
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:PRKG1-SLC24A3 (FusionGDB2 ID:68859) |
Fusion Gene Summary for PRKG1-SLC24A3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: PRKG1-SLC24A3 | Fusion gene ID: 68859 | Hgene | Tgene | Gene symbol | PRKG1 | SLC24A3 | Gene ID | 5592 | 57419 |
| Gene name | protein kinase cGMP-dependent 1 | solute carrier family 24 member 3 | |
| Synonyms | AAT8|PKG|PKG1|PRKG1B|PRKGR1B|cGK|cGK 1|cGK1|cGKI|cGKI-BETA|cGKI-alpha | NCKX3 | |
| Cytomap | 10q11.23-q21.1 | 20p11.23 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cGMP-dependent protein kinase 1protein kinase, cGMP-dependent, regulatory, type I, betaprotein kinase, cGMP-dependent, type I | sodium/potassium/calcium exchanger 3Na(+)/K(+)/Ca(2+)-exchange protein 3sodium calcium exchangersolute carrier family 24 (sodium/potassium/calcium exchanger), member 3 | |
| Modification date | 20200322 | 20200327 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000373980, ENST00000373985, ENST00000401604, ENST00000373975, | ENST00000328041, | |
| Fusion gene scores | * DoF score | 10 X 10 X 2=200 | 12 X 12 X 11=1584 |
| # samples | 10 | 15 | |
| ** MAII score | log2(10/200*10)=-1 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(15/1584*10)=-3.40053792958373 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: PRKG1 [Title/Abstract] AND SLC24A3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | PRKG1(52913090)-SLC24A3(19496132), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | PRKG1 | GO:0006468 | protein phosphorylation | 15905169 |
| Hgene | PRKG1 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation | 25447536 |
| Hgene | PRKG1 | GO:1904753 | negative regulation of vascular associated smooth muscle cell migration | 25447536 |
 Fusion gene breakpoints across PRKG1 (5'-gene) Fusion gene breakpoints across PRKG1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
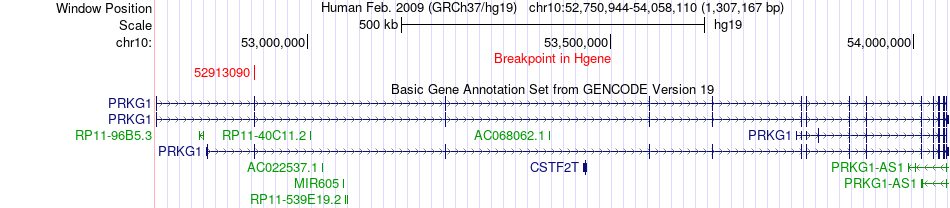 |
 Fusion gene breakpoints across SLC24A3 (3'-gene) Fusion gene breakpoints across SLC24A3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
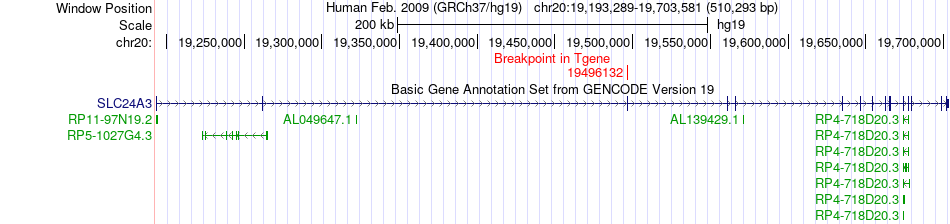 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-BR-8365-01A | PRKG1 | chr10 | 52913090 | + | SLC24A3 | chr20 | 19496132 | + |
Top |
Fusion Gene ORF analysis for PRKG1-SLC24A3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000373980 | ENST00000328041 | PRKG1 | chr10 | 52913090 | + | SLC24A3 | chr20 | 19496132 | + |
| In-frame | ENST00000373985 | ENST00000328041 | PRKG1 | chr10 | 52913090 | + | SLC24A3 | chr20 | 19496132 | + |
| In-frame | ENST00000401604 | ENST00000328041 | PRKG1 | chr10 | 52913090 | + | SLC24A3 | chr20 | 19496132 | + |
| intron-3CDS | ENST00000373975 | ENST00000328041 | PRKG1 | chr10 | 52913090 | + | SLC24A3 | chr20 | 19496132 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000401604 | PRKG1 | chr10 | 52913090 | + | ENST00000328041 | SLC24A3 | chr20 | 19496132 | + | 4088 | 627 | 194 | 2290 | 698 |
| ENST00000373985 | PRKG1 | chr10 | 52913090 | + | ENST00000328041 | SLC24A3 | chr20 | 19496132 | + | 3915 | 454 | 21 | 2117 | 698 |
