
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:PSME3-TM9SF2 (FusionGDB2 ID:69753) |
Fusion Gene Summary for PSME3-TM9SF2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: PSME3-TM9SF2 | Fusion gene ID: 69753 | Hgene | Tgene | Gene symbol | PSME3 | TM9SF2 | Gene ID | 10197 | 9375 |
| Gene name | proteasome activator subunit 3 | transmembrane 9 superfamily member 2 | |
| Synonyms | HEL-S-283|Ki|PA28-gamma|PA28G|PA28gamma|REG-GAMMA | P76 | |
| Cytomap | 17q21.31 | 13q32.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | proteasome activator complex subunit 311S regulator complex gamma subunit11S regulator complex subunit gammaKi antigenKi nuclear autoantigenPA28 gamma variant 5REG gamma-3activator of multicatalytic protease subunit 3epididymis secretory protein L | transmembrane 9 superfamily member 276 kDa membrane proteindinucleotide oxidase disulfide thiol exchanger 3 superfamily member 2epididymis secretory sperm binding proteintransmembrane protein 9 superfamily member 2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000541124, ENST00000293362, ENST00000441946, ENST00000545225, ENST00000590720, ENST00000592169, ENST00000592578, | ENST00000376387, ENST00000463709, | |
| Fusion gene scores | * DoF score | 7 X 6 X 4=168 | 9 X 8 X 3=216 |
| # samples | 7 | 9 | |
| ** MAII score | log2(7/168*10)=-1.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/216*10)=-1.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: PSME3 [Title/Abstract] AND TM9SF2 [Title/Abstract] AND fusion [Title/Abstract] | ||
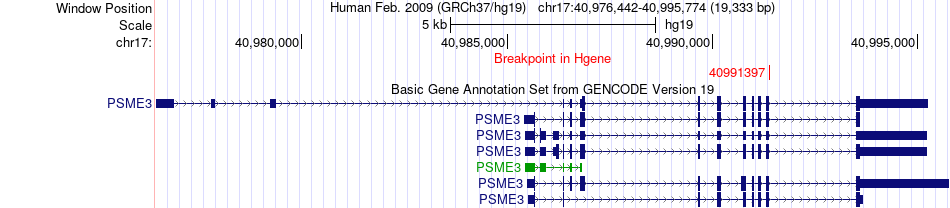
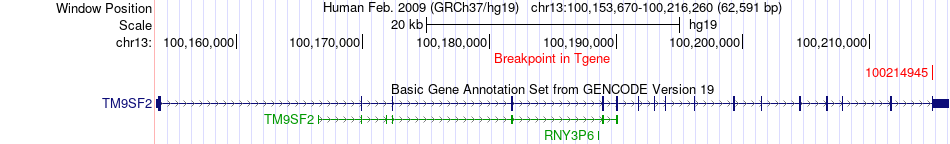
| Most frequent breakpoint | PSME3(40991397)-TM9SF2(100214945), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | PSME3 | GO:0061136 | regulation of proteasomal protein catabolic process | 18309296 |
| Hgene | PSME3 | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | 18309296 |
 Fusion gene breakpoints across PSME3 (5'-gene) Fusion gene breakpoints across PSME3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across TM9SF2 (3'-gene) Fusion gene breakpoints across TM9SF2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-V1-A9ZR-01A | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + |
Top |
Fusion Gene ORF analysis for PSME3-TM9SF2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000541124 | ENST00000376387 | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + |
| 3UTR-intron | ENST00000541124 | ENST00000463709 | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + |
| 5CDS-intron | ENST00000293362 | ENST00000463709 | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + |
| 5CDS-intron | ENST00000441946 | ENST00000463709 | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + |
| 5CDS-intron | ENST00000545225 | ENST00000463709 | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + |
| 5CDS-intron | ENST00000590720 | ENST00000463709 | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + |
| 5CDS-intron | ENST00000592169 | ENST00000463709 | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + |
| In-frame | ENST00000293362 | ENST00000376387 | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + |
| In-frame | ENST00000441946 | ENST00000376387 | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + |
| In-frame | ENST00000545225 | ENST00000376387 | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + |
| In-frame | ENST00000590720 | ENST00000376387 | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + |
| In-frame | ENST00000592169 | ENST00000376387 | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + |
| intron-3CDS | ENST00000592578 | ENST00000376387 | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + |
| intron-intron | ENST00000592578 | ENST00000463709 | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000545225 | PSME3 | chr17 | 40991397 | + | ENST00000376387 | TM9SF2 | chr13 | 100214945 | + | 2656 | 1340 | 740 | 1396 | 218 |
| ENST00000590720 | PSME3 | chr17 | 40991397 | + | ENST00000376387 | TM9SF2 | chr13 | 100214945 | + | 2233 | 917 | 233 | 973 | 246 |
| ENST00000441946 | PSME3 | chr17 | 40991397 | + | ENST00000376387 | TM9SF2 | chr13 | 100214945 | + | 2541 | 1225 | 493 | 1281 | 262 |
| ENST00000293362 | PSME3 | chr17 | 40991397 | + | ENST00000376387 | TM9SF2 | chr13 | 100214945 | + | 2200 | 884 | 161 | 940 | 259 |
| ENST00000592169 | PSME3 | chr17 | 40991397 | + | ENST00000376387 | TM9SF2 | chr13 | 100214945 | + | 1968 | 652 | 136 | 708 | 190 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000545225 | ENST00000376387 | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + | 0.001348016 | 0.99865204 |
| ENST00000590720 | ENST00000376387 | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + | 0.000536689 | 0.9994634 |
| ENST00000441946 | ENST00000376387 | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + | 0.002271839 | 0.99772817 |
| ENST00000293362 | ENST00000376387 | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + | 0.000551459 | 0.9994485 |
| ENST00000592169 | ENST00000376387 | PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214945 | + | 0.00045637 | 0.9995436 |
Top |
Fusion Genomic Features for PSME3-TM9SF2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214944 | + | 7.94E-07 | 0.99999917 |
| PSME3 | chr17 | 40991397 | + | TM9SF2 | chr13 | 100214944 | + | 7.94E-07 | 0.99999917 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for PSME3-TM9SF2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:40991397/chr13:100214945) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | TM9SF2 | chr17:40991397 | chr13:100214945 | ENST00000376387 | 15 | 17 | 653_663 | 641 | 664.0 | Topological domain | Cytoplasmic |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | TM9SF2 | chr17:40991397 | chr13:100214945 | ENST00000376387 | 15 | 17 | 29_300 | 641 | 664.0 | Topological domain | Lumenal | |
