
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:PLXNC1-APPL2 (FusionGDB2 ID:66582) |
Fusion Gene Summary for PLXNC1-APPL2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: PLXNC1-APPL2 | Fusion gene ID: 66582 | Hgene | Tgene | Gene symbol | PLXNC1 | APPL2 | Gene ID | 10154 | 55198 |
| Gene name | plexin C1 | adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 2 | |
| Synonyms | CD232|PLXN-C1|VESPR | DIP13B | |
| Cytomap | 12q22 | 12q23.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | plexin-C1plexin (semaphorin receptor)receptor for viral semaphorin proteinreceptor for virally-encoded semaphorinvirus-encoded semaphorin protein receptor | DCC-interacting protein 13-betaDIP13 betaadapter protein containing PH domain, PTB domain and leucine zipper motif 2adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q8NEU8 | |
| Ensembl transtripts involved in fusion gene | ENST00000258526, ENST00000547057, ENST00000545312, ENST00000551495, | ENST00000539978, ENST00000546731, ENST00000549573, ENST00000551662, ENST00000258530, | |
| Fusion gene scores | * DoF score | 7 X 6 X 5=210 | 8 X 6 X 5=240 |
| # samples | 10 | 9 | |
| ** MAII score | log2(10/210*10)=-1.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/240*10)=-1.41503749927884 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: PLXNC1 [Title/Abstract] AND APPL2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | PLXNC1(94659001)-APPL2(105623001), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | APPL2 | GO:0006606 | protein import into nucleus | 26583432 |
| Tgene | APPL2 | GO:0051289 | protein homotetramerization | 23055524 |
| Tgene | APPL2 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 15016378 |
 Fusion gene breakpoints across PLXNC1 (5'-gene) Fusion gene breakpoints across PLXNC1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
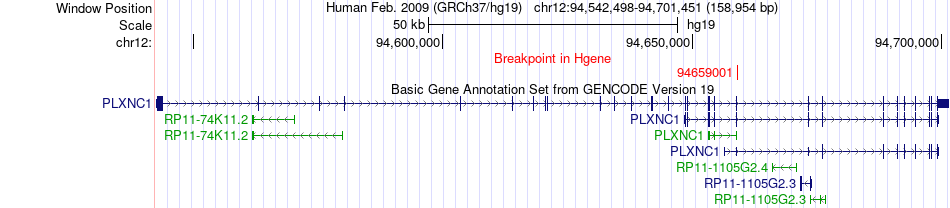 |
 Fusion gene breakpoints across APPL2 (3'-gene) Fusion gene breakpoints across APPL2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
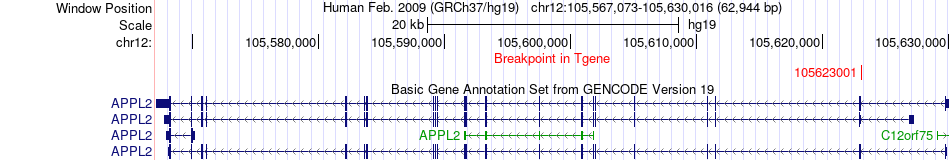 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | GBM | TCGA-06-0129-01A | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
Top |
Fusion Gene ORF analysis for PLXNC1-APPL2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000258526 | ENST00000539978 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
| 5CDS-5UTR | ENST00000547057 | ENST00000539978 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
| 5CDS-intron | ENST00000258526 | ENST00000546731 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
| 5CDS-intron | ENST00000258526 | ENST00000549573 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
| 5CDS-intron | ENST00000258526 | ENST00000551662 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
| 5CDS-intron | ENST00000547057 | ENST00000546731 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
| 5CDS-intron | ENST00000547057 | ENST00000549573 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
| 5CDS-intron | ENST00000547057 | ENST00000551662 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