| ENST00000373980 | PRKG1 | chr10 | 52913090 | + | ENST00000328041 | SLC24A3 | chr20 | 19496132 | + | 4356 | 895 | 417 | 2558 | 713 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000401604 | ENST00000328041 | PRKG1 | chr10 | 52913090 | + | SLC24A3 | chr20 | 19496132 | + | 0.001326368 | 0.9986737 |
| ENST00000373985 | ENST00000328041 | PRKG1 | chr10 | 52913090 | + | SLC24A3 | chr20 | 19496132 | + | 0.001000817 | 0.9989992 |
| ENST00000373980 | ENST00000328041 | PRKG1 | chr10 | 52913090 | + | SLC24A3 | chr20 | 19496132 | + | 0.001553854 | 0.99844617 |
Top |
Fusion Genomic Features for PRKG1-SLC24A3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| PRKG1 | chr10 | 52913090 | + | SLC24A3 | chr20 | 19496131 | + | 1.19E-06 | 0.9999988 |
| PRKG1 | chr10 | 52913090 | + | SLC24A3 | chr20 | 19496131 | + | 1.19E-06 | 0.9999988 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
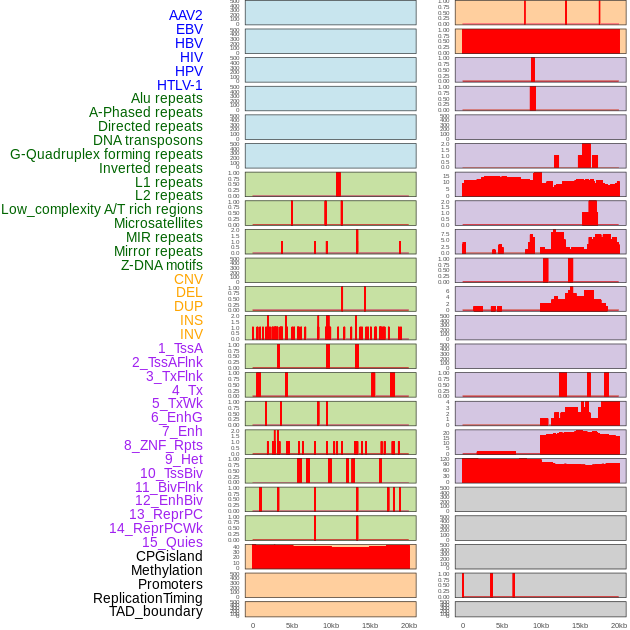 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
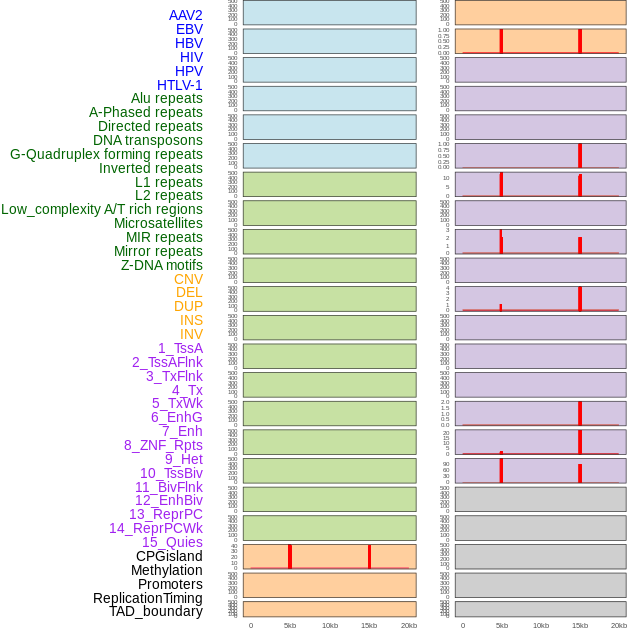 |
Top |
Fusion Protein Features for PRKG1-SLC24A3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:52913090/chr20:19496132) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000373980 | + | 2 | 18 | 2_59 | 159 | 687.0 | Coiled coil | Ontology_term=ECO:0000269 |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000401604 | + | 2 | 18 | 2_59 | 144 | 672.0 | Coiled coil | Ontology_term=ECO:0000269 |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000373980 | + | 2 | 18 | 2_102 | 159 | 687.0 | Region | Note=Required for dimerization |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000373980 | + | 2 | 18 | 50_75 | 159 | 687.0 | Region | Autoinhibitory domain |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000373980 | + | 2 | 18 | 9_44 | 159 | 687.0 | Region | Note=Leucine-zipper |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000401604 | + | 2 | 18 | 2_102 | 144 | 672.0 | Region | Note=Required for dimerization |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000401604 | + | 2 | 18 | 50_75 | 144 | 672.0 | Region | Autoinhibitory domain |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000401604 | + | 2 | 18 | 9_44 | 144 | 672.0 | Region | Note=Leucine-zipper |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 422_428 | 90 | 645.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 149_189 | 90 | 645.0 | Repeat | Note=Alpha-1 | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 518_549 | 90 | 645.0 | Repeat | Note=Alpha-2 | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 129_153 | 90 | 645.0 | Topological domain | Cytoplasmic | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 175_182 | 90 | 645.0 | Topological domain | Extracellular | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 204_210 | 90 | 645.0 | Topological domain | Cytoplasmic | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 232_234 | 90 | 645.0 | Topological domain | Extracellular | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 256_484 | 90 | 645.0 | Topological domain | Cytoplasmic | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 506_510 | 90 | 645.0 | Topological domain | Extracellular | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 532_549 | 90 | 645.0 | Topological domain | Cytoplasmic | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 571_580 | 90 | 645.0 | Topological domain | Extracellular | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 602_615 | 90 | 645.0 | Topological domain | Cytoplasmic | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 637_644 | 90 | 645.0 | Topological domain | Extracellular | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 108_128 | 90 | 645.0 | Transmembrane | Helical | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 154_174 | 90 | 645.0 | Transmembrane | Helical | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 183_203 | 90 | 645.0 | Transmembrane | Helical | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 211_231 | 90 | 645.0 | Transmembrane | Helical | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 235_255 | 90 | 645.0 | Transmembrane | Helical | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 485_505 | 90 | 645.0 | Transmembrane | Helical | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 511_531 | 90 | 645.0 | Transmembrane | Helical | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 550_570 | 90 | 645.0 | Transmembrane | Helical | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 581_601 | 90 | 645.0 | Transmembrane | Helical | |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 616_636 | 90 | 645.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000373980 | + | 2 | 18 | 360_619 | 159 | 687.0 | Domain | Protein kinase |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000373980 | + | 2 | 18 | 620_671 | 159 | 687.0 | Domain | AGC-kinase C-terminal |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000401604 | + | 2 | 18 | 360_619 | 144 | 672.0 | Domain | Protein kinase |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000401604 | + | 2 | 18 | 620_671 | 144 | 672.0 | Domain | AGC-kinase C-terminal |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000373980 | + | 2 | 18 | 167_170 | 159 | 687.0 | Nucleotide binding | 3'%2C5'-cGMP 1 |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000373980 | + | 2 | 18 | 177_178 | 159 | 687.0 | Nucleotide binding | 3'%2C5'-cGMP 1 |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000373980 | + | 2 | 18 | 291_294 | 159 | 687.0 | Nucleotide binding | 3'%2C5'-cGMP 2 |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000373980 | + | 2 | 18 | 301_302 | 159 | 687.0 | Nucleotide binding | 3'%2C5'-cGMP 2 |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000373980 | + | 2 | 18 | 366_374 | 159 | 687.0 | Nucleotide binding | ATP |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000401604 | + | 2 | 18 | 167_170 | 144 | 672.0 | Nucleotide binding | 3'%2C5'-cGMP 1 |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000401604 | + | 2 | 18 | 177_178 | 144 | 672.0 | Nucleotide binding | 3'%2C5'-cGMP 1 |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000401604 | + | 2 | 18 | 291_294 | 144 | 672.0 | Nucleotide binding | 3'%2C5'-cGMP 2 |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000401604 | + | 2 | 18 | 301_302 | 144 | 672.0 | Nucleotide binding | 3'%2C5'-cGMP 2 |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000401604 | + | 2 | 18 | 366_374 | 144 | 672.0 | Nucleotide binding | ATP |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000373980 | + | 2 | 18 | 103_220 | 159 | 687.0 | Region | Note=cGMP-binding%2C high affinity |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000373980 | + | 2 | 18 | 221_341 | 159 | 687.0 | Region | Note=cGMP-binding%2C low affinity |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000401604 | + | 2 | 18 | 103_220 | 144 | 672.0 | Region | Note=cGMP-binding%2C high affinity |
| Hgene | PRKG1 | chr10:52913090 | chr20:19496132 | ENST00000401604 | + | 2 | 18 | 221_341 | 144 | 672.0 | Region | Note=cGMP-binding%2C low affinity |
| Tgene | SLC24A3 | chr10:52913090 | chr20:19496132 | ENST00000328041 | 1 | 17 | 45_107 | 90 | 645.0 | Topological domain | Extracellular |
Top |
Fusion Gene Sequence for PRKG1-SLC24A3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >68859_68859_1_PRKG1-SLC24A3_PRKG1_chr10_52913090_ENST00000373980_SLC24A3_chr20_19496132_ENST00000328041_length(transcript)=4356nt_BP=895nt GCCCCGCAAGCTTCCTTGGGCGCCCGGCGCGAGGGACTCTGAGAGCCCAGCCGCTGCCTCGGAGCCCCCATTCACGTCCCCTCCTCCCGC TCCCCCGCCACCGCGCCTCCGACTCTTCTCCCCCGCGCCGCGGCGCCCACCGCGTCTCAGGTTTAATCCCGGGTTGGACCTGCTGGTTTG CTCTCCCCGCTCTGAGCCTCACAGCCAGCCCAGAGAAGTGGAATCTGCACTGAAACTCTGGGTGGCTGGAGCCGGCGGACTGGGCATGCT CAGAAGCCAGGGCTGGCTTTGGTCTCAAGTAGGAAGCTTTGGCACTCGGGAGGCAGCGGCGACTTTGGGGAAAGTTGATCGGAGAGGGGA GGAAGCCTCAAGACGCGGAGCAGCGGCAGGAAGGAGCCCCCGGCAGCCCGGAGGAGCATGGGCACCTTGCGGGATTTACAGTACGCGCTC CAGGAGAAGATCGAGGAGCTGAGGCAGCGGGATGCTCTCATCGACGAGCTGGAGCTGGAGTTGGATCAGAAGGACGAACTGATCCAGAAG CTGCAGAACGAGCTGGACAAGTACCGCTCGGTGATCCGACCAGCCACCCAGCAGGCGCAGAAGCAGAGCGCGAGCACCTTGCAGGGCGAG CCGCGCACCAAGCGGCAGGCGATCTCCGCCGAGCCCACCGCCTTCGACATCCAGGATCTCAGCCATGTGACCCTGCCCTTCTACCCCAAG AGCCCACAGTCCAAGGATCTTATAAAGGAAGCTATCCTTGACAATGACTTTATGAAGAACTTGGAGCTGTCGCAGATCCAGGAGATTGTG GATTGTATGTACCCGGTGGAGTATGGCAAGGACAGTTGCATCATCAAAGAAGGAGACGTGGGGTCACTGGTGTATGTCATGGAAGCCCTG CATGAATTCCCCAATGACATCTTCACAAACGAGGATAGAAGACAAGGTGCGGTGGTCCTCCATGTGCTCTGTGCCATATACATGTTCTAT GCGCTGGCCATTGTGTGTGATGACTTCTTCGTCCCTTCCTTGGAAAAGATCTGTGAGCGCCTGCACCTCAGTGAAGATGTGGCTGGGGCC ACATTCATGGCAGCGGGAAGTTCGGCCCCAGAGCTGTTCACATCGGTCATAGGGGTCTTCATCACCAAAGGCGATGTGGGAGTTGGCACC ATCGTGGGCTCAGCGGTATTCAACATCCTGTGCATCATTGGTGTCTGTGGGCTCTTTGCTGGGCAGGTTGTGGCTCTTTCCTCCTGGTGC CTGCTGAGGGATTCTATTTACTACACGCTGTCTGTGATCGCGCTCATCGTGTTTATTTATGATGAAAAAGTTTCCTGGTGGGAGTCTTTA GTCCTTGTGCTGATGTATCTTATCTACATTGTCATCATGAAATATAACGCTTGCATACATCAGTGCTTTGAGAGGAGGACAAAAGGTGCC GGGAACATGGTCAACGGATTGGCCAACAATGCTGAAATTGATGACAGCAGCAACTGCGACGCAACTGTGGTGCTACTTAAGAAAGCAAAT TTCCACCGCAAAGCATCAGTGATCATGGTAGACGAGCTGCTGTCAGCCTACCCACACCAGCTTTCCTTCTCTGAGGCTGGCCTTCGAATC ATGATAACCAGCCACTTTCCCCCCAAGACCCGGCTCTCCATGGCCAGTCGCATGTTGATCAATGAGAGACAAAGATTGATAAACAGCAGG GCTTATACCAACGGGGAATCTGAGGTGGCCATCAAAATCCCAATTAAGCACACCGTGGAGAATGGGACAGGGCCCAGCAGTGCCCCAGAC AGGGGCGTGAATGGGACACGGAGGGACGATGTTGTGGCTGAGGCTGGCAACGAAACAGAGAATGAAAATGAGGACAATGAGAATGATGAG GAGGAAGAGGAGGACGAGGATGATGATGAAGGACCGTACACACCATTCGACACCCCCTCGGGTAAACTGGAAACAGTGAAATGGGCGTTC ACCTGGCCGCTGAGTTTCGTCTTATACTTCACTGTACCCAACTGCAACAAGCCGCGCTGGGAGAAATGGTTCATGGTGACGTTTGCTTCC TCCACGCTGTGGATCGCAGCCTTCTCCTACATGATGGTGTGGATGGTCACAATCATTGGTTACACCCTGGGGATTCCTGACGTCATCATG GGGATCACCTTCCTGGCTGCTGGGACCAGCGTGCCTGACTGCATGGCCAGCCTCATTGTGGCCAGACAAGGGATGGGGGACATGGCTGTG TCCAACTCCATTGGGAGCAACGTGTTTGACATCCTGATTGGCCTCGGTCTCCCCTGGGCTCTGCAGACCCTGGCTGTGGATTACGGATCC