| Tgene | TM9SF2 | chr17:40991397 | chr13:100214945 | ENST00000376387 | 15 | 17 | 322_374 | 641 | 664.0 | Topological domain | Cytoplasmic | |
| Tgene | TM9SF2 | chr17:40991397 | chr13:100214945 | ENST00000376387 | 15 | 17 | 396_398 | 641 | 664.0 | Topological domain | Lumenal | |
| Tgene | TM9SF2 | chr17:40991397 | chr13:100214945 | ENST00000376387 | 15 | 17 | 420_437 | 641 | 664.0 | Topological domain | Cytoplasmic | |
| Tgene | TM9SF2 | chr17:40991397 | chr13:100214945 | ENST00000376387 | 15 | 17 | 459_466 | 641 | 664.0 | Topological domain | Lumenal | |
| Tgene | TM9SF2 | chr17:40991397 | chr13:100214945 | ENST00000376387 | 15 | 17 | 488_522 | 641 | 664.0 | Topological domain | Cytoplasmic | |
| Tgene | TM9SF2 | chr17:40991397 | chr13:100214945 | ENST00000376387 | 15 | 17 | 544_554 | 641 | 664.0 | Topological domain | Lumenal | |
| Tgene | TM9SF2 | chr17:40991397 | chr13:100214945 | ENST00000376387 | 15 | 17 | 576_591 | 641 | 664.0 | Topological domain | Cytoplasmic | |
| Tgene | TM9SF2 | chr17:40991397 | chr13:100214945 | ENST00000376387 | 15 | 17 | 613_631 | 641 | 664.0 | Topological domain | Lumenal | |
| Tgene | TM9SF2 | chr17:40991397 | chr13:100214945 | ENST00000376387 | 15 | 17 | 301_321 | 641 | 664.0 | Transmembrane | Helical | |
| Tgene | TM9SF2 | chr17:40991397 | chr13:100214945 | ENST00000376387 | 15 | 17 | 375_395 | 641 | 664.0 | Transmembrane | Helical | |
| Tgene | TM9SF2 | chr17:40991397 | chr13:100214945 | ENST00000376387 | 15 | 17 | 399_419 | 641 | 664.0 | Transmembrane | Helical | |
| Tgene | TM9SF2 | chr17:40991397 | chr13:100214945 | ENST00000376387 | 15 | 17 | 438_458 | 641 | 664.0 | Transmembrane | Helical | |
| Tgene | TM9SF2 | chr17:40991397 | chr13:100214945 | ENST00000376387 | 15 | 17 | 467_487 | 641 | 664.0 | Transmembrane | Helical | |
| Tgene | TM9SF2 | chr17:40991397 | chr13:100214945 | ENST00000376387 | 15 | 17 | 523_543 | 641 | 664.0 | Transmembrane | Helical | |
| Tgene | TM9SF2 | chr17:40991397 | chr13:100214945 | ENST00000376387 | 15 | 17 | 555_575 | 641 | 664.0 | Transmembrane | Helical | |
| Tgene | TM9SF2 | chr17:40991397 | chr13:100214945 | ENST00000376387 | 15 | 17 | 592_612 | 641 | 664.0 | Transmembrane | Helical | |
| Tgene | TM9SF2 | chr17:40991397 | chr13:100214945 | ENST00000376387 | 15 | 17 | 632_652 | 641 | 664.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for PSME3-TM9SF2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >69753_69753_1_PSME3-TM9SF2_PSME3_chr17_40991397_ENST00000293362_TM9SF2_chr13_100214945_ENST00000376387_length(transcript)=2200nt_BP=884nt GGGCGGACGGCACAGAGGGAGGGAGCGAGCGAGCAGTGAGTAAGCCAGCAAGGGCGGTCGGGTCCCGAGGTCAGCCGAGATTTCTCAGGT CCCTCCGGCCCCCTCCCTGGAGTCCACAGCGCCTCCGGTGTCCAGAGGATCGGACACGGCCCGGCCCGGCCATGGCCTCGTTGCTGAAGG TGGATCAGGAAGTGAAGCTCAAGGTTGATTCTTTCAGGGAGCGGATCACAAGTGAGGCAGAAGACTTGGTGGCAAATTTTTTCCCAAAGA AGTTATTAGAACTTGATAGTTTTCTGAAGGAACCAATCTTAAACATCCATGACCTAACTCAGATCCACTCTGACATGAATCTCCCAGTCC