
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ASAH1-HGSNAT (FusionGDB2 ID:6936) |
Fusion Gene Summary for ASAH1-HGSNAT |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ASAH1-HGSNAT | Fusion gene ID: 6936 | Hgene | Tgene | Gene symbol | ASAH1 | HGSNAT | Gene ID | 427 | 138050 |
| Gene name | N-acylsphingosine amidohydrolase 1 | heparan-alpha-glucosaminide N-acetyltransferase | |
| Synonyms | AC|ACDase|ASAH|PHP|PHP32|SMAPME | HGNAT|MPS3C|RP73|TMEM76 | |
| Cytomap | 8p22 | 8p11.21-p11.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | acid ceramidaseN-acylethanolamine hydrolase ASAH1N-acylsphingosine amidohydrolase (acid ceramidase) 1acid CDaseacylsphingosine deacylaseputative 32 kDa heart protein | heparan-alpha-glucosaminide N-acetyltransferasetransmembrane protein 76 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q13510 | Q68CP4 | |
| Ensembl transtripts involved in fusion gene | ENST00000417108, ENST00000520051, ENST00000262097, ENST00000314146, ENST00000381733, ENST00000520781, | ENST00000297798, ENST00000379644, ENST00000458501, ENST00000521576, | |
| Fusion gene scores | * DoF score | 15 X 16 X 7=1680 | 12 X 13 X 4=624 |
| # samples | 16 | 13 | |
| ** MAII score | log2(16/1680*10)=-3.39231742277876 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(13/624*10)=-2.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ASAH1 [Title/Abstract] AND HGSNAT [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ASAH1(17933049)-HGSNAT(43054530), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ASAH1-HGSNAT seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. ASAH1-HGSNAT seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ASAH1 | GO:0046512 | sphingosine biosynthetic process | 12815059 |
| Hgene | ASAH1 | GO:0046513 | ceramide biosynthetic process | 12764132|12815059 |
| Hgene | ASAH1 | GO:0046514 | ceramide catabolic process | 12815059 |
| Tgene | HGSNAT | GO:0007041 | lysosomal transport | 20650889 |
| Tgene | HGSNAT | GO:0051259 | protein complex oligomerization | 20650889 |
 Fusion gene breakpoints across ASAH1 (5'-gene) Fusion gene breakpoints across ASAH1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
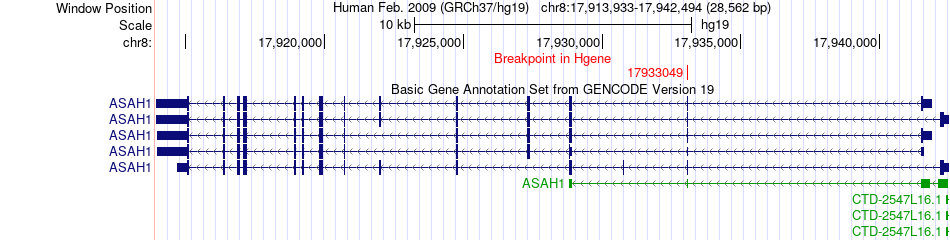 |
 Fusion gene breakpoints across HGSNAT (3'-gene) Fusion gene breakpoints across HGSNAT (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
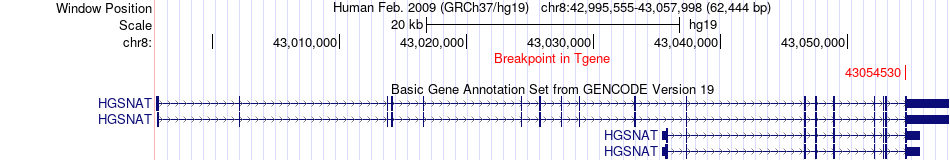 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-BR-A452 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
Top |
Fusion Gene ORF analysis for ASAH1-HGSNAT |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5UTR-3CDS | ENST00000417108 | ENST00000297798 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| 5UTR-3CDS | ENST00000417108 | ENST00000379644 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| 5UTR-3CDS | ENST00000417108 | ENST00000458501 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| 5UTR-3CDS | ENST00000417108 | ENST00000521576 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| 5UTR-3CDS | ENST00000520051 | ENST00000297798 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| 5UTR-3CDS | ENST00000520051 | ENST00000379644 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| 5UTR-3CDS | ENST00000520051 | ENST00000458501 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| 5UTR-3CDS | ENST00000520051 | ENST00000521576 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| Frame-shift | ENST00000262097 | ENST00000297798 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| Frame-shift | ENST00000262097 | ENST00000458501 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| Frame-shift | ENST00000262097 | ENST00000521576 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| Frame-shift | ENST00000314146 | ENST00000297798 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| Frame-shift | ENST00000314146 | ENST00000458501 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| Frame-shift | ENST00000314146 | ENST00000521576 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| Frame-shift | ENST00000381733 | ENST00000297798 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| Frame-shift | ENST00000381733 | ENST00000458501 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| Frame-shift | ENST00000381733 | ENST00000521576 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| Frame-shift | ENST00000520781 | ENST00000297798 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| Frame-shift | ENST00000520781 | ENST00000458501 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| Frame-shift | ENST00000520781 | ENST00000521576 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| In-frame | ENST00000262097 | ENST00000379644 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| In-frame | ENST00000314146 | ENST00000379644 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| In-frame | ENST00000381733 | ENST00000379644 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
| In-frame | ENST00000520781 | ENST00000379644 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000262097 | ASAH1 | chr8 | 17933049 | - | ENST00000379644 | HGSNAT | chr8 | 43054530 | + | 3905 | 437 | 1816 | 2460 | 214 |
| ENST00000381733 | ASAH1 | chr8 | 17933049 | - | ENST00000379644 | HGSNAT | chr8 | 43054530 | + | 3808 | 340 | 1719 | 2363 | 214 |
| ENST00000520781 | ASAH1 | chr8 | 17933049 | - | ENST00000379644 | HGSNAT | chr8 | 43054530 | + | 3905 | 437 | 1816 | 2460 | 214 |
| ENST00000314146 | ASAH1 | chr8 | 17933049 | - | ENST00000379644 | HGSNAT | chr8 | 43054530 | + | 3825 | 357 | 1736 | 2380 | 214 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000262097 | ENST00000379644 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + | 0.7123546 | 0.28764543 |
| ENST00000381733 | ENST00000379644 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + | 0.8276688 | 0.17233115 |
| ENST00000520781 | ENST00000379644 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + | 0.7123546 | 0.28764543 |
| ENST00000314146 | ENST00000379644 | ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + | 0.82275367 | 0.1772463 |
Top |
Fusion Genomic Features for ASAH1-HGSNAT |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + | 4.46E-11 | 1 |
| ASAH1 | chr8 | 17933049 | - | HGSNAT | chr8 | 43054530 | + | 4.46E-11 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for ASAH1-HGSNAT |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:17933049/chr8:43054530) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ASAH1 | HGSNAT |
| FUNCTION: Lysosomal ceramidase that hydrolyzes sphingolipid ceramides into sphingosine and free fatty acids at acidic pH (PubMed:10610716, PubMed:7744740, PubMed:15655246, PubMed:11451951). Ceramides, sphingosine, and its phosphorylated form sphingosine-1-phosphate are bioactive lipids that mediate cellular signaling pathways regulating several biological processes including cell proliferation, apoptosis and differentiation (PubMed:10610716). Has a higher catalytic efficiency towards C12-ceramides versus other ceramides (PubMed:7744740, PubMed:15655246). Also catalyzes the reverse reaction allowing the synthesis of ceramides from fatty acids and sphingosine (PubMed:12764132, PubMed:12815059). For the reverse synthetic reaction, the natural sphingosine D-erythro isomer is more efficiently utilized as a substrate compared to D-erythro-dihydrosphingosine and D-erythro-phytosphingosine, while the fatty acids with chain lengths of 12 or 14 carbons are the most efficiently used (PubMed:12764132). Has also an N-acylethanolamine hydrolase activity (PubMed:15655246). By regulating the levels of ceramides, sphingosine and sphingosine-1-phosphate in the epidermis, mediates the calcium-induced differentiation of epidermal keratinocytes (PubMed:17713573). Also indirectly regulates tumor necrosis factor/TNF-induced apoptosis (By similarity). By regulating the intracellular balance