TACATCCGGCTGAATAGCAGGGGGCTGATCTACTCCGTAGGCTTGCTCCTGGCCTCTGTTTTTGTCACGGTGTTCGGCGTCCACCTGAAC AAGTGGCAGCTGGACAAGAAGCTGGGCTGTGGGTGCCTCCTCCTGTATGGTGTGTTCCTGTGCTTCTCCATCATGACTGAGTTCAACGTG TTCACCTTTGTGAACCTGCCCATGTGCGGGGACCACTGAGCCGCCGGGTGCCCACAGAGGCTCAGCTCCTTCTTTTCTGTGCAATACGAG ACCCGGCCGCACCCCGAGTCACACAGGCCCCCGGGGCCACGGCGTTCGTCTCTCCTGTGCTGTCCTCAGGCCTCCGCTCCTGTTTTGGTG GCCCAGGCTCTCCCCTGACCCATCCTCGCTCCCCCACCTCCTTGGGTCATGCCCACCCACCCTTTCCTGCCTCCTCCGTGTGAAGACATC CAACATCCACGTGACTTTTCCAGCTCCATTTTTGAACAGTGACTGAGATTCTAGAAAAACTGGCTGCTAACTGGCCTGAGCCAGGCAACA CTGATTCCAATCCCTCCTCCTTTTTTAAGTTATTTGATGGAAGACTCACCTAATTTGTGACCTGAGACTGTTGAAGAAATAGAGAGGAGG GGGCCCGTTGATTACAGAGAGCATTTGGGATTTTGTTTGGTTTGGAGATGATGCCTAGGTTACTGGGTTTGGGGGGATTGTTTTCTTTTG GGGGCCTTCCCCTTTTACTCCTTTTCTTCCAGAGATCAAGAGCTTCTCTTGCATCTTCTTCCACTGGGCTCTGGATTAATCAATTACCCA AAGGCTGCACCTGCCGTGTTGTCTGGGCTTGCATCCCAGATGTGTTGGAGTATGCATGGATGTAGTGCTTTTTAGAGGAGCCACTGGGCA AGGCCACCAAGAACAAATGCATGACATTTTATAGCCAAGGACGCCTCGCTAAAGTCTTATGGGCGTCCCCTGGGGTTGGGGGGGCACAAG GTTTTGGAGGAAGAAGACAACTTCCCTCATTCCATCATCACCATCTCTTTCTCACTAGGTTCTTTCTAGTTTTCAAGCAATAGTTCTAGC CTGCCTTGGACAAGGGGGCCCCAGTTAAACAAACTACCCATCCATGAGCTGCCAGGCAGTCAAAAACAGAAGCTTCCCCGACTTGTGAGT CCCTGAGATGTGCTCTTGTTGTTTGGCATTTGGGGTGACAGGGAGTGACCCAGAGGCCACCACTGCTTTTCATGCAGGAGTTACAGACAC TGGTTTCTTGGAAAATGGAGAGAAGCGCACTTTGCACAGACGTCGTCAATTAAGTCCCAATTTGCCACTTGGTATTGAGTACACTGGACC CTGACCACTGGCTCTTGGGCAAACGTCCTTCCTCACGGGGCGCCTCCGCCAAGCCGGCCCAGCTGCACCCCTCCCTTCCTGGAGGGATGG CCAGGGAAGGAGAAAACAGAGAACTGACACCTTTGAAACCACAGAATGTGTTACATGCAGACTCGCTCAAGGGCATAAGTTATTGTGAAC GTTTTTGCCAATCACTGCTCAACAGCCCTGCTAGATTTTGTATGATGCTGAATTATTATGCAGACTAATTCCACCCAGTTGAGACACACC ATGCTTGTTCACTTGTATTTATTGAAACTGTGGATTCTTGCCCGTGCTGTCCCTTGTATTTACTTTAAGCACTGATCACTTATCATTCAT TCGGTATGGTTTTCCCTGTCCCTTGTACACATTCTGGTATGAATTTGTAAAAATAACCTGCTACAAATTGGTTGAATGTTTCTGTCTGTG GTGCGAACCAGCATTAACGGATGGGGCACGTGCCCAACTGAGGAACAGGAGAAGAAATCACCAATTTGGGCTCTCAGAGCTAAGACACAC TTATTGATTCTGTTGCACATTTTGCACTGGTTTATGGCGATTGTTTTCTTGGACGGATAGTGTAAAATAAACTTCTCTGTTCTCTATCCT >68859_68859_1_PRKG1-SLC24A3_PRKG1_chr10_52913090_ENST00000373980_SLC24A3_chr20_19496132_ENST00000328041_length(amino acids)=713AA_BP=159 MGTLRDLQYALQEKIEELRQRDALIDELELELDQKDELIQKLQNELDKYRSVIRPATQQAQKQSASTLQGEPRTKRQAISAEPTAFDIQD LSHVTLPFYPKSPQSKDLIKEAILDNDFMKNLELSQIQEIVDCMYPVEYGKDSCIIKEGDVGSLVYVMEALHEFPNDIFTNEDRRQGAVV LHVLCAIYMFYALAIVCDDFFVPSLEKICERLHLSEDVAGATFMAAGSSAPELFTSVIGVFITKGDVGVGTIVGSAVFNILCIIGVCGLF AGQVVALSSWCLLRDSIYYTLSVIALIVFIYDEKVSWWESLVLVLMYLIYIVIMKYNACIHQCFERRTKGAGNMVNGLANNAEIDDSSNC DATVVLLKKANFHRKASVIMVDELLSAYPHQLSFSEAGLRIMITSHFPPKTRLSMASRMLINERQRLINSRAYTNGESEVAIKIPIKHTV ENGTGPSSAPDRGVNGTRRDDVVAEAGNETENENEDNENDEEEEEDEDDDEGPYTPFDTPSGKLETVKWAFTWPLSFVLYFTVPNCNKPR WEKWFMVTFASSTLWIAAFSYMMVWMVTIIGYTLGIPDVIMGITFLAAGTSVPDCMASLIVARQGMGDMAVSNSIGSNVFDILIGLGLPW -------------------------------------------------------------- >68859_68859_2_PRKG1-SLC24A3_PRKG1_chr10_52913090_ENST00000373985_SLC24A3_chr20_19496132_ENST00000328041_length(transcript)=3915nt_BP=454nt GCGGAGGGGCTCAGTGAAAAAATGAGCGAGCTAGAGGAAGACTTTGCCAAGATTCTCATGCTCAAGGAGGAGAGGATCAAAGAGCTGGAG AAGCGGCTGTCAGAGAAGGAGGAAGAAATTCAGGAGCTGAAGAGGAAACTCCACAAATGCCAGTCGGTGCTCCCAGTGCCCTCGACCCAC ATCGGCCCCCGGACCACCCGGGCGCAGGGCATCTCGGCCGAGCCGCAGACGTACAGGTCCTTCCACGACCTCCGACAGGCATTCCGGAAG