| 5UTR-3CDS | ENST00000545312 | ENST00000258530 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
| 5UTR-5UTR | ENST00000545312 | ENST00000539978 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
| 5UTR-intron | ENST00000545312 | ENST00000546731 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
| 5UTR-intron | ENST00000545312 | ENST00000549573 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
| 5UTR-intron | ENST00000545312 | ENST00000551662 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
| In-frame | ENST00000258526 | ENST00000258530 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
| In-frame | ENST00000547057 | ENST00000258530 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
| intron-3CDS | ENST00000551495 | ENST00000258530 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
| intron-5UTR | ENST00000551495 | ENST00000539978 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
| intron-intron | ENST00000551495 | ENST00000546731 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
| intron-intron | ENST00000551495 | ENST00000549573 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
| intron-intron | ENST00000551495 | ENST00000551662 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000258526 | PLXNC1 | chr12 | 94659001 | + | ENST00000258530 | APPL2 | chr12 | 105623001 | - | 6805 | 3846 | 33 | 5786 | 1917 |
| ENST00000547057 | PLXNC1 | chr12 | 94659001 | + | ENST00000258530 | APPL2 | chr12 | 105623001 | - | 3934 | 975 | 141 | 2915 | 924 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000258526 | ENST00000258530 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - | 0.000678508 | 0.99932146 |
| ENST00000547057 | ENST00000258530 | PLXNC1 | chr12 | 94659001 | + | APPL2 | chr12 | 105623001 | - | 0.000603577 | 0.99939644 |
Top |
Fusion Genomic Features for PLXNC1-APPL2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
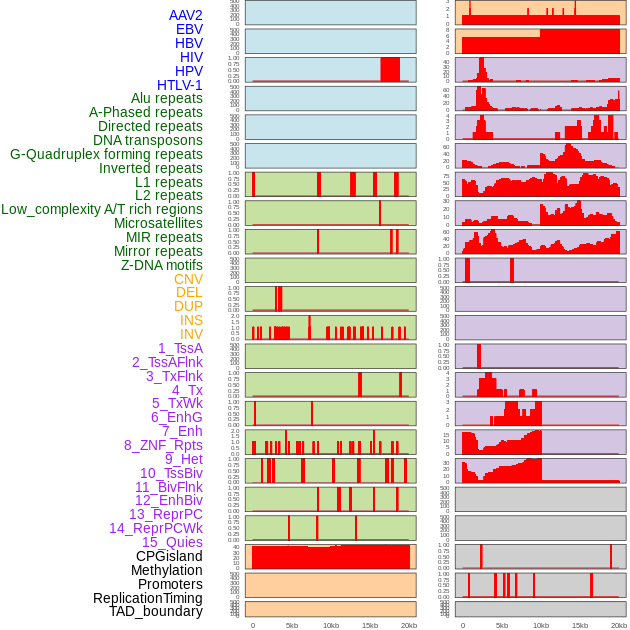 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for PLXNC1-APPL2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:94659001/chr12:105623001) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | APPL2 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Multifunctional adapter protein that binds to various membrane receptors, nuclear factors and signaling proteins to regulate many processes, such as cell proliferation, immune response, endosomal trafficking and cell metabolism (PubMed:26583432, PubMed:15016378, PubMed:24879834). Regulates signaling pathway leading to cell proliferation through interaction with RAB5A and subunits of the NuRD/MeCP1 complex (PubMed:15016378). Plays a role in immune response by modulating phagocytosis, inflammatory and innate immune responses. In macrophages, enhances Fc-gamma receptor-mediated phagocytosis through interaction with RAB31 leading to activation of PI3K/Akt signaling. In response to LPS, modulates inflammatory responses by playing a key role on the regulation of TLR4 signaling and in the nuclear translocation of RELA/NF-kappa-B p65 and the secretion of pro- and