between ceramides and sphingosine, in adrenocortical cells, probably also acts as a regulator of steroidogenesis (PubMed:22261821). {ECO:0000250|UniProtKB:Q9WV54, ECO:0000269|PubMed:10610716, ECO:0000269|PubMed:11451951, ECO:0000269|PubMed:12764132, ECO:0000269|PubMed:12815059, ECO:0000269|PubMed:15655246, ECO:0000269|PubMed:17713573, ECO:0000269|PubMed:22261821, ECO:0000269|PubMed:7744740, ECO:0000303|PubMed:10610716}.; FUNCTION: [Isoform 2]: May directly regulate steroidogenesis by binding the nuclear receptor NR5A1 and negatively regulating its transcriptional activity. {ECO:0000305|PubMed:22927646}. | FUNCTION: Lysosomal acetyltransferase that acetylates the non-reducing terminal alpha-glucosamine residue of intralysosomal heparin or heparan sulfate, converting it into a substrate for luminal alpha-N-acetyl glucosaminidase. {ECO:0000269|PubMed:16960811, ECO:0000269|PubMed:17033958, ECO:0000269|PubMed:19823584, ECO:0000269|PubMed:20650889}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 624_635 | 575 | 636.0 | Region | Note=Lysosomal targeting region | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 624_635 | 603 | 664.0 | Region | Note=Lysosomal targeting region | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 586_592 | 575 | 636.0 | Topological domain | Cytoplasmic | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 614_634 | 575 | 636.0 | Topological domain | Lumenal%2C vesicle | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 656_663 | 575 | 636.0 | Topological domain | Cytoplasmic | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 614_634 | 603 | 664.0 | Topological domain | Lumenal%2C vesicle | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 656_663 | 603 | 664.0 | Topological domain | Cytoplasmic | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 593_613 | 575 | 636.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 635_655 | 575 | 636.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 635_655 | 603 | 664.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 4_64 | 575 | 636.0 | Compositional bias | Note=Ala-rich | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 4_64 | 603 | 664.0 | Compositional bias | Note=Ala-rich | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 1_190 | 575 | 636.0 | Topological domain | Lumenal%2C vesicle | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 212_275 | 575 | 636.0 | Topological domain | Cytoplasmic | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 297_302 | 575 | 636.0 | Topological domain | Lumenal%2C vesicle | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 324_345 | 575 | 636.0 | Topological domain | Cytoplasmic | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 367_374 | 575 | 636.0 | Topological domain | Lumenal%2C vesicle | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 396_420 | 575 | 636.0 | Topological domain | Cytoplasmic | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 442_500 | 575 | 636.0 | Topological domain | Lumenal%2C vesicle | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 522_529 | 575 | 636.0 | Topological domain | Cytoplasmic | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 551_564 | 575 | 636.0 | Topological domain | Lumenal%2C vesicle | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 1_190 | 603 | 664.0 | Topological domain | Lumenal%2C vesicle | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 212_275 | 603 | 664.0 | Topological domain | Cytoplasmic | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 297_302 | 603 | 664.0 | Topological domain | Lumenal%2C vesicle | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 324_345 | 603 | 664.0 | Topological domain | Cytoplasmic | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 367_374 | 603 | 664.0 | Topological domain | Lumenal%2C vesicle | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 396_420 | 603 | 664.0 | Topological domain | Cytoplasmic | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 442_500 | 603 | 664.0 | Topological domain | Lumenal%2C vesicle | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 522_529 | 603 | 664.0 | Topological domain | Cytoplasmic | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 551_564 | 603 | 664.0 | Topological domain | Lumenal%2C vesicle | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 586_592 | 603 | 664.0 | Topological domain | Cytoplasmic | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 191_211 | 575 | 636.