TTCACCAAGTCCGAAAGGTCCAAGGATCTTATAAAGGAAGCTATCCTTGACAATGACTTTATGAAGAACTTGGAGCTGTCGCAGATCCAG GAGATTGTGGATTGTATGTACCCGGTGGAGTATGGCAAGGACAGTTGCATCATCAAAGAAGGAGACGTGGGGTCACTGGTGTATGTCATG GAAGCCCTGCATGAATTCCCCAATGACATCTTCACAAACGAGGATAGAAGACAAGGTGCGGTGGTCCTCCATGTGCTCTGTGCCATATAC ATGTTCTATGCGCTGGCCATTGTGTGTGATGACTTCTTCGTCCCTTCCTTGGAAAAGATCTGTGAGCGCCTGCACCTCAGTGAAGATGTG GCTGGGGCCACATTCATGGCAGCGGGAAGTTCGGCCCCAGAGCTGTTCACATCGGTCATAGGGGTCTTCATCACCAAAGGCGATGTGGGA GTTGGCACCATCGTGGGCTCAGCGGTATTCAACATCCTGTGCATCATTGGTGTCTGTGGGCTCTTTGCTGGGCAGGTTGTGGCTCTTTCC TCCTGGTGCCTGCTGAGGGATTCTATTTACTACACGCTGTCTGTGATCGCGCTCATCGTGTTTATTTATGATGAAAAAGTTTCCTGGTGG GAGTCTTTAGTCCTTGTGCTGATGTATCTTATCTACATTGTCATCATGAAATATAACGCTTGCATACATCAGTGCTTTGAGAGGAGGACA AAAGGTGCCGGGAACATGGTCAACGGATTGGCCAACAATGCTGAAATTGATGACAGCAGCAACTGCGACGCAACTGTGGTGCTACTTAAG AAAGCAAATTTCCACCGCAAAGCATCAGTGATCATGGTAGACGAGCTGCTGTCAGCCTACCCACACCAGCTTTCCTTCTCTGAGGCTGGC CTTCGAATCATGATAACCAGCCACTTTCCCCCCAAGACCCGGCTCTCCATGGCCAGTCGCATGTTGATCAATGAGAGACAAAGATTGATA AACAGCAGGGCTTATACCAACGGGGAATCTGAGGTGGCCATCAAAATCCCAATTAAGCACACCGTGGAGAATGGGACAGGGCCCAGCAGT GCCCCAGACAGGGGCGTGAATGGGACACGGAGGGACGATGTTGTGGCTGAGGCTGGCAACGAAACAGAGAATGAAAATGAGGACAATGAG AATGATGAGGAGGAAGAGGAGGACGAGGATGATGATGAAGGACCGTACACACCATTCGACACCCCCTCGGGTAAACTGGAAACAGTGAAA TGGGCGTTCACCTGGCCGCTGAGTTTCGTCTTATACTTCACTGTACCCAACTGCAACAAGCCGCGCTGGGAGAAATGGTTCATGGTGACG TTTGCTTCCTCCACGCTGTGGATCGCAGCCTTCTCCTACATGATGGTGTGGATGGTCACAATCATTGGTTACACCCTGGGGATTCCTGAC GTCATCATGGGGATCACCTTCCTGGCTGCTGGGACCAGCGTGCCTGACTGCATGGCCAGCCTCATTGTGGCCAGACAAGGGATGGGGGAC ATGGCTGTGTCCAACTCCATTGGGAGCAACGTGTTTGACATCCTGATTGGCCTCGGTCTCCCCTGGGCTCTGCAGACCCTGGCTGTGGAT TACGGATCCTACATCCGGCTGAATAGCAGGGGGCTGATCTACTCCGTAGGCTTGCTCCTGGCCTCTGTTTTTGTCACGGTGTTCGGCGTC CACCTGAACAAGTGGCAGCTGGACAAGAAGCTGGGCTGTGGGTGCCTCCTCCTGTATGGTGTGTTCCTGTGCTTCTCCATCATGACTGAG TTCAACGTGTTCACCTTTGTGAACCTGCCCATGTGCGGGGACCACTGAGCCGCCGGGTGCCCACAGAGGCTCAGCTCCTTCTTTTCTGTG CAATACGAGACCCGGCCGCACCCCGAGTCACACAGGCCCCCGGGGCCACGGCGTTCGTCTCTCCTGTGCTGTCCTCAGGCCTCCGCTCCT GTTTTGGTGGCCCAGGCTCTCCCCTGACCCATCCTCGCTCCCCCACCTCCTTGGGTCATGCCCACCCACCCTTTCCTGCCTCCTCCGTGT GAAGACATCCAACATCCACGTGACTTTTCCAGCTCCATTTTTGAACAGTGACTGAGATTCTAGAAAAACTGGCTGCTAACTGGCCTGAGC CAGGCAACACTGATTCCAATCCCTCCTCCTTTTTTAAGTTATTTGATGGAAGACTCACCTAATTTGTGACCTGAGACTGTTGAAGAAATA GAGAGGAGGGGGCCCGTTGATTACAGAGAGCATTTGGGATTTTGTTTGGTTTGGAGATGATGCCTAGGTTACTGGGTTTGGGGGGATTGT TTTCTTTTGGGGGCCTTCCCCTTTTACTCCTTTTCTTCCAGAGATCAAGAGCTTCTCTTGCATCTTCTTCCACTGGGCTCTGGATTAATC AATTACCCAAAGGCTGCACCTGCCGTGTTGTCTGGGCTTGCATCCCAGATGTGTTGGAGTATGCATGGATGTAGTGCTTTTTAGAGGAGC CACTGGGCAAGGCCACCAAGAACAAATGCATGACATTTTATAGCCAAGGACGCCTCGCTAAAGTCTTATGGGCGTCCCCTGGGGTTGGGG GGGCACAAGGTTTTGGAGGAAGAAGACAACTTCCCTCATTCCATCATCACCATCTCTTTCTCACTAGGTTCTTTCTAGTTTTCAAGCAAT AGTTCTAGCCTGCCTTGGACAAGGGGGCCCCAGTTAAACAAACTACCCATCCATGAGCTGCCAGGCAGTCAAAAACAGAAGCTTCCCCGA CTTGTGAGTCCCTGAGATGTGCTCTTGTTGTTTGGCATTTGGGGTGACAGGGAGTGACCCAGAGGCCACCACTGCTTTTCATGCAGGAGT TACAGACACTGGTTTCTTGGAAAATGGAGAGAAGCGCACTTTGCACAGACGTCGTCAATTAAGTCCCAATTTGCCACTTGGTATTGAGTA CACTGGACCCTGACCACTGGCTCTTGGGCAAACGTCCTTCCTCACGGGGCGCCTCCGCCAAGCCGGCCCAGCTGCACCCCTCCCTTCCTG GAGGGATGGCCAGGGAAGGAGAAAACAGAGAACTGACACCTTTGAAACCACAGAATGTGTTACATGCAGACTCGCTCAAGGGCATAAGTT ATTGTGAACGTTTTTGCCAATCACTGCTCAACAGCCCTGCTAGATTTTGTATGATGCTGAATTATTATGCAGACTAATTCCACCCAGTTG