anti-inflammatory cytokines. Also functions as a negative regulator of innate immune response via inhibition of AKT1 signaling pathway by forming a complex with APPL1 and PIK3R1 (By similarity). Plays a role in endosomal trafficking of TGFBR1 from the endosomes to the nucleus (PubMed:26583432). Plays a role in cell metabolism by regulating adiponecting ans insulin signaling pathways and adaptative thermogenesis (PubMed:24879834) (By similarity). In muscle, negatively regulates adiponectin-simulated glucose uptake and fatty acid oxidation by inhibiting adiponectin signaling pathway through APPL1 sequestration thereby antagonizing APPL1 action (By similarity). In muscles, negativeliy regulates insulin-induced plasma membrane recruitment of GLUT4 and glucose uptake through interaction with TBC1D1 (PubMed:24879834). Plays a role in cold and diet-induced adaptive thermogenesis by activating ventromedial hypothalamus (VMH) neurons throught AMPK inhibition which enhances sympathetic outflow to subcutaneous white adipose tissue (sWAT), sWAT beiging and cold tolerance (By similarity). Also plays a role in other signaling pathways namely Wnt/beta-catenin, HGF and glucocorticoid receptor signaling (PubMed:19433865) (By similarity). Positive regulator of beta-catenin/TCF-dependent transcription through direct interaction with RUVBL2/reptin resulting in the relief of RUVBL2-mediated repression of beta-catenin/TCF target genes by modulating the interactions within the beta-catenin-reptin-HDAC complex (PubMed:19433865). May affect adult neurogenesis in hippocampus and olfactory system via regulating the sensitivity of glucocorticoid receptor. Required for fibroblast migration through HGF cell signaling (By similarity). {ECO:0000250|UniProtKB:Q8K3G9, ECO:0000269|PubMed:15016378, ECO:0000269|PubMed:19433865, ECO:0000269|PubMed:24879834, ECO:0000269|PubMed:26583432}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PLXNC1 | chr12:94659001 | chr12:105623001 | ENST00000258526 | + | 21 | 31 | 35_452 | 1199 | 1569.0 | Domain | Sema |
| Hgene | PLXNC1 | chr12:94659001 | chr12:105623001 | ENST00000258526 | + | 21 | 31 | 35_944 | 1199 | 1569.0 | Topological domain | Extracellular |
| Hgene | PLXNC1 | chr12:94659001 | chr12:105623001 | ENST00000258526 | + | 21 | 31 | 945_965 | 1199 | 1569.0 | Transmembrane | Helical |
| Tgene | APPL2 | chr12:94659001 | chr12:105623001 | ENST00000258530 | 0 | 21 | 277_375 | 18 | 665.0 | Domain | PH | |
| Tgene | APPL2 | chr12:94659001 | chr12:105623001 | ENST00000258530 | 0 | 21 | 488_637 | 18 | 665.0 | Domain | PID | |
| Tgene | APPL2 | chr12:94659001 | chr12:105623001 | ENST00000539978 | 0 | 21 | 277_375 | 0 | 622.0 | Domain | PH | |
| Tgene | APPL2 | chr12:94659001 | chr12:105623001 | ENST00000539978 | 0 | 21 | 3_268 | 0 | 622.0 | Domain | BAR | |
| Tgene | APPL2 | chr12:94659001 | chr12:105623001 | ENST00000539978 | 0 | 21 | 488_637 | 0 | 622.0 | Domain | PID | |
| Tgene | APPL2 | chr12:94659001 | chr12:105623001 | ENST00000551662 | 0 | 21 | 277_375 | 18 | 671.0 | Domain | PH | |
| Tgene | APPL2 | chr12:94659001 | chr12:105623001 | ENST00000551662 | 0 | 21 | 488_637 | 18 | 671.0 | Domain | PID | |
| Tgene | APPL2 | chr12:94659001 | chr12:105623001 | ENST00000539978 | 0 | 21 | 1_428 | 0 | 622.0 | Region | Required for RAB5A binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PLXNC1 | chr12:94659001 | chr12:105623001 | ENST00000258526 | + | 21 | 31 | 966_1568 | 1199 | 1569.0 | Topological domain | Cytoplasmic |
| Tgene | APPL2 | chr12:94659001 | chr12:105623001 | ENST00000258530 | 0 | 21 | 3_268 | 18 | 665.0 | Domain | BAR | |
| Tgene | APPL2 | chr12:94659001 | chr12:105623001 | ENST00000551662 | 0 | 21 | 3_268 | 18 | 671.0 | Domain | BAR | |
| Tgene | APPL2 | chr12:94659001 | chr12:105623001 | ENST00000258530 | 0 | 21 | 1_428 | 18 | 665.0 | Region | Required for RAB5A binding | |
| Tgene | APPL2 | chr12:94659001 | chr12:105623001 | ENST00000551662 | 0 | 21 | 1_428 | 18 | 671.0 | Region | Required for RAB5A binding |
Top |
Fusion Gene Sequence for PLXNC1-APPL2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >66582_66582_1_PLXNC1-APPL2_PLXNC1_chr12_94659001_ENST00000258526_APPL2_chr12_105623001_ENST00000258530_length(transcript)=6805nt_BP=3846nt GCGAGGAGGAAACGGTGCCGGAGCGCGCAGGGCTTGCTGCCGCCACCGCCGCTGCACAGGCTGCCGGAGCGAGCCTGCCGCGCGCCGCCC TCCCCGCTCTCCTTCCTGGGCGAGCTGCGGGGATGGGGCGGCCGCGGGAGCCCGAGCGCGCGCAGGAACCGCCGCCGCCGCCGCCCGCGT CTCCGTTGCCGCGCGCCTGAGCCGCCGTCGCCGCCGCGCGCCCTGCCCGGGGGCGGCCCCCCCAGCCCCATGGAGGTCTCCCGGAGGAAG GCGCCGCCGCGCCCCCCGCGCCCCGCAGCGCCACTGCCCCTGCTCGCCTATCTGCTGGCACTGGCGGCTCCCGGCCGGGGCGCGGACGAG CCCGTGTGGCGGTCGGAGCAAGCCATCGGAGCCATCGCGGCGAGCCAGGAGGACGGCGTGTTTGTGGCGAGCGGCAGCTGCCTGGACCAG CTGGACTACAGCCTGGAGCACAGCCTCTCGCGCCTGTACCGGGACCAAGCGGGCAACTGCACAGAGCCGGTCTCGCTGGCGCCCCCCGCG CGGCCCCGGCCCGGGAGCAGCTTCAGCAAGCTGCTGCTGCCCTACCGCGAGGGGGCGGCCGGCCTCGGGGGGCTGCTGCTCACCGGCTGG ACCTTCGACCGGGGCGCCTGCGAGGTGCGGCCCCTGGGCAACCTGAGCCGCAACTCCCTGCGCAACGGCACCGAGGTGGTGTCGTGCCAC CCGCAGGGCTCGACGGCCGGCGTGGTGTACCGCGCGGGCCGGAACAACCGCTGGTACCTGGCGGTGGCCGCCACCTACGTGCTGCCTGAG CCGGAGACGGCGAGCCGCTGCAACCCCGCGGCATCCGACCACGACACGGCCATCGCGCTCAAGGACACGGAGGGGCGCAGCCTGGCCACG CAGGAGCTGGGGCGCCTCAAGCTGTGCGAGGGCGCGGGCAGCCTGCACTTCGTGGACGCCTTTCTCTGGAACGGCAGCATCTACTTCCCC TACTACCCCTACAACTACACGAGCGGCGCTGCCACCGGCTGGCCCAGCATGGCGCGCATCGCGCAGAGCACCGAGGTGCTGTTCCAGGGC CAGGCATCCCTCGACTGCGGCCACGGCCACCCCGACGGCCGCCGCCTGCTCCTCTCCTCCAGCCTAGTGGAGGCCCTGGACGTCTGGGCG GGAGTGTTCAGCGCGGCCGCTGGAGAGGGCCAGGAGCGGCGCTCCCCCACCACCACGGCGCTCTGCCTCTTCAGAATGAGTGAGATCCAG GCGCGCGCCAAGAGGGTCAGCTGGGACTTCAAGACGGCCGAGAGCCACTGCAAAGAAGGGGATCAACCTGAAAGAGTCCAACCAATCGCA TCATCTACCTTGATCCATTCCGACCTGACATCCGTTTATGGCACCGTGGTAATGAACAGGACTGTTTTATTCTTGGGGACTGGAGATGGC CAGTTACTTAAGGTTATTCTTGGTGAGAATTTGACTTCAAATTGTCCAGAGGTTATCTATGAAATTAAAGAAGAGACACCTGTTTTCTAC AAACTCGTTCCTGATCCTGTGAAGAATATCTACATTTATCTAACAGCTGGGAAAGAGGTGAGGAGAATTCGTGTTGCAAACTGCAATAAA CATAAATCCTGTTCGGAGTGTTTAACAGCCACAGACCCTCACTGCGGTTGGTGCCATTCGCTACAAAGGTGCACTTTTCAAGGAGATTGT GTACATTCAGAGAACTTAGAAAACTGGCTGGATATTTCGTCTGGAGCAAAAAAGTGCCCTAAAATTCAGATAATTCGAAGCAGTAAAGAA AAGACTACAGTGACTATGGTGGGAAGCTTCTCTCCAAGACACTCAAAGTGCATGGTGAAGAATGTGGACTCTAGCAGGGAGCTCTGCCAG