CTGACCCCATTCTTCTCACCAATAGCCATGATGGACTGGATGGTCCCACTTATAAGAAGCGAAGGTTGGATGAGTGTGAAGAAGCCTTCC AAGGAACCAAGGTGTTTGTGATGCCCAATGGGATGCTGAAAAGCAACCAGCAGCTGGTGGACATTATTGAGAAAGTGAAACCTGAGATCC GGCTGTTGATTGAGAAATGTAACACGCCTTCAGGCAAAGGTCCTCATATATGTTTTGACCTCCAGGTCAAAATGTGGGTACAGCTCCTGA TTCCCAGGATAGAAGATGGAAACAACTTTGGGGTGTCCATTCAGGAGGAAACAGTTGCAGAGCTAAGAACTGTTGAGAGTGAAGCTGCAT CTTATCTGGACCAGATTTCTAGATATTATATTACAAGAGCCAAATTGGTTTCTAAAATAGCTAAATATCCCCATGTGGAGGACTATCGCC GCACCGTGACAGAGATTGATGAGAAAGAATATATCAGCCTTCGGCTCATCATATCAGAGCTGAGGAATCAATATGAACAATTGGCTTCTT TGCATGCTTTTGGTTTGTTACCAAAATATACAGTGTGGTGAAGGTTGACTGAAGAAGTCCAGTGTGTCCAGTTAAAACAGAAATAAATTA AACTCTTCATCAACAAAGACCTGTTTTTGTGACTGCCTTGAGTTTTATCAGAATTATTGGCCTAGTAATCCTTCAGAAACACCGTAATTC TAAATAAACCTCTTCCCATACACCTTTCCCCCATAAGATGTGTCTTCAACACTATAAAGCATTTGTATTGTGATTTGATTAAGTATATAT TTGGTTGTTCTCAATGAAGAGCAAATTTAAATATTATGTGCATTTGTAAATACAGTAGCTATAAAATTTTCCATACTTCTAATGGCAGAA TAGAGGAGGCCATATTAAATAATACTGATGAAAGGCAGGACACTGCATTGTAAATAGGATTTTCTAGGCTCGGTAGGCAGAAAGAATTAT TTTTCTTTGAAGGAAATAACTTTTTATCATGGTAATTTTGAAGGATGATTCCTATGATGTGTTCACCAGGGGAATGTGGCTTTTAAAGAA AATCTTCTATTGGTTGTAACTGTTCATATCTTCTTACTTTTCTGTGTTGACTTCATTATTCCCATGGTATTGGCCTTTTAAACTATGTGC CTCTGAGTCTTTCAATTTATAAATTTGTTATCTTAATAAATATTATAAAAATGTCTTCATTGCATCATTACCTATTATATATTCAAGGAG GTTCTGTAAATACAGATCTATTTACCAAGGTGTTTTGAAATGATTTATACTATTTTATCAGATTTTACTGGATTTTGTTAATGTAACACT TTCTGAGTTTTGATCTGAATGGAATTTCACTGTGTAAGGCATTGTCTAAAATGTTCACTGTGAGGCAGTAGAGGAGGTTGAAGTCATGCA TATCTAGACAGATGAATTATCTGCAAGTACTTAAGGTGGAGCCTTACTACATTGTCATTAAGGAGTTTCTATTTTTGACTTGTTTTTCAA AATACAGTGTTTTAAGTCAGGTTTTATTACCTGGACATTAGCAATCACTAGCAAGTCTTCATCTTTATGATTTCATTGTTTTAATTTAGA TTTCACAAACAGTTTTACTTCTAAGAGTTAAGAAATGCACTGTCTTTCACATTCATCTGACTAATTGAAATCCATTTATGTAGAATATAG GAATGGGGGGTGATTCTGAAGTATCGGTCTGCTTGTATCTGTGAGCATATTCATTTGTATTTTAGGGTCTCTCTGCTTGCTGCTTAGGAG >69753_69753_1_PSME3-TM9SF2_PSME3_chr17_40991397_ENST00000293362_TM9SF2_chr13_100214945_ENST00000376387_length(amino acids)=259AA_BP=241 MASLLKVDQEVKLKVDSFRERITSEAEDLVANFFPKKLLELDSFLKEPILNIHDLTQIHSDMNLPVPDPILLTNSHDGLDGPTYKKRRLD ECEEAFQGTKVFVMPNGMLKSNQQLVDIIEKVKPEIRLLIEKCNTPSGKGPHICFDLQVKMWVQLLIPRIEDGNNFGVSIQEETVAELRT -------------------------------------------------------------- >69753_69753_2_PSME3-TM9SF2_PSME3_chr17_40991397_ENST00000441946_TM9SF2_chr13_100214945_ENST00000376387_length(transcript)=2541nt_BP=1225nt GAAAGCCGGGAGGGCGAGCGAGAGAGCAAGCAGGCAGCAGGCTGCCGGCGGGCGGGCGGACGGCACAGAGGGAGGGAGCGAGCGAGCAGT GAGTAAGCCAGCAAGGGCGGTCGGGTCCCGAGGTCAGCCGAGATTTCTCAGGTCCCTCCGGCCCCCTCCCTGGAGTCCACAGCGCCTCCG GTGTCCAGAGGATCGGACACGGCCCGGCCCGGCCATGGCCTCGTTGCTGAAGGTGGATCAGGAAGTGAAGCTCAAGCTCTGAGCTTCCGC TCGGCTCAGCCCAGCCCCCAACTCCCGGGGTCGGCTTCGCGGGAGAGGAAAATATAGGATGGCTTTCTGTACCGCGTCTGATGATGGAGA GTCCCGGGGACACCTCCGCGGACACGTGTTGGACTCAGGGCTTTAGGCCCGTGACAGCTGACAGCTTCAGACACAGGAGGAAGGGAGGGA AGGGGGAGGACAGAGGAGACAGGCAGGTGCTGTCCTCAAAGCCCTGCGTTCTTCTGAGATGGAAAAATGGATCCTCAAAAAAATAAAGTA TTTGCAGTCTGGGGGCCTCTCAGCTTCTTATTACAGTTACAAGