCCCTGGCCCTGCCTCATCCACCAATCTCTAAGCCAAACCCCTAAACTCAGGAGTCAACGTGTTTACC >65773_65773_11_PKN1-LDLR_PKN1_chr19_14557373_ENST00000342216_LDLR_chr19_11227535_ENST00000558013_length(amino acids)=499AA_BP=209 MAEANNPSEQELESEPRSWSLLEQLGLAGADLAAPGVQQQLELERERLRREIRKELKLKEGAENLRRATTDLGRSLGPVELLLRGSSRRL DLLHQQLQELHAHVVLPDPAATHDGPQSPGAGGPTCSATNLSRVAGLEKQLAIELKVKQGAENMIQTYSNGSTKDRKLLLTAQQMLQDSK TKIDIIRMQLRRALQAGQLENQAAPDDTQDLLSGRLYWVDSKLHSISSIDVNGGNRKTILEDEKRLAHPFSLAVFEDKVFWTDIINEAIF SANRLTGSDVNLLAENLLSPEDMVLFHNLTQPRGVNWCERTTLSNGGCQYLCLPAPQINPHSPKFTCACPDGMLLARDMRSCLTEAEAAV ATQETSTVRLKVSSTAVRTQHTTTRPVPDTSRLPGATPGLTTVEIVTMSHQALGDVAGRGNEKKPSSVRALSIVLPIVLLVFLCLGVFLL -------------------------------------------------------------- >65773_65773_12_PKN1-LDLR_PKN1_chr19_14557373_ENST00000342216_LDLR_chr19_11227535_ENST00000558518_length(transcript)=2384nt_BP=659nt AAGAGGGCGGGGCCCTCAGCCCGGACCCAGGATGGCGGAGGCCAATAACCCCTCGGAGCAGGAGCTGGAGAGTGAGCCTCGCAGCTGGTC CCTGCTAGAGCAGCTGGGCCTGGCCGGGGCAGACCTGGCGGCCCCCGGGGTACAGCAGCAGCTGGAGCTGGAGCGGGAGCGGCTGCGGCG GGAAATCCGCAAGGAGCTGAAGCTGAAGGAGGGTGCTGAGAACCTGCGGCGGGCCACCACTGACCTGGGCCGCAGCCTGGGCCCCGTAGA GCTGCTGCTGCGGGGCTCCTCGCGCCGCCTCGACCTGCTGCACCAGCAGCTGCAGGAGCTGCACGCCCACGTGGTGCTTCCCGACCCGGC GGCCACCCACGATGGCCCCCAGTCCCCTGGTGCGGGTGGCCCCACCTGCTCGGCCACCAACCTGAGCCGCGTGGCGGGCCTGGAGAAGCA GTTGGCCATTGAGCTGAAGGTGAAGCAGGGGGCGGAGAACATGATCCAGACCTACAGCAATGGCAGCACCAAGGACCGGAAGCTGCTGCT GACAGCCCAGCAGATGTTGCAGGACAGTAAGACCAAGATTGACATCATCCGCATGCAACTCCGCCGGGCGCTGCAGGCCGGCCAGCTGGA GAACCAGGCAGCCCCGGATGACACCCAAGATCTCCTCAGTGGCCGCCTCTACTGGGTTGACTCCAAACTTCACTCCATCTCAAGCATCGA TGTCAACGGGGGCAACCGGAAGACCATCTTGGAGGATGAAAAGAGGCTGGCCCACCCCTTCTCCTTGGCCGTCTTTGAGGACAAAGTATT TTGGACAGATATCATCAACGAAGCCATTTTCAGTGCCAACCGCCTCACAGGTTCCGATGTCAACTTGTTGGCTGAAAACCTACTGTCCCC AGAGGATATGGTTCTCTTCCACAACCTCACCCAGCCAAGAGGAGTGAACTGGTGTGAGAGGACCACCCTGAGCAATGGCGGCTGCCAGTA TCTGTGCCTCCCTGCCCCGCAGATCAACCCCCACTCGCCCAAGTTTACCTGCGCCTGCCCGGACGGCATGCTGCTGGCCAGGGACATGAG GAGCTGCCTCACAGAGGCTGAGGCTGCAGTGGCCACCCAGGAGACATCCACCGTCAGGCTAAAGGTCAGCTCCACAGCCGTAAGGACACA