AATAAAAGTCAGCCCAACCGGACCTGCACCTGTAGCATCCCAACCAGAGCAACCTACAAAGATGTTTCAGTTGTCAACGTGATGTTCTCC TTCGGTTCTTGGAATTTATCAGACAGATTCAACTTTACCAACTGCTCATCATTAAAAGAATGCCCAGCATGCGTAGAAACTGGCTGCGCG TGGTGTAAAAGTGCAAGAAGGTGTATCCACCCCTTCACAGCTTGCGACCCTTCTGATTATGAGAGAAACCAGGAACAGTGTCCAGTGGCT GTCGAGAAGACATCAGGAGGAGGAAGACCCAAGGAGAACAAGGGGAACAGAACCAACCAGGCTTTACAGGTCTTCTACATTAAGTCCATT GAGCCACAGAAAGTATCGACATTAGGGAAAAGCAACGTGATAGTAACGGGAGCAAACTTTACCCGGGCATCGAACATCACAATGATCCTG AAAGGAACCAGTACCTGTGATAAGGATGTGATACAGGTTAGCCATGTGCTAAATGACACCCACATGAAATTCTCTCTTCCATCAAGCCGG AAAGAAATGAAGGATGTGTGTATCCAGTTTGATGGTGGGAACTGCTCTTCTGTGGGATCCTTATCCTACATTGCTCTGCCACATTGTTCC CTTATATTTCCTGCTACCACCTGGATCAGTGGTGGTCAAAATATAACCATGATGGGCAGAAATTTTGATGTAATTGACAACTTAATCATT TCACATGAATTAAAAGGAAACATAAATGTCTCTGAATATTGTGTGGCGACTTACTGCGGGTTTTTAGCCCCCAGTTTAAAGAGTTCAAAA GTGCGCACGAATGTCACTGTGAAGCTGAGAGTACAAGACACCTACTTGGATTGTGGAACCCTGCAGTATCGGGAGGACCCCAGATTCACG GGGTATCGGGTGGAATCCGAGGTGGACACAGAACTGGAAGTGAAAATTCAAAAAGAAAATGACAACTTCAACATTTCCAAAAAAGACATT GAAATTACTCTCTTCCATGGGGAAAATGGGCAATTAAATTGCAGTTTTGAAAATATTACTAGAAATCAAGATCTTACCACCATCCTTTGC AAAATTAAAGGCATCAAGACTGCAAGCACCATTGCCAACTCTTCTAAGAAAGTTCGGGTCAAGCTGGGAAACCTGGAGCTCTACGTCGAG CAGGAGTCAGTTCCTTCCACATGGTATTTTCTGATTGTGCTCCCTGTCTTGCTAGTGATTGTCATTTTTGCGGCCGTGGGGGTGACCAGG CACAAATCGAAGGAGCTGAGTCGCAAACAGAGTCAACAACTAGAATTGCTGGAAAGCGAGCTCCGGAAAGAGATACGTGACGGCTTTGCT GAGCTGCAGATGGATAAATTGGATGTGGTTGATAGTTTTGGAACTGTTCCCTTCCTTGACTACAAACATTTTGCTCTGAGAACTTTCTTC CCTGAGTCAGGTGGCTTCACCCACATCTTCACTGAAGATATGCATAACAGAGACGCCAACGACAAGAATGAAAGTCTCACAGCTTTGGAT GCCCTAATCTGTAATAAAAGCTTTCTTGTTACTGTCATCCACACCCTTGAAAAGCAGAAGAACTTTTCTGTGAAGGACAGGTGTCTGTTT GCCTCCTTCCTAACCATTGCACTGCAAACCAAGCTGGTCTACCTGACCAGCATCCTAGAGGTGCTGACCAGGGACTTGATGGAACAGTGT AGTAACATGCAGCCGAAACTCATGCTGAGACGCACGGAGTCCGTCGTCGAAAAACTCCTCACAAACTGGATGTCCGTCTGCCTTTCTGGA TTTCTCCGGGAGACTGTCGGAGAGCCCTTCTATTTGCTGGTGACGACTCTGAACCAGAAAATTAACAAGGGTCCCGTGGATGTAATCACT TGCAAAGCCCTGTACACACTTAATGAAGACTGGCTGTTGTGGCAGGTTCCGGAATTCAGTACTGTGACTCGCTCTTTACTGAGCGTGTTT GAAGAAGATGCTGGCACCCTCACAGACTATACCAACCAGCTGCTCCAGGCAATGCAGCGCGTCTATGGAGCCCAGAATGAGATGTGCCTG GCCACACAACAGCTTTCTAAGCAACTGCTGGCATATGAAAAACAGAACTTTGCTCTTGGCAAAGGTGATGAAGAAGTAATTTCAACACTC CACTATTTTTCCAAAGTGGTGGATGAGCTTAATCTTCTCCATACAGAGCTGGCTAAACAGTTGGCAGACACAATGGTTCTACCTATCATA CAATTCCGAGAAAAGGATCTCACAGAAGTAAGCACTTTAAAGGATCTATTTGGACTCGCTAGCAATGAGCATGACCTCTCAATGGCAAAA TACAGCAGGCTGCCTAAGAAAAAGGAGAATGAGAAGGTGAAGACCGAAGTCGGAAAAGAGGTGGCCGCGGCCCGGCGGAAGCAGCACCTC TCCTCCCTTCAGTACTACTGTGCCCTCAACGCGCTGCAGTACAGAAAGCAAATGGCCATGATGGAGCCCATGATAGGCTTTGCCCATGGA CAGATTAACTTTTTTAAGAAGGGAGCAGAGATGTTTTCCAAACGTATGGACAGCTTTTTATCCTCCGTTGCAGACATGGTTCAAAGCATT CAGGTAGAACTGGAAGCCGAGGCGGAAAAGATGCGGGTGTCCCAGCAAGAATTACTTTCTGTTGATGAATCTGTTTACACTCCAGACTCT GATGTGGCCGCACCACAGATCAACAGGAACCTCATCCAGAAGGCTGGTTACCTTAATCTTAGAAACAAAACAGGGCTGGTCACCACCACC TGGGAGAGGCTTTATTTCTTCACCCAAGGCGGGAATCTCATGTGTCAGCCCAGGGGAGCCGTGGCTGGAGGTTTGATCCAGGACCTGGAC AACTGCTCAGTGATGGCCGTGGATTGCGAAGACCGGCGCTACTGCTTCCAGATCACCACGCCCAATGGAAAATCGGGAATAATCCTCCAG GCTGAGAGCAGAAAGGAAAATGAAGAGTGGATATGTGCAATAAACAACATCTCCAGACAGATCTACCTGACCGACAACCCTGAGGCAGTC GCGATCAAGTTGAATCAGACCGCTCTGCAAGCAGTGACTCCCATTACAAGTTTTGGAAAAAAACAAGAAAGCTCATGCCCCAGCCAGAAC CTGAAAAATTCAGAGATGGAAAATGAAAATGACAAGATTGTTCCCAAAGCAACAGCCAGTCTACCTGAAGCAGAGGAGCTGATCGCGCCT GGAACGCCGATTCAATTCGATATTGTGCTTCCTGCTACAGAATTCCTTGATCAGAACAGAGGGAGCAGGCGTACCAACCCTTTTGGTGAA ACTGAGGATGAATCATTTCCAGAAGCAGAAGATTCTCTTTTGCAGCAGATGTTTATAGTTCGGTTTTTGGGATCAATGGCAGTTAAAACA GACAGCACTACTGAAGTGATTTATGAAGCGATGAGACAAGTATTGGCTGCTCGGGCTATTCATAACATCTTCCGCATGACAGAATCCCAT CTGATGGTCACCAGTCAATCTTTGAGGTTGATAGATCCACAGACTCAAGTATCAAGGGCCAATTTTGAACTTACCAGTGTCACACAATTT GCTGCTCATCAAGAAAACAAGAGACTGGTTGGTTTTGTCATCCGTGTTCCTGAATCCACTGGAGAAGAATCTCTGAGTACATACATTTTT GAAAGCAACTCAGAAGGCGAAAAGATATGTTATGCTATTAATTTGGGAAAAGAAATTATTGAGGTTCAGAAGGATCCAGAAGCACTGGCT CAATTAATGCTGTCCATACCACTAACCAATGACGGAAAATATGTACTGTTAAACGATCAACCAGATGACGATGATGGAAATCCAAACGAA CATAGAGGCGCAGAATCCGAAGCATAACTCACTTGCGCCTGTGGGGGAAGAGCAAACAGGAAGGAGAGCTACCTCCTAAGGGTTTTAACG TCTCTGACATACAGGCACACTGACCTGATTTCCGAAGGCTGACAATCGTTTGTGGAATGTAATCTTGATGCCTTGATACTGAGACTTGGG AGGGAAACTAAGAAATGGTTGACAGCGTTCCCACCCATCTACAATGTTATTTTAGGTGCTTTGTGGTAAGTCTTTTTTCTTAGATTGCGC TAAAATTTCTTAGATTGTTCAGCGCTCAGAACAAAAGTTTGAAAAATGCATTGTTCATATGAATGTCATCTCTTTTCAGTTTCCAGTATC CTTTTTAAAAAATGGCAAAAGCCTAGATTTACAATTTGATGAACACTAAATATTTCTTATTAATATAATCTATTTTTGTATTTTACTTAA TGAGCTTTAAGTGCCTGTCGTTCTGAAAATTGTGTATTTATAATTCAGCTTATCTCATAATTGGACCTAATAGCATTTCTTTGTGCAGTT AGGTGATGAGCACTGCTTTGAGGCCCAAGCACTAGTAGAGATGCGCGATACAGGTCTAGTTTCGGTAACTGTTCCAGACATCAAGCAATA AAAAAATGAATACCACAAAAGATGTTTGATTTTACAGTGGAGCCTTACTGAACCAGCATTCAGAAGTTTAAGGTCCTCCTAGGTATGAGT ATTTTTAGTAGTGGATCACTGTGGACAGGGTGCAGCTCTACCAGTTCCTGTTTCTTCTGAGCCAGACCCTCTTCAGGGAAGGGACCAATT AATTTTAAAACTCACTTGAAGCACAGCTGGTCATGGGGCTTGGTATAAAGTTCCTATTTCCACCCTGATACTTCCAATTCCTGGAACCCC AGCCCACTCCCCCATCCCTCCTCCCTATCAAACTAGTATAATGATTTTGAATCGGTACAGTGTGTTTAACTGTAACTAAGTTCAACAGAC >66582_66582_1_PLXNC1-APPL2_PLXNC1_chr12_94659001_ENST00000258526_APPL2_chr12_105623001_ENST00000258530_length(amino acids)=1917AA_BP=1271 MLPPPPLHRLPERACRAPPSPLSFLGELRGWGGRGSPSARRNRRRRRPRLRCRAPEPPSPPRALPGGGPPSPMEVSRRKAPPRPPRPAAP LPLLAYLLALAAPGRGADEPVWRSEQAIGAIAASQEDGVFVASGSCLDQLDYSLEHSLSRLYRDQAGNCTEPVSLAPPARPRPGSSFSKL LLPYREGAAGLGGLLLTGWTFDRGACEVRPLGNLSRNSLRNGTEVVSCHPQGSTAGVVYRAGRNNRWYLAVAATYVLPEPETASRCNPAA SDHDTAIALKDTEGRSLATQELGRLKLCEGAGSLHFVDAFLWNGSIYFPYYPYNYTSGAATGWPSMARIAQSTEVLFQGQASLDCGHGHP DGRRLLLSSSLVEALDVWAGVFSAAAGEGQERRSPTTTALCLFRMSEIQARAKRVSWDFKTAESHCKEGDQPERVQPIASSTLIHSDLTS VYGTVVMNRTVLFLGTGDGQLLKVILGENLTSNCPEVIYEIKEETPVFYKLVPDPVKNIYIYLTAGKEVRRIRVANCNKHKSCSECLTAT DPHCGWCHSLQRCTFQGDCVHSENLENWLDISSGAKKCPKIQIIRSSKEKTTVTMVGSFSPRHSKCMVKNVDSSRELCQNKSQPNRTCTC SIPTRATYKDVSVVNVMFSFGSWNLSDRFNFTNCSSLKECPACVETGCAWCKSARRCIHPFTACDPSDYERNQEQCPVAVEKTSGGGRPK ENKGNRTNQALQVFYIKSIEPQKVSTLGKSNVIVTGANFTRASNITMILKGTSTCDKDVIQVSHVLNDTHMKFSLPSSRKEMKDVCIQFD GGNCSSVGSLSYIALPHCSLIFPATTWISGGQNITMMGRNFDVIDNLIISHELKGNINVSEYCVATYCGFLAPSLKSSKVRTNVTVKLRV QDTYLDCGTLQYREDPRFTGYRVESEVDTELEVKIQKENDNFNISKKDIEITLFHGENGQLNCSFENITRNQDLTTILCKIKGIKTASTI ANSSKKVRVKLGNLELYVEQESVPSTWYFLIVLPVLLVIVIFAAVGVTRHKSKELSRKQSQQLELLESELRKEIRDGFAELQMDKLDVVD SFGTVPFLDYKHFALRTFFPESGGFTHIFTEDMHNRDANDKNESLTALDALICNKSFLVTVIHTLEKQKNFSVKDRCLFASFLTIALQTK LVYLTSILEVLTRDLMEQCSNMQPKLMLRRTESVVEKLLTNWMSVCLSGFLRETVGEPFYLLVTTLNQKINKGPVDVITCKALYTLNEDW LLWQVPEFSTVTRSLLSVFEEDAGTLTDYTNQLLQAMQRVYGAQNEMCLATQQLSKQLLAYEKQNFALGKGDEEVISTLHYFSKVVDELN LLHTELAKQLADTMVLPIIQFREKDLTEVSTLKDLFGLASNEHDLSMAKYSRLPKKKENEKVKTEVGKEVAAARRKQHLSSLQYYCALNA LQYRKQMAMMEPMIGFAHGQINFFKKGAEMFSKRMDSFLSSVADMVQSIQVELEAEAEKMRVSQQELLSVDESVYTPDSDVAAPQINRNL IQKAGYLNLRNKTGLVTTTWERLYFFTQGGNLMCQPRGAVAGGLIQDLDNCSVMAVDCEDRRYCFQITTPNGKSGIILQAESRKENEEWI CAINNISRQIYLTDNPEAVAIKLNQTALQAVTPITSFGKKQESSCPSQNLKNSEMENENDKIVPKATASLPEAEELIAPGTPIQFDIVLP ATEFLDQNRGSRRTNPFGETEDESFPEAEDSLLQQMFIVRFLGSMAVKTDSTTEVIYEAMRQVLAARAIHNIFRMTESHLMVTSQSLRLI DPQTQVSRANFELTSVTQFAAHQENKRLVGFVIRVPESTGEESLSTYIFESNSEGEKICYAINLGKEIIEVQKDPEALAQLMLSIPLTND -------------------------------------------------------------- >66582_66582_2_PLXNC1-APPL2_PLXNC1_chr12_94659001_ENST00000547057_APPL2_chr12_105623001_ENST00000258530_length(transcript)=3934nt_BP=975nt ACTTGTAAAATGAGTAAGGTTTTGGTTTTTTTCTGCTGCATTTTGGCATCTTCCTCTTCAGAGTGACCCCTCACTGGGAAATTTTCTTAT AAGATCCAGCTTTTTAATGCTATTTTCTAGTTTCATTCTAGCTCCTGCTCTCTGTCCCTATCTGTTTTGTTTTTGTTTTTGTTTTTAAGA ACTGTAAAGATTGCTATTTGTTTTCATTTTTGGTTTTTGAGTTACCATGTCCCTCTGATGCAGCATCTCTGTCTCTTAGCGGCCGTGGGG GTGACCAGGCACAAATCGAAGGAGCTGAGTCGCAAACAGAGTCAACAACTAGAATTGCTGGAAAGCGAGCTCCGGAAAGAGATACGTGAC GGCTTTGCTGAGCTGCAGATGGATAAATTGGATGTGGTTGATAGTTTTGGAACTGTTCCCTTCCTTGACTACAAACATTTTGCTCTGAGA ACTTTCTTCCCTGAGTCAGGTGGCTTCACCCACATCTTCACTGAAGATATGCATAACAGAGACGCCAACGACAAGAATGAAAGTCTCACA GCTTTGGATGCCCTAATCTGTAATAAAAGCTTTCTTGTTACTGTCATCCACACCCTTGAAAAGCAGAAGAACTTTTCTGTGAAGGACAGG TGTCTGTTTGCCTCCTTCCTAACCATTGCACTGCAAACCAAGCTGGTCTACCTGACCAGCATCCTAGAGGTGCTGACCAGGGACTTGATG GAACAGTGTAGTAACATGCAGCCGAAACTCATGCTGAGACGCACGGAGTCCGTCGTCGAAAAACTCCTCACAAACTGGATGTCCGTCTGC CTTTCTGGATTTCTCCGGGAGACTGTCGGAGAGCCCTTCTATTTGCTGGTGACGACTCTGAACCAGAAAATTAACAAGGGTCCCGTGGAT GTAATCACTTGCAAAGCCCTGTACACACTTAATGAAGACTGGCTGTTGTGGCAGGTTCCGGAATTCAGTACTGTGACTCGCTCTTTACTG AGCGTGTTTGAAGAAGATGCTGGCACCCTCACAGACTATACCAACCAGCTGCTCCAGGCAATGCAGCGCGTCTATGGAGCCCAGAATGAG ATGTGCCTGGCCACACAACAGCTTTCTAAGCAACTGCTGGCATATGAAAAACAGAACTTTGCTCTTGGCAAAGGTGATGAAGAAGTAATT TCAACACTCCACTATTTTTCCAAAGTGGTGGATGAGCTTAATCTTCTCCATACAGAGCTGGCTAAACAGTTGGCAGACACAATGGTTCTA CCTATCATACAATTCCGAGAAAAGGATCTCACAGAAGTAAGCACTTTAAAGGATCTATTTGGACTCGCTAGCAATGAGCATGACCTCTCA ATGGCAAAATACAGCAGGCTGCCTAAGAAAAAGGAGAATGAGAAGGTGAAGACCGAAGTCGGAAAAGAGGTGGCCGCGGCCCGGCGGAAG CAGCACCTCTCCTCCCTTCAGTACTACTGTGCCCTCAACGCGCTGCAGTACAGAAAGCAAATGGCCATGATGGAGCCCATGATAGGCTTT GCCCATGGACAGATTAACTTTTTTAAGAAGGGAGCAGAGATGTTTTCCAAACGTATGGACAGCTTTTTATCCTCCGTTGCAGACATGGTT CAAAGCATTCAGGTAGAACTGGAAGCCGAGGCGGAAAAGATGCGGGTGTCCCAGCAAGAATTACTTTCTGTTGATGAATCTGTTTACACT CCAGACTCTGATGTGGCCGCACCACAGATCAACAGGAACCTCATCCAGAAGGCTGGTTACCTTAATCTTAGAAACAAAACAGGGCTGGTC ACCACCACCTGGGAGAGGCTTTATTTCTTCACCCAAGGCGGGAATCTCATGTGTCAGCCCAGGGGAGCCGTGGCTGGAGGTTTGATCCAG GACCTGGACAACTGCTCAGTGATGGCCGTGGATTGCGAAGACCGGCGCTACTGCTTCCAGATCACCACGCCCAATGGAAAATCGGGAATA ATCCTCCAGGCTGAGAGCAGAAAGGAAAATGAAGAGTGGATATGTGCAATAAACAACATCTCCAGACAGATCTACCTGACCGACAACCCT