AGACACACCATGCTTGTTCACTTGTATTTATTGAAACTGTGGATTCTTGCCCGTGCTGTCCCTTGTATTTACTTTAAGCACTGATCACTT ATCATTCATTCGGTATGGTTTTCCCTGTCCCTTGTACACATTCTGGTATGAATTTGTAAAAATAACCTGCTACAAATTGGTTGAATGTTT CTGTCTGTGGTGCGAACCAGCATTAACGGATGGGGCACGTGCCCAACTGAGGAACAGGAGAAGAAATCACCAATTTGGGCTCTCAGAGCT AAGACACACTTATTGATTCTGTTGCACATTTTGCACTGGTTTATGGCGATTGTTTTCTTGGACGGATAGTGTAAAATAAACTTCTCTGTT >68859_68859_2_PRKG1-SLC24A3_PRKG1_chr10_52913090_ENST00000373985_SLC24A3_chr20_19496132_ENST00000328041_length(amino acids)=698AA_BP=144 MSELEEDFAKILMLKEERIKELEKRLSEKEEEIQELKRKLHKCQSVLPVPSTHIGPRTTRAQGISAEPQTYRSFHDLRQAFRKFTKSERS KDLIKEAILDNDFMKNLELSQIQEIVDCMYPVEYGKDSCIIKEGDVGSLVYVMEALHEFPNDIFTNEDRRQGAVVLHVLCAIYMFYALAI VCDDFFVPSLEKICERLHLSEDVAGATFMAAGSSAPELFTSVIGVFITKGDVGVGTIVGSAVFNILCIIGVCGLFAGQVVALSSWCLLRD SIYYTLSVIALIVFIYDEKVSWWESLVLVLMYLIYIVIMKYNACIHQCFERRTKGAGNMVNGLANNAEIDDSSNCDATVVLLKKANFHRK ASVIMVDELLSAYPHQLSFSEAGLRIMITSHFPPKTRLSMASRMLINERQRLINSRAYTNGESEVAIKIPIKHTVENGTGPSSAPDRGVN GTRRDDVVAEAGNETENENEDNENDEEEEEDEDDDEGPYTPFDTPSGKLETVKWAFTWPLSFVLYFTVPNCNKPRWEKWFMVTFASSTLW IAAFSYMMVWMVTIIGYTLGIPDVIMGITFLAAGTSVPDCMASLIVARQGMGDMAVSNSIGSNVFDILIGLGLPWALQTLAVDYGSYIRL -------------------------------------------------------------- >68859_68859_3_PRKG1-SLC24A3_PRKG1_chr10_52913090_ENST00000401604_SLC24A3_chr20_19496132_ENST00000328041_length(transcript)=4088nt_BP=627nt ATTCAGCAGAAGCGGATCGAAGCAGGAGGCTCCCCGCGCCGCATTAGGGGCGCACTCCGCCGCGCTCGAGTACTTAGCGCCCATTCACTC GCTCACCCGCGCTCTCCGCTGCCGGCTGCCGTCCCAGCCGCCGCCGCCGCCGCCGCCGCCGCCGCCGCCCGAGAAAAAGTTTCGCGGAGG GGCTCAGTGAAAAAATGAGCGAGCTAGAGGAAGACTTTGCCAAGATTCTCATGCTCAAGGAGGAGAGGATCAAAGAGCTGGAGAAGCGGC TGTCAGAGAAGGAGGAAGAAATTCAGGAGCTGAAGAGGAAACTCCACAAATGCCAGTCGGTGCTCCCAGTGCCCTCGACCCACATCGGCC CCCGGACCACCCGGGCGCAGGGCATCTCGGCCGAGCCGCAGACGTACAGGTCCTTCCACGACCTCCGACAGGCATTCCGGAAGTTCACCA AGTCCGAAAGGTCCAAGGATCTTATAAAGGAAGCTATCCTTGACAATGACTTTATGAAGAACTTGGAGCTGTCGCAGATCCAGGAGATTG TGGATTGTATGTACCCGGTGGAGTATGGCAAGGACAGTTGCATCATCAAAGAAGGAGACGTGGGGTCACTGGTGTATGTCATGGAAGCCC TGCATGAATTCCCCAATGACATCTTCACAAACGAGGATAGAAGACAAGGTGCGGTGGTCCTCCATGTGCTCTGTGCCATATACATGTTCT ATGCGCTGGCCATTGTGTGTGATGACTTCTTCGTCCCTTCCTTGGAAAAGATCTGTGAGCGCCTGCACCTCAGTGAAGATGTGGCTGGGG CCACATTCATGGCAGCGGGAAGTTCGGCCCCAGAGCTGTTCACATCGGTCATAGGGGTCTTCATCACCAAAGGCGATGTGGGAGTTGGCA CCATCGTGGGCTCAGCGGTATTCAACATCCTGTGCATCATTGGTGTCTGTGGGCTCTTTGCTGGGCAGGTTGTGGCTCTTTCCTCCTGGT GCCTGCTGAGGGATTCTATTTACTACACGCTGTCTGTGATCGCGCTCATCGTGTTTATTTATGATGAAAAAGTTTCCTGGTGGGAGTCTT TAGTCCTTGTGCTGATGTATCTTATCTACATTGTCATCATGAAATATAACGCTTGCATACATCAGTGCTTTGAGAGGAGGACAAAAGGTG CCGGGAACATGGTCAACGGATTGGCCAACAATGCTGAAATTGATGACAGCAGCAACTGCGACGCAACTGTGGTGCTACTTAAGAAAGCAA ATTTCCACCGCAAAGCATCAGTGATCATGGTAGACGAGCTGCTGTCAGCCTACCCACACCAGCTTTCCTTCTCTGAGGCTGGCCTTCGAA TCATGATAACCAGCCACTTTCCCCCCAAGACCCGGCTCTCCATGGCCAGTCGCATGTTGATCAATGAGAGACAAAGATTGATAAACAGCA GGGCTTATACCAACGGGGAATCTGAGGTGGCCATCAAAATCCCAATTAAGCACACCGTGGAGAATGGGACAGGGCCCAGCAGTGCCCCAG ACAGGGGCGTGAATGGGACACGGAGGGACGATGTTGTGGCTGAGGCTGGCAACGAAACAGAGAATGAAAATGAGGACAATGAGAATGATG AGGAGGAAGAGGAGGACGAGGATGATGATGAAGGACCGTACACACCATTCGACACCCCCTCGGGTAAACTGGAAACAGTGAAATGGGCGT TCACCTGGCCGCTGAGTTTCGTCTTATACTTCACTGTACCCAACTGCAACAAGCCGCGCTGGGAGAAATGGTTCATGGTGACGTTTGCTT CCTCCACGCTGTGGATCGCAGCCTTCTCCTACATGATGGTGTGGATGGTCACAATCATTGGTTACACCCTGGGGATTCCTGACGTCATCA