GCACACAACCACCCGACCTGTTCCCGACACCTCCCGGCTGCCTGGGGCCACCCCTGGGCTCACCACGGTGGAGATAGTGACAATGTCTCA CCAAGCTCTGGGCGACGTTGCTGGCAGAGGAAATGAGAAGAAGCCCAGTAGCGTGAGGGCTCTGTCCATTGTCCTCCCCATCGTGCTCCT CGTCTTCCTTTGCCTGGGGGTCTTCCTTCTATGGAAGAACTGGCGGCTTAAGAACATCAACAGCATCAACTTTGACAACCCCGTCTATCA GAAGACCACAGAGGATGAGGTCCACATTTGCCACAACCAGGACGGCTACAGCTACCCCTCGAGACAGATGGTCAGTCTGGAGGATGACGT GGCGTGAACATCTGCCTGGAGTCCCGTCCCTGCCCAGAACCCTTCCTGAGACCTCGCCGGCCTTGTTTTATTCAAAGACAGAGAAGACCA AAGCATTGCCTGCCAGAGCTTTGTTTTATATATTTATTCATCTGGGAGGCAGAACAGGCTTCGGACAGTGCCCATGCAATGGCTTGGGTT GGGATTTTGGTTTCTTCCTTTCCTCGTGAAGGATAAGAGAAACAGGCCCGGGGGGACCAGGATGACACCTCCATTTCTCTCCAGGAAGTT TTGAGTTTCTCTCCACCGTGACACAATCCTCAAACATGGAAGATGAAAGGGGAGGGGATGTCAGGCCCAGAGAAGCAAGTGGCTTTCAAC ACACAACAGCAGATGGCACCAACGGGACCCCCTGGCCCTGCCTCATCCACCAATCTCTAAGCCAAACCCCTAAACTCAGGAGTCAACGTG TTTACCTCTTCTATGCAAGCCTTGCTAGACAGCCAGGTTAGCCTTTGCCCTGTCACCCCCGAATCATGACCCACCCAGTGTCTTTCGAGG TGGGTTTGTACCTTCCTTAAGCCAGGAAAGGGATTCATGGCGTCGGAAATGATCTGGCTGAATCCGTGGTGGCACCGAGACCAAACTCAT TCACCAAATGATGCCACTTCCCAGAGGCAGAGCCTGAGTCACTGGTCACCCTTAATATTTATTAAGTGCCTGAGACACCCGGTTACCTTG GCCGTGAGGACACGTGGCCTGCACCCAGGTGTGGCTGTCAGGACACCAGCCTGGTGCCCATCCTCCCGACCCCTACCCACTTCCATTCCC >65773_65773_12_PKN1-LDLR_PKN1_chr19_14557373_ENST00000342216_LDLR_chr19_11227535_ENST00000558518_length(amino acids)=501AA_BP=209 MAEANNPSEQELESEPRSWSLLEQLGLAGADLAAPGVQQQLELERERLRREIRKELKLKEGAENLRRATTDLGRSLGPVELLLRGSSRRL DLLHQQLQELHAHVVLPDPAATHDGPQSPGAGGPTCSATNLSRVAGLEKQLAIELKVKQGAENMIQTYSNGSTKDRKLLLTAQQMLQDSK TKIDIIRMQLRRALQAGQLENQAAPDDTQDLLSGRLYWVDSKLHSISSIDVNGGNRKTILEDEKRLAHPFSLAVFEDKVFWTDIINEAIF SANRLTGSDVNLLAENLLSPEDMVLFHNLTQPRGVNWCERTTLSNGGCQYLCLPAPQINPHSPKFTCACPDGMLLARDMRSCLTEAEAAV ATQETSTVRLKVSSTAVRTQHTTTRPVPDTSRLPGATPGLTTVEIVTMSHQALGDVAGRGNEKKPSSVRALSIVLPIVLLVFLCLGVFLL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for PKN1-LDLR |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for PKN1-LDLR |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for PKN1-LDLR |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