GAGGCAGTCGCGATCAAGTTGAATCAGACCGCTCTGCAAGCAGTGACTCCCATTACAAGTTTTGGAAAAAAACAAGAAAGCTCATGCCCC AGCCAGAACCTGAAAAATTCAGAGATGGAAAATGAAAATGACAAGATTGTTCCCAAAGCAACAGCCAGTCTACCTGAAGCAGAGGAGCTG ATCGCGCCTGGAACGCCGATTCAATTCGATATTGTGCTTCCTGCTACAGAATTCCTTGATCAGAACAGAGGGAGCAGGCGTACCAACCCT TTTGGTGAAACTGAGGATGAATCATTTCCAGAAGCAGAAGATTCTCTTTTGCAGCAGATGTTTATAGTTCGGTTTTTGGGATCAATGGCA GTTAAAACAGACAGCACTACTGAAGTGATTTATGAAGCGATGAGACAAGTATTGGCTGCTCGGGCTATTCATAACATCTTCCGCATGACA GAATCCCATCTGATGGTCACCAGTCAATCTTTGAGGTTGATAGATCCACAGACTCAAGTATCAAGGGCCAATTTTGAACTTACCAGTGTC ACACAATTTGCTGCTCATCAAGAAAACAAGAGACTGGTTGGTTTTGTCATCCGTGTTCCTGAATCCACTGGAGAAGAATCTCTGAGTACA TACATTTTTGAAAGCAACTCAGAAGGCGAAAAGATATGTTATGCTATTAATTTGGGAAAAGAAATTATTGAGGTTCAGAAGGATCCAGAA GCACTGGCTCAATTAATGCTGTCCATACCACTAACCAATGACGGAAAATATGTACTGTTAAACGATCAACCAGATGACGATGATGGAAAT CCAAACGAACATAGAGGCGCAGAATCCGAAGCATAACTCACTTGCGCCTGTGGGGGAAGAGCAAACAGGAAGGAGAGCTACCTCCTAAGG GTTTTAACGTCTCTGACATACAGGCACACTGACCTGATTTCCGAAGGCTGACAATCGTTTGTGGAATGTAATCTTGATGCCTTGATACTG AGACTTGGGAGGGAAACTAAGAAATGGTTGACAGCGTTCCCACCCATCTACAATGTTATTTTAGGTGCTTTGTGGTAAGTCTTTTTTCTT AGATTGCGCTAAAATTTCTTAGATTGTTCAGCGCTCAGAACAAAAGTTTGAAAAATGCATTGTTCATATGAATGTCATCTCTTTTCAGTT TCCAGTATCCTTTTTAAAAAATGGCAAAAGCCTAGATTTACAATTTGATGAACACTAAATATTTCTTATTAATATAATCTATTTTTGTAT TTTACTTAATGAGCTTTAAGTGCCTGTCGTTCTGAAAATTGTGTATTTATAATTCAGCTTATCTCATAATTGGACCTAATAGCATTTCTT TGTGCAGTTAGGTGATGAGCACTGCTTTGAGGCCCAAGCACTAGTAGAGATGCGCGATACAGGTCTAGTTTCGGTAACTGTTCCAGACAT CAAGCAATAAAAAAATGAATACCACAAAAGATGTTTGATTTTACAGTGGAGCCTTACTGAACCAGCATTCAGAAGTTTAAGGTCCTCCTA GGTATGAGTATTTTTAGTAGTGGATCACTGTGGACAGGGTGCAGCTCTACCAGTTCCTGTTTCTTCTGAGCCAGACCCTCTTCAGGGAAG GGACCAATTAATTTTAAAACTCACTTGAAGCACAGCTGGTCATGGGGCTTGGTATAAAGTTCCTATTTCCACCCTGATACTTCCAATTCC TGGAACCCCAGCCCACTCCCCCATCCCTCCTCCCTATCAAACTAGTATAATGATTTTGAATCGGTACAGTGTGTTTAACTGTAACTAAGT >66582_66582_2_PLXNC1-APPL2_PLXNC1_chr12_94659001_ENST00000547057_APPL2_chr12_105623001_ENST00000258530_length(amino acids)=924AA_BP=278 MSLSVLFLFLFLRTVKIAICFHFWFLSYHVPLMQHLCLLAAVGVTRHKSKELSRKQSQQLELLESELRKEIRDGFAELQMDKLDVVDSFG TVPFLDYKHFALRTFFPESGGFTHIFTEDMHNRDANDKNESLTALDALICNKSFLVTVIHTLEKQKNFSVKDRCLFASFLTIALQTKLVY LTSILEVLTRDLMEQCSNMQPKLMLRRTESVVEKLLTNWMSVCLSGFLRETVGEPFYLLVTTLNQKINKGPVDVITCKALYTLNEDWLLW QVPEFSTVTRSLLSVFEEDAGTLTDYTNQLLQAMQRVYGAQNEMCLATQQLSKQLLAYEKQNFALGKGDEEVISTLHYFSKVVDELNLLH TELAKQLADTMVLPIIQFREKDLTEVSTLKDLFGLASNEHDLSMAKYSRLPKKKENEKVKTEVGKEVAAARRKQHLSSLQYYCALNALQY RKQMAMMEPMIGFAHGQINFFKKGAEMFSKRMDSFLSSVADMVQSIQVELEAEAEKMRVSQQELLSVDESVYTPDSDVAAPQINRNLIQK AGYLNLRNKTGLVTTTWERLYFFTQGGNLMCQPRGAVAGGLIQDLDNCSVMAVDCEDRRYCFQITTPNGKSGIILQAESRKENEEWICAI NNISRQIYLTDNPEAVAIKLNQTALQAVTPITSFGKKQESSCPSQNLKNSEMENENDKIVPKATASLPEAEELIAPGTPIQFDIVLPATE FLDQNRGSRRTNPFGETEDESFPEAEDSLLQQMFIVRFLGSMAVKTDSTTEVIYEAMRQVLAARAIHNIFRMTESHLMVTSQSLRLIDPQ TQVSRANFELTSVTQFAAHQENKRLVGFVIRVPESTGEESLSTYIFESNSEGEKICYAINLGKEIIEVQKDPEALAQLMLSIPLTNDGKY -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for PLXNC1-APPL2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for PLXNC1-APPL2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for PLXNC1-APPL2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