TGGGGATCACCTTCCTGGCTGCTGGGACCAGCGTGCCTGACTGCATGGCCAGCCTCATTGTGGCCAGACAAGGGATGGGGGACATGGCTG TGTCCAACTCCATTGGGAGCAACGTGTTTGACATCCTGATTGGCCTCGGTCTCCCCTGGGCTCTGCAGACCCTGGCTGTGGATTACGGAT CCTACATCCGGCTGAATAGCAGGGGGCTGATCTACTCCGTAGGCTTGCTCCTGGCCTCTGTTTTTGTCACGGTGTTCGGCGTCCACCTGA ACAAGTGGCAGCTGGACAAGAAGCTGGGCTGTGGGTGCCTCCTCCTGTATGGTGTGTTCCTGTGCTTCTCCATCATGACTGAGTTCAACG TGTTCACCTTTGTGAACCTGCCCATGTGCGGGGACCACTGAGCCGCCGGGTGCCCACAGAGGCTCAGCTCCTTCTTTTCTGTGCAATACG AGACCCGGCCGCACCCCGAGTCACACAGGCCCCCGGGGCCACGGCGTTCGTCTCTCCTGTGCTGTCCTCAGGCCTCCGCTCCTGTTTTGG TGGCCCAGGCTCTCCCCTGACCCATCCTCGCTCCCCCACCTCCTTGGGTCATGCCCACCCACCCTTTCCTGCCTCCTCCGTGTGAAGACA TCCAACATCCACGTGACTTTTCCAGCTCCATTTTTGAACAGTGACTGAGATTCTAGAAAAACTGGCTGCTAACTGGCCTGAGCCAGGCAA CACTGATTCCAATCCCTCCTCCTTTTTTAAGTTATTTGATGGAAGACTCACCTAATTTGTGACCTGAGACTGTTGAAGAAATAGAGAGGA GGGGGCCCGTTGATTACAGAGAGCATTTGGGATTTTGTTTGGTTTGGAGATGATGCCTAGGTTACTGGGTTTGGGGGGATTGTTTTCTTT TGGGGGCCTTCCCCTTTTACTCCTTTTCTTCCAGAGATCAAGAGCTTCTCTTGCATCTTCTTCCACTGGGCTCTGGATTAATCAATTACC CAAAGGCTGCACCTGCCGTGTTGTCTGGGCTTGCATCCCAGATGTGTTGGAGTATGCATGGATGTAGTGCTTTTTAGAGGAGCCACTGGG CAAGGCCACCAAGAACAAATGCATGACATTTTATAGCCAAGGACGCCTCGCTAAAGTCTTATGGGCGTCCCCTGGGGTTGGGGGGGCACA AGGTTTTGGAGGAAGAAGACAACTTCCCTCATTCCATCATCACCATCTCTTTCTCACTAGGTTCTTTCTAGTTTTCAAGCAATAGTTCTA GCCTGCCTTGGACAAGGGGGCCCCAGTTAAACAAACTACCCATCCATGAGCTGCCAGGCAGTCAAAAACAGAAGCTTCCCCGACTTGTGA GTCCCTGAGATGTGCTCTTGTTGTTTGGCATTTGGGGTGACAGGGAGTGACCCAGAGGCCACCACTGCTTTTCATGCAGGAGTTACAGAC ACTGGTTTCTTGGAAAATGGAGAGAAGCGCACTTTGCACAGACGTCGTCAATTAAGTCCCAATTTGCCACTTGGTATTGAGTACACTGGA CCCTGACCACTGGCTCTTGGGCAAACGTCCTTCCTCACGGGGCGCCTCCGCCAAGCCGGCCCAGCTGCACCCCTCCCTTCCTGGAGGGAT GGCCAGGGAAGGAGAAAACAGAGAACTGACACCTTTGAAACCACAGAATGTGTTACATGCAGACTCGCTCAAGGGCATAAGTTATTGTGA ACGTTTTTGCCAATCACTGCTCAACAGCCCTGCTAGATTTTGTATGATGCTGAATTATTATGCAGACTAATTCCACCCAGTTGAGACACA CCATGCTTGTTCACTTGTATTTATTGAAACTGTGGATTCTTGCCCGTGCTGTCCCTTGTATTTACTTTAAGCACTGATCACTTATCATTC ATTCGGTATGGTTTTCCCTGTCCCTTGTACACATTCTGGTATGAATTTGTAAAAATAACCTGCTACAAATTGGTTGAATGTTTCTGTCTG TGGTGCGAACCAGCATTAACGGATGGGGCACGTGCCCAACTGAGGAACAGGAGAAGAAATCACCAATTTGGGCTCTCAGAGCTAAGACAC ACTTATTGATTCTGTTGCACATTTTGCACTGGTTTATGGCGATTGTTTTCTTGGACGGATAGTGTAAAATAAACTTCTCTGTTCTCTATC >68859_68859_3_PRKG1-SLC24A3_PRKG1_chr10_52913090_ENST00000401604_SLC24A3_chr20_19496132_ENST00000328041_length(amino acids)=698AA_BP=144 MSELEEDFAKILMLKEERIKELEKRLSEKEEEIQELKRKLHKCQSVLPVPSTHIGPRTTRAQGISAEPQTYRSFHDLRQAFRKFTKSERS KDLIKEAILDNDFMKNLELSQIQEIVDCMYPVEYGKDSCIIKEGDVGSLVYVMEALHEFPNDIFTNEDRRQGAVVLHVLCAIYMFYALAI VCDDFFVPSLEKICERLHLSEDVAGATFMAAGSSAPELFTSVIGVFITKGDVGVGTIVGSAVFNILCIIGVCGLFAGQVVALSSWCLLRD SIYYTLSVIALIVFIYDEKVSWWESLVLVLMYLIYIVIMKYNACIHQCFERRTKGAGNMVNGLANNAEIDDSSNCDATVVLLKKANFHRK ASVIMVDELLSAYPHQLSFSEAGLRIMITSHFPPKTRLSMASRMLINERQRLINSRAYTNGESEVAIKIPIKHTVENGTGPSSAPDRGVN GTRRDDVVAEAGNETENENEDNENDEEEEEDEDDDEGPYTPFDTPSGKLETVKWAFTWPLSFVLYFTVPNCNKPRWEKWFMVTFASSTLW IAAFSYMMVWMVTIIGYTLGIPDVIMGITFLAAGTSVPDCMASLIVARQGMGDMAVSNSIGSNVFDILIGLGLPWALQTLAVDYGSYIRL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for PRKG1-SLC24A3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for PRKG1-SLC24A3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for PRKG1-SLC24A3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