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 276_296 | 575 | 636.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 303_323 | 575 | 636.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 346_366 | 575 | 636.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 375_395 | 575 | 636.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 421_441 | 575 | 636.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 501_521 | 575 | 636.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 530_550 | 575 | 636.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000379644 | 16 | 18 | 565_585 | 575 | 636.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 191_211 | 603 | 664.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 276_296 | 603 | 664.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 303_323 | 603 | 664.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 346_366 | 603 | 664.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 375_395 | 603 | 664.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 421_441 | 603 | 664.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 501_521 | 603 | 664.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 530_550 | 603 | 664.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 565_585 | 603 | 664.0 | Transmembrane | Helical | |
| Tgene | HGSNAT | chr8:17933049 | chr8:43054530 | ENST00000458501 | 16 | 18 | 593_613 | 603 | 664.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for ASAH1-HGSNAT |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >6936_6936_1_ASAH1-HGSNAT_ASAH1_chr8_17933049_ENST00000262097_HGSNAT_chr8_43054530_ENST00000379644_length(transcript)=3905nt_BP=437nt GGCTCGGTCCGACTATTGCCCGCGGTGGGGGAGGGGGATGGATCACGCCACGCGCCAAAGGCGATCGCGACTCTCCTTCTGCAGGTAGCC TGGAAGGCTCTCTCTCTTTCTCTACGCCACCCTTTTCGTGGCACTGAAAAGCCCCGTCCTCTCCTCCCAGTCCCGCCTCCTCCGAGCGTT CCCCCTACTGCCTGGAATGGTGCGGTCCCAGGTCGCGGGTCACGCGGCGGAGGGGGCGTGGCCTGCCCCCGGCCCAGCCGGCTCTTCTTT GCCTCTGCTGGAGTCCGGGGAGTGGCGTTGGCTGCTAGAGCGATGCCGGGCCGGAGTTGCGTCGCCTTAGTCCTCCTGGCTGCCGCCGTC AGCTGTGCCGTCGCGCAGCACGCGCCGCCGTGGACAGAGGACTGCAGAAAATCAACCTATCCTCCTTCAGGACCAACGAATGAATTCCAT TCTGGTATATGTCGGCCACGAGGTGTTTGAGAACTACTTCCCCTTTCAGTGGAAGCTGAAGGACAACCAGTCCCACAAGGAGCACCTGAC TCAGAACATCGTCGCCACTGCCCTCTGGGTGCTCATTGCCTACATCCTCTATAGAAAGAAGATTTTTTGGAAAATCTGATGGCTCCCACT GAGATGTGCTGCTGGAAGACTCTAGTAGGCCTGCAGGGAGGACTGAAGCAGCCTTTGTTAAAGGGAAGCATTCATTAGGAAATTGACTGG CTGCGTGTTTACAGACTCTGGGGGAAGACACTGATGTCCTCAAACTGGTTAACTGTGACACGGCTCGCCAGAACTCTGCCTGTCTATTTG TGACTTACAGATTTGAAATGTAATTGTCTTTTTTCCTCCATCTTCTGTGGAAATGGATGTCTTTGGAACTTCATTCCGAGGAGATAAGCT TTAACTTTCCAAAAGGGAATTGCCATGGGTGTTTTTCTTCTGTGGTGAGTGAAACAATCTGAGGTCTGGTTCTTGCTGACCTTGTTGCCC TGCAAACTTCCTTTCCACGTGTACGCGCACACCAACACGAAATGCCATCACTCCTACTGCGGCTGCTATGAAGCTTACTGGTTGTGATGT GTTATAATTTAGTCTGTTTTTTTGATTGAATGCAGTTTAATGTTTCCAGAAAGCCAAAGTAATTTTCTTTTCAGATATGCAAGGCTTTGG TGGGTCCAAAAAATGTCTATCACAAGCCATTTTTTCCTTTTCCTCTCTCGAAAAGTTAAAATATCTATGTGTTATTCCCAAACCCTCTTA CCTATGTATCTGCCTGTCTGTCCATCATCTTCCTTCCTCCCTATCTCTGTGTATCTGGATGGCAGCCGCTGCCCAGGGGAGTGGCTGTGG GGAGGGCAGGTACTGTCTTTGCCTGTGGGTCCAGCTGAGCCATCCCTGCTGGGTGATGCTGGGCAAGACCCTTGGCCCGTCTGGGCCTTG GCTTCCTCACTTGTGAAATGAGCGGGAAGATGACTCTCAGTTCCTTCCACCTCTTAGACATGGTGAGGTAACAGACATCAAAAGCTTTTC TGAAATCTTCAGAAGAAATAGTTCCATTACAGAAAACTCTTCAAAATAAATAGTAGTGAAAACTTTTAAAAACTCTCATTGGAGTAAGTC TTTTCAAGATGATCCTCCACAATGGAGGCAGCGTTCCTACTTGTCATCACACAGCTGAAGACATTGTTTCTTAGGTGTGAAATCGGGGAC AAAGGACAAACAGAGACACACGGCATTGTTCATGGGAGGCATCGTCACCCTCCTGGGTGTTCTGTGGGAATTTCCTGTGTGAGGAAAACG TGGCCACAGGGTTGTGCTGTACCCACCCTTCCCCGGCGAGATGGCCCTCGGCCTGTGCCGCTGCTTCCACCCTCGCCACTCCATGGCAGC TTTTGGTCTGTTTCCGGCTCTGCCCTCTGCCCTGAACTCTCATCCGGCTTGTACCTGCCTGCTGGACCCCTCCACCTGGAGGCCAGCCCA TGTCTCAGGCCCAGCCCTAGCCTCTTCTCCTCAAATTCTAAGTGTTTTCTCTTTAGGTTTCCCTGGCTTTGTGAATGGATCATGTGTCTC TAGGTATAAACCTGACATCATCTTTCCACCCGGCTTACCTCCACCAGATCTCCCCAGTTCTGTCTCCATCTTCTACCTGCAGCTGCTCTG TTCTCATGGTCACTGCTGCATCACTGAGTCTGGACCCTTGTTATCATTTTCAAACTGGCCTCCTTCCCTCGTTCCCCACTTCTTAAAGTC ACCTGTCCATTGCCACCAGATTAAGCTTTCTCCAGCCAGATCACCTCTCTCTGAGAAACCTCCATTGACATGGAAACACCATTGTCTGGC ACACATACTCACATACTCACCTTCCCGTCTTGATCCCCACACATCTTTCCAGCCTCCCCTCCCACTCCACTCCCTGCTCTCTCCTCCACC TCCCCATCCTCTTGTCTCCCCTCCCCTCTGAATCCAGCCCAGCGGGGCTTCTCCTGCCTCCATCACATCACAGAAGTACCTCCTGCTTCT GGTTTTAATTAGAGCCTTCCCCGATTACATTTTCCTCTGAATTTTTTCCTATCTACATTTGATCTGTCATGTTTAAACCCCCTACTTCTA AGGGAACTTCTCTAATCTCTTATCCTCATCCCCAAATAGTGTTTTCTTCCTCTGGGTTCTTATAATGTTGGTATCAATCTCACAGCATTT AGTGCTTCCTGCCTGGTGTGACAGTTACCTGTGTGCATGTGCAATTTCTAATTTCCCACGCTAGACTGTGAGCTTCCTAAGGCAAGAATC ATGCCTTGTTGGTTTCTGTATTCCTCATGGTGCCAAACACAGTGCCTTCTACATTGCAGGCGCTGAATAAACATTTTTAAAGCAAAATGA TGTGGATTTTTAAAATAAATATTTAAGTGCTGGTAAGATGAGCATGTATCCGGGGTGCCCATGAAATGTTCTTGGGGCCGTGTGGGGACA GTCGTCATTCCTCCTCCTGCCACCCTTTTCTTTCAGTGAGTCACTGTGGATGGTCCCAGCTGTGTCATCCCAAAGTTCAGCAGGGAAAGC TGAGCTGGGCCTCTCCAGGTGAGTTTTCTAGAAGCATTTCTCAAACTGTGGGTTACATCAACTTGGGTGTCTTGAGCTGTAAGGAAGGAA CTCCGGAGTCAGCTGGGCTACAGGGGAGCTTCTCTAAGTCCTGCGGGAGGCCAGACCCAGCCTGAGCTTGCTGTTAGCTAGCGGAGGCAG CTGCTGGTGGCCCAGGTGCTCGACACCAGGCATCCCCTCTCCTCCCACGAAGGGTGTGCCATAATCCCCTTCAACAGGAAATGCTTCCCA GAAGCCTCTCAGCAGCCTCCCCTCCTGTCCTATCAGCTAGAAGCGCCTCGCTTGTCCCAAGACCAGCAGGGACAGGGAACTGTCCGAGCC CGTGGCTGTGTGGAGGAAGGCGACCCCCAGCACAAGATTGGTTTCCTTTGGGAAGGGAAGAGGGAGTGTGTTGGGGTAAGGGGTAGAGCA GAGGAATGGTCAGGGGGCAACAACCGCTGACAGCTGCAACAGGTGCATGGCATCTCACAGGGAGGCAGGGAGGTGCGAGCTCCTAAGTAA TGGAGCAAAAAAATTCTATTCTGTAGAATGGGGAGAGAAAATGTGACATTTTAATTTTTTTTTGCATTTATATTCCTAATTCCTACTTAA AGTGAATATACTGCCGCTGTAGATCATAAAATGTATCTTTTCCATGGCCAACAAGGGGCATCTTTTATAAATGCATAATAACCCAGTTTG TATCAAAGGGTATCGACTTAAGTGAAATTTCAACATGCTGTTACTTTTTCCTTTTAATGTAATTCTGTTTTCCAAATAAATGGGGGAGAC >6936_6936_1_ASAH1-HGSNAT_ASAH1_chr8_17933049_ENST00000262097_HGSNAT_chr8_43054530_ENST00000379644_length(amino acids)=214AA_BP= MYPPFPGEMALGLCRCFHPRHSMAAFGLFPALPSALNSHPACTCLLDPSTWRPAHVSGPALASSPQILSVFSLGFPGFVNGSCVSRYKPD IIFPPGLPPPDLPSSVSIFYLQLLCSHGHCCITESGPLLSFSNWPPSLVPHFLKSPVHCHQIKLSPARSPLSEKPPLTWKHHCLAHILTY -------------------------------------------------------------- >6936_6936_2_ASAH1-HGSNAT_ASAH1_chr8_17933049_ENST00000314146_HGSNAT_chr8_43054530_ENST00000379644_length(transcript)=3825nt_BP=357nt GGGCAGGATTTCCTGCTGGACTTTGAAATCCAACCCGGTCACCTACCCGCGCGACTGTGTCCACGGATGGCACGAAAGCCAAGCGAGTCC CCCTGCCGAGCTACTCGCGTCCGCCTCCTCCCAAGCTGAGCTCTGCTCCGCCCACCTGAGTCCTTCGCCAGTTAGGAGGAAACACAGCCG CTTAATGAACTGCTGCATCGGGCTGGGAGAGAAAGCTCGCGGGTCCCACCGGGCCTCCTACCCAAGTCTCAGCGCGCTTTTCACCGAGGC CTCAATTCTGGGATTTGGCAGCTTTGCTGTGAAAGCCCAATGGACAGAGGACTGCAGAAAATCAACCTATCCTCCTTCAGGACCAACGAA TGAATTCCATTCTGGTATATGTCGGCCACGAGGTGTTTGAGAACTACTTCCCCTTTCAGTGGAAGCTGAAGGACAACCAGTCCCACAAGG AGCACCTGACTCAGAACATCGTCGCCACTGCCCTCTGGGTGCTCATTGCCTACATCCTCTATAGAAAGAAGATTTTTTGGAAAATCTGAT GGCTCCCACTGAGATGTGCTGCTGGAAGACTCTAGTAGGCCTGCAGGGAGGACTGAAGCAGCCTTTGTTAAAGGGAAGCATTCATTAGGA AATTGACTGGCTGCGTGTTTACAGACTCTGGGGGAAGACACTGATGTCCTCAAACTGGTTAACTGTGACACGGCTCGCCAGAACTCTGCC TGTCTATTTGTGACTTACAGATTTGAAATGTAATTGTCTTTTTTCCTCCATCTTCTGTGGAAATGGATGTCTTTGGAACTTCATTCCGAG GAGATAAGCTTTAACTTTCCAAAAGGGAATTGCCATGGGTGTTTTTCTTCTGTGGTGAGTGAAACAATCTGAGGTCTGGTTCTTGCTGAC CTTGTTGCCCTGCAAACTTCCTTTCCACGTGTACGCGCACACCAACACGAAATGCCATCACTCCTACTGCGGCTGCTATGAAGCTTACTG GTTGTGATGTGTTATAATTTAGTCTGTTTTTTTGATTGAATGCAGTTTAATGTTTCCAGAAAGCCAAAGTAATTTTCTTTTCAGATATGC AAGGCTTTGGTGGGTCCAAAAAATGTCTATCACAAGCCATTTTTTCCTTTTCCTCTCTCGAAAAGTTAAAATATCTATGTGTTATTCCCA AACCCTCTTACCTATGTATCTGCCTGTCTGTCCATCATCTTCCTTCCTCCCTATCTCTGTGTATCTGGATGGCAGCCGCTGCCCAGGGGA GTGGCTGTGGGGAGGGCAGGTACTGTCTTTGCCTGTGGGTCCAGCTGAGCCATCCCTGCTGGGTGATGCTGGGCAAGACCCTTGGCCCGT CTGGGCCTTGGCTTCCTCACTTGTGAAATGAGCGGGAAGATGACTCTCAGTTCCTTCCACCTCTTAGACATGGTGAGGTAACAGACATCA AAAGCTTTTCTGAAATCTTCAGAAGAAATAGTTCCATTACAGAAAACTCTTCAAAATAAATAGTAGTGAAAACTTTTAAAAACTCTCATT GGAGTAAGTCTTTTCAAGATGATCCTCCACAATGGAGGCAGCGTTCCTACTTGTCATCACACAGCTGAAGACATTGTTTCTTAGGTGTGA AATCGGGGACAAAGGACAAACAGAGACACACGGCATTGTTCATGGGAGGCATCGTCACCCTCCTGGGTGTTCTGTGGGAATTTCCTGTGT GAGGAAAACGTGGCCACAGGGTTGTGCTGTACCCACCCTTCCCCGGCGAGATGGCCCTCGGCCTGTGCCGCTGCTTCCACCCTCGCCACT CCATGGCAGCTTTTGGTCTGTTTCCGGCTCTGCCCTCTGCCCTGAACTCTCATCCGGCTTGTACCTGCCTGCTGGACCCCTCCACCTGGA GGCCAGCCCATGTCTCAGGCCCAGCCCTAGCCTCTTCTCCTCAAATTCTAAGTGTTTTCTCTTTAGGTTTCCCTGGCTTTGTGAATGGAT CATGTGTCTCTAGGTATAAACCTGACATCATCTTTCCACCCGGCTTACCTCCACCAGATCTCCCCAGTTCTGTCTCCATCTTCTACCTGC AGCTGCTCTGTTCTCATGGTCACTGCTGCATCACTGAGTCTGGACCCTTGTTATCATTTTCAAACTGGCCTCCTTCCCTCGTTCCCCACT TCTTAAAGTCACCTGTCCATTGCCACCAGATTAAGCTTTCTCCAGCCAGATCACCTCTCTCTGAGAAACCTCCATTGACATGGAAACACC ATTGTCTGGCACACATACTCACATACTCACCTTCCCGTCTTGATCCCCACACATCTTTCCAGCCTCCCCTCCCACTCCACTCCCTGCTCT CTCCTCCACCTCCCCATCCTCTTGTCTCCCCTCCCCTCTGAATCCAGCCCAGCGGGGCTTCTCCTGCCTCCATCACATCACAGAAGTACC TCCTGCTTCTGGTTTTAATTAGAGCCTTCCCCGATTACATTTTCCTCTGAATTTTTTCCTATCTACATTTGATCTGTCATGTTTAAACCC CCTACTTCTAAGGGAACTTCTCTAATCTCTTATCCTCATCCCCAAATAGTGTTTTCTTCCTCTGGGTTCTTATAATGTTGGTATCAATCT CACAGCATTTAGTGCTTCCTGCCTGGTGTGACAGTTACCTGTGTGCATGTGCAATTTCTAATTTCCCACGCTAGACTGTGAGCTTCCTAA GGCAAGAATCATGCCTTGTTGGTTTCTGTATTCCTCATGGTGCCAAACACAGTGCCTTCTACATTGCAGGCGCTGAATAAACATTTTTAA AGCAAAATGATGTGGATTTTTAAAATAAATATTTAAGTGCTGGTAAGATGAGCATGTATCCGGGGTGCCCATGAAATGTTCTTGGGGCCG TGTGGGGACAGTCGTCATTCCTCCTCCTGCCACCCTTTTCTTTCAGTGAGTCACTGTGGATGGTCCCAGCTGTGTCATCCCAAAGTTCAG CAGGGAAAGCTGAGCTGGGCCTCTCCAGGTGAGTTTTCTAGAAGCATTTCTCAAACTGTGGGTTACATCAACTTGGGTGTCTTGAGCTGT AAGGAAGGAACTCCGGAGTCAGCTGGGCTACAGGGGAGCTTCTCTAAGTCCTGCGGGAGGCCAGACCCAGCCTGAGCTTGCTGTTAGCTA GCGGAGGCAGCTGCTGGTGGCCCAGGTGCTCGACACCAGGCATCCCCTCTCCTCCCACGAAGGGTGTGCCATAATCCCCTTCAACAGGAA ATGCTTCCCAGAAGCCTCTCAGCAGCCTCCCCTCCTGTCCTATCAGCTAGAAGCGCCTCGCTTGTCCCAAGACCAGCAGGGACAGGGAAC TGTCCGAGCCCGTGGCTGTGTGGAGGAAGGCGACCCCCAGCACAAGATTGGTTTCCTTTGGGAAGGGAAGAGGGAGTGTGTTGGGGTAAG GGGTAGAGCAGAGGAATGGTCAGGGGGCAACAACCGCTGACAGCTGCAACAGGTGCATGGCATCTCACAGGGAGGCAGGGAGGTGCGAGC TCCTAAGTAATGGAGCAAAAAAATTCTATTCTGTAGAATGGGGAGAGAAAATGTGACATTTTAATTTTTTTTTGCATTTATATTCCTAAT TCCTACTTAAAGTGAATATACTGCCGCTGTAGATCATAAAATGTATCTTTTCCATGGCCAACAAGGGGCATCTTTTATAAATGCATAATA ACCCAGTTTGTATCAAAGGGTATCGACTTAAGTGAAATTTCAACATGCTGTTACTTTTTCCTTTTAATGTAATTCTGTTTTCCAAATAAA >6936_6936_2_ASAH1-HGSNAT_ASAH1_chr8_17933049_ENST00000314146_HGSNAT_chr8_43054530_ENST00000379644_length(amino acids)=214AA_BP= MYPPFPGEMALGLCRCFHPRHSMAAFGLFPALPSALNSHPACTCLLDPSTWRPAHVSGPALASSPQILSVFSLGFPGFVNGSCVSRYKPD IIFPPGLPPPDLPSSVSIFYLQLLCSHGHCCITESGPLLSFSNWPPSLVPHFLKSPVHCHQIKLSPARSPLSEKPPLTWKHHCLAHILTY -------------------------------------------------------------- >6936_6936_3_ASAH1-HGSNAT_ASAH1_chr8_17933049_ENST00000381733_HGSNAT_chr8_43054530_ENST00000379644_length(transcript)=3808nt_BP=340nt GGACTTTGAAATCCAACCCGGTCACCTACCCGCGCGACTGTGTCCACGGATGGCACGAAAGCCAAGCGAGTCCCCCTGCCGAGCTACTCG CGTCCGCCTCCTCCCAAGCTGAGCTCTGCTCCGCCCACCTGAGTCCTTCGCCAGTTAGGAGGAAACACAGCCGCTTAATGAACTGCTGCA TCGGGCTGGGAGAGAAAGCTCGCGGGTCCCACCGGGCCTCCTACCCAAGTCTCAGCGCGCTTTTCACCGAGGCCTCAATTCTGGGATTTG GCAGCTTTGCTGTGAAAGCCCAATGGACAGAGGACTGCAGAAAATCAACCTATCCTCCTTCAGGACCAACGAATGAATTCCATTCTGGTA TATGTCGGCCACGAGGTGTTTGAGAACTACTTCCCCTTTCAGTGGAAGCTGAAGGACAACCAGTCCCACAAGGAGCACCTGACTCAGAAC ATCGTCGCCACTGCCCTCTGGGTGCTCATTGCCTACATCCTCTATAGAAAGAAGATTTTTTGGAAAATCTGATGGCTCCCACTGAGATGT GCTGCTGGAAGACTCTAGTAGGCCTGCAGGGAGGACTGAAGCAGCCTTTGTTAAAGGGAAGCATTCATTAGGAAATTGACTGGCTGCGTG TTTACAGACTCTGGGGGAAGACACTGATGTCCTCAAACTGGTTAACTGTGACACGGCTCGCCAGAACTCTGCCTGTCTATTTGTGACTTA CAGATTTGAAATGTAATTGTCTTTTTTCCTCCATCTTCTGTGGAAATGGATGTCTTTGGAACTTCATTCCGAGGAGATAAGCTTTAACTT TCCAAAAGGGAATTGCCATGGGTGTTTTTCTTCTGTGGTGAGTGAAACAATCTGAGGTCTGGTTCTTGCTGACCTTGTTGCCCTGCAAAC TTCCTTTCCACGTGTACGCGCACACCAACACGAAATGCCATCACTCCTACTGCGGCTGCTATGAAGCTTACTGGTTGTGATGTGTTATAA TTTAGTCTGTTTTTTTGATTGAATGCAGTTTAATGTTTCCAGAAAGCCAAAGTAATTTTCTTTTCAGATATGCAAGGCTTTGGTGGGTCC AAAAAATGTCTATCACAAGCCATTTTTTCCTTTTCCTCTCTCGAAAAGTTAAAATATCTATGTGTTATTCCCAAACCCTCTTACCTATGT ATCTGCCTGTCTGTCCATCATCTTCCTTCCTCCCTATCTCTGTGTATCTGGATGGCAGCCGCTGCCCAGGGGAGTGGCTGTGGGGAGGGC AGGTACTGTCTTTGCCTGTGGGTCCAGCTGAGCCATCCCTGCTGGGTGATGCTGGGCAAGACCCTTGGCCCGTCTGGGCCTTGGCTTCCT CACTTGTGAAATGAGCGGGAAGATGACTCTCAGTTCCTTCCACCTCTTAGACATGGTGAGGTAACAGACATCAAAAGCTTTTCTGAAATC TTCAGAAGAAATAGTTCCATTACAGAAAACTCTTCAAAATAAATAGTAGTGAAAACTTTTAAAAACTCTCATTGGAGTAAGTCTTTTCAA GATGATCCTCCACAATGGAGGCAGCGTTCCTACTTGTCATCACACAGCTGAAGACATTGTTTCTTAGGTGTGAAATCGGGGACAAAGGAC AAACAGAGACACACGGCATTGTTCATGGGAGGCATCGTCACCCTCCTGGGTGTTCTGTGGGAATTTCCTGTGTGAGGAAAACGTGGCCAC AGGGTTGTGCTGTACCCACCCTTCCCCGGCGAGATGGCCCTCGGCCTGTGCCGCTGCTTCCACCCTCGCCACTCCATGGCAGCTTTTGGT CTGTTTCCGGCTCTGCCCTCTGCCCTGAACTCTCATCCGGCTTGTACCTGCCTGCTGGACCCCTCCACCTGGAGGCCAGCCCATGTCTCA GGCCCAGCCCTAGCCTCTTCTCCTCAAATTCTAAGTGTTTTCTCTTTAGGTTTCCCTGGCTTTGTGAATGGATCATGTGTCTCTAGGTAT AAACCTGACATCATCTTTCCACCCGGCTTACCTCCACCAGATCTCCCCAGTTCTGTCTCCATCTTCTACCTGCAGCTGCTCTGTTCTCAT GGTCACTGCTGCATCACTGAGTCTGGACCCTTGTTATCATTTTCAAACTGGCCTCCTTCCCTCGTTCCCCACTTCTTAAAGTCACCTGTC CATTGCCACCAGATTAAGCTTTCTCCAGCCAGATCACCTCTCTCTGAGAAACCTCCATTGACATGGAAACACCATTGTCTGGCACACATA CTCACATACTCACCTTCCCGTCTTGATCCCCACACATCTTTCCAGCCTCCCCTCCCACTCCACTCCCTGCTCTCTCCTCCACCTCCCCAT CCTCTTGTCTCCCCTCCCCTCTGAATCCAGCCCAGCGGGGCTTCTCCTGCCTCCATCACATCACAGAAGTACCTCCTGCTTCTGGTTTTA ATTAGAGCCTTCCCCGATTACATTTTCCTCTGAATTTTTTCCTATCTACATTTGATCTGTCATGTTTAAACCCCCTACTTCTAAGGGAAC TTCTCTAATCTCTTATCCTCATCCCCAAATAGTGTTTTCTTCCTCTGGGTTCTTATAATGTTGGTATCAATCTCACAGCATTTAGTGCTT CCTGCCTGGTGTGACAGTTACCTGTGTGCATGTGCAATTTCTAATTTCCCACGCTAGACTGTGAGCTTCCTAAGGCAAGAATCATGCCTT GTTGGTTTCTGTATTCCTCATGGTGCCAAACACAGTGCCTTCTACATTGCAGGCGCTGAATAAACATTTTTAAAGCAAAATGATGTGGAT TTTTAAAATAAATATTTAAGTGCTGGTAAGATGAGCATGTATCCGGGGTGCCCATGAAATGTTCTTGGGGCCGTGTGGGGACAGTCGTCA TTCCTCCTCCTGCCACCCTTTTCTTTCAGTGAGTCACTGTGGATGGTCCCAGCTGTGTCATCCCAAAGTTCAGCAGGGAAAGCTGAGCTG GGCCTCTCCAGGTGAGTTTTCTAGAAGCATTTCTCAAACTGTGGGTTACATCAACTTGGGTGTCTTGAGCTGTAAGGAAGGAACTCCGGA GTCAGCTGGGCTACAGGGGAGCTTCTCTAAGTCCTGCGGGAGGCCAGACCCAGCCTGAGCTTGCTGTTAGCTAGCGGAGGCAGCTGCTGG TGGCCCAGGTGCTCGACACCAGGCATCCCCTCTCCTCCCACGAAGGGTGTGCCATAATCCCCTTCAACAGGAAATGCTTCCCAGAAGCCT CTCAGCAGCCTCCCCTCCTGTCCTATCAGCTAGAAGCGCCTCGCTTGTCCCAAGACCAGCAGGGACAGGGAACTGTCCGAGCCCGTGGCT GTGTGGAGGAAGGCGACCCCCAGCACAAGATTGGTTTCCTTTGGGAAGGGAAGAGGGAGTGTGTTGGGGTAAGGGGTAGAGCAGAGGAAT GGTCAGGGGGCAACAACCGCTGACAGCTGCAACAGGTGCATGGCATCTCACAGGGAGGCAGGGAGGTGCGAGCTCCTAAGTAATGGAGCA AAAAAATTCTATTCTGTAGAATGGGGAGAGAAAATGTGACATTTTAATTTTTTTTTGCATTTATATTCCTAATTCCTACTTAAAGTGAAT ATACTGCCGCTGTAGATCATAAAATGTATCTTTTCCATGGCCAACAAGGGGCATCTTTTATAAATGCATAATAACCCAGTTTGTATCAAA GGGTATCGACTTAAGTGAAATTTCAACATGCTGTTACTTTTTCCTTTTAATGTAATTCTGTTTTCCAAATAAATGGGGGAGACAAATGGA >6936_6936_3_ASAH1-HGSNAT_ASAH1_chr8_17933049_ENST00000381733_HGSNAT_chr8_43054530_ENST00000379644_length(amino acids)=214AA_BP= MYPPFPGEMALGLCRCFHPRHSMAAFGLFPALPSALNSHPACTCLLDPSTWRPAHVSGPALASSPQILSVFSLGFPGFVNGSCVSRYKPD IIFPPGLPPPDLPSSVSIFYLQLLCSHGHCCITESGPLLSFSNWPPSLVPHFLKSPVHCHQIKLSPARSPLSEKPPLTWKHHCLAHILTY -------------------------------------------------------------- >6936_6936_4_ASAH1-HGSNAT_ASAH1_chr8_17933049_ENST00000520781_HGSNAT_chr8_43054530_ENST00000379644_length(transcript)=3905nt_BP=437nt GGCTCGGTCCGACTATTGCCCGCGGTGGGGGAGGGGGATGGATCACGCCACGCGCCAAAGGCGATCGCGACTCTCCTTCTGCAGGTAGCC TGGAAGGCTCTCTCTCTTTCTCTACGCCACCCTTTTCGTGGCACTGAAAAGCCCCGTCCTCTCCTCCCAGTCCCGCCTCCTCCGAGCGTT CCCCCTACTGCCTGGAATGGTGCGGTCCCAGGTCGCGGGTCACGCGGCGGAGGGGGCGTGGCCTGCCCCCGGCCCAGCCGGCTCTTCTTT GCCTCTGCTGGAGTCCGGGGAGTGGCGTTGGCTGCTAGAGCGATGCCGGGCCGGAGTTGCGTCGCCTTAGTCCTCCTGGCTGCCGCCGTC AGCTGTGCCGTCGCGCAGCACGCGCCGCCGTGGACAGAGGACTGCAGAAAATCAACCTATCCTCCTTCAGGACCAACGAATGAATTCCAT TCTGGTATATGTCGGCCACGAGGTGTTTGAGAACTACTTCCCCTTTCAGTGGAAGCTGAAGGACAACCAGTCCCACAAGGAGCACCTGAC TCAGAACATCGTCGCCACTGCCCTCTGGGTGCTCATTGCCTACATCCTCTATAGAAAGAAGATTTTTTGGAAAATCTGATGGCTCCCACT GAGATGTGCTGCTGGAAGACTCTAGTAGGCCTGCAGGGAGGACTGAAGCAGCCTTTGTTAAAGGGAAGCATTCATTAGGAAATTGACTGG CTGCGTGTTTACAGACTCTGGGGGAAGACACTGATGTCCTCAAACTGGTTAACTGTGACACGGCTCGCCAGAACTCTGCCTGTCTATTTG TGACTTACAGATTTGAAATGTAATTGTCTTTTTTCCTCCATCTTCTGTGGAAATGGATGTCTTTGGAACTTCATTCCGAGGAGATAAGCT TTAACTTTCCAAAAGGGAATTGCCATGGGTGTTTTTCTTCTGTGGTGAGTGAAACAATCTGAGGTCTGGTTCTTGCTGACCTTGTTGCCC TGCAAACTTCCTTTCCACGTGTACGCGCACACCAACACGAAATGCCATCACTCCTACTGCGGCTGCTATGAAGCTTACTGGTTGTGATGT GTTATAATTTAGTCTGTTTTTTTGATTGAATGCAGTTTAATGTTTCCAGAAAGCCAAAGTAATTTTCTTTTCAGATATGCAAGGCTTTGG TGGGTCCAAAAAATGTCTATCACAAGCCATTTTTTCCTTTTCCTCTCTCGAAAAGTTAAAATATCTATGTGTTATTCCCAAACCCTCTTA CCTATGTATCTGCCTGTCTGTCCATCATCTTCCTTCCTCCCTATCTCTGTGTATCTGGATGGCAGCCGCTGCCCAGGGGAGTGGCTGTGG GGAGGGCAGGTACTGTCTTTGCCTGTGGGTCCAGCTGAGCCATCCCTGCTGGGTGATGCTGGGCAAGACCCTTGGCCCGTCTGGGCCTTG GCTTCCTCACTTGTGAAATGAGCGGGAAGATGACTCTCAGTTCCTTCCACCTCTTAGACATGGTGAGGTAACAGACATCAAAAGCTTTTC TGAAATCTTCAGAAGAAATAGTTCCATTACAGAAAACTCTTCAAAATAAATAGTAGTGAAAACTTTTAAAAACTCTCATTGGAGTAAGTC TTTTCAAGATGATCCTCCACAATGGAGGCAGCGTTCCTACTTGTCATCACACAGCTGAAGACATTGTTTCTTAGGTGTGAAATCGGGGAC AAAGGACAAACAGAGACACACGGCATTGTTCATGGGAGGCATCGTCACCCTCCTGGGTGTTCTGTGGGAATTTCCTGTGTGAGGAAAACG TGGCCACAGGGTTGTGCTGTACCCACCCTTCCCCGGCGAGATGGCCCTCGGCCTGTGCCGCTGCTTCCACCCTCGCCACTCCATGGCAGC TTTTGGTCTGTTTCCGGCTCTGCCCTCTGCCCTGAACTCTCATCCGGCTTGTACCTGCCTGCTGGACCCCTCCACCTGGAGGCCAGCCCA TGTCTCAGGCCCAGCCCTAGCCTCTTCTCCTCAAATTCTAAGTGTTTTCTCTTTAGGTTTCCCTGGCTTTGTGAATGGATCATGTGTCTC TAGGTATAAACCTGACATCATCTTTCCACCCGGCTTACCTCCACCAGATCTCCCCAGTTCTGTCTCCATCTTCTACCTGCAGCTGCTCTG TTCTCATGGTCACTGCTGCATCACTGAGTCTGGACCCTTGTTATCATTTTCAAACTGGCCTCCTTCCCTCGTTCCCCACTTCTTAAAGTC ACCTGTCCATTGCCACCAGATTAAGCTTTCTCCAGCCAGATCACCTCTCTCTGAGAAACCTCCATTGACATGGAAACACCATTGTCTGGC ACACATACTCACATACTCACCTTCCCGTCTTGATCCCCACACATCTTTCCAGCCTCCCCTCCCACTCCACTCCCTGCTCTCTCCTCCACC TCCCCATCCTCTTGTCTCCCCTCCCCTCTGAATCCAGCCCAGCGGGGCTTCTCCTGCCTCCATCACATCACAGAAGTACCTCCTGCTTCT GGTTTTAATTAGAGCCTTCCCCGATTACATTTTCCTCTGAATTTTTTCCTATCTACATTTGATCTGTCATGTTTAAACCCCCTACTTCTA AGGGAACTTCTCTAATCTCTTATCCTCATCCCCAAATAGTGTTTTCTTCCTCTGGGTTCTTATAATGTTGGTATCAATCTCACAGCATTT AGTGCTTCCTGCCTGGTGTGACAGTTACCTGTGTGCATGTGCAATTTCTAATTTCCCACGCTAGACTGTGAGCTTCCTAAGGCAAGAATC ATGCCTTGTTGGTTTCTGTATTCCTCATGGTGCCAAACACAGTGCCTTCTACATTGCAGGCGCTGAATAAACATTTTTAAAGCAAAATGA TGTGGATTTTTAAAATAAATATTTAAGTGCTGGTAAGATGAGCATGTATCCGGGGTGCCCATGAAATGTTCTTGGGGCCGTGTGGGGACA GTCGTCATTCCTCCTCCTGCCACCCTTTTCTTTCAGTGAGTCACTGTGGATGGTCCCAGCTGTGTCATCCCAAAGTTCAGCAGGGAAAGC TGAGCTGGGCCTCTCCAGGTGAGTTTTCTAGAAGCATTTCTCAAACTGTGGGTTACATCAACTTGGGTGTCTTGAGCTGTAAGGAAGGAA CTCCGGAGTCAGCTGGGCTACAGGGGAGCTTCTCTAAGTCCTGCGGGAGGCCAGACCCAGCCTGAGCTTGCTGTTAGCTAGCGGAGGCAG CTGCTGGTGGCCCAGGTGCTCGACACCAGGCATCCCCTCTCCTCCCACGAAGGGTGTGCCATAATCCCCTTCAACAGGAAATGCTTCCCA GAAGCCTCTCAGCAGCCTCCCCTCCTGTCCTATCAGCTAGAAGCGCCTCGCTTGTCCCAAGACCAGCAGGGACAGGGAACTGTCCGAGCC CGTGGCTGTGTGGAGGAAGGCGACCCCCAGCACAAGATTGGTTTCCTTTGGGAAGGGAAGAGGGAGTGTGTTGGGGTAAGGGGTAGAGCA GAGGAATGGTCAGGGGGCAACAACCGCTGACAGCTGCAACAGGTGCATGGCATCTCACAGGGAGGCAGGGAGGTGCGAGCTCCTAAGTAA TGGAGCAAAAAAATTCTATTCTGTAGAATGGGGAGAGAAAATGTGACATTTTAATTTTTTTTTGCATTTATATTCCTAATTCCTACTTAA AGTGAATATACTGCCGCTGTAGATCATAAAATGTATCTTTTCCATGGCCAACAAGGGGCATCTTTTATAAATGCATAATAACCCAGTTTG TATCAAAGGGTATCGACTTAAGTGAAATTTCAACATGCTGTTACTTTTTCCTTTTAATGTAATTCTGTTTTCCAAATAAATGGGGGAGAC >6936_6936_4_ASAH1-HGSNAT_ASAH1_chr8_17933049_ENST00000520781_HGSNAT_chr8_43054530_ENST00000379644_length(amino acids)=214AA_BP= MYPPFPGEMALGLCRCFHPRHSMAAFGLFPALPSALNSHPACTCLLDPSTWRPAHVSGPALASSPQILSVFSLGFPGFVNGSCVSRYKPD IIFPPGLPPPDLPSSVSIFYLQLLCSHGHCCITESGPLLSFSNWPPSLVPHFLKSPVHCHQIKLSPARSPLSEKPPLTWKHHCLAHILTY -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ASAH1-HGSNAT |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ASAH1-HGSNAT |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ASAH1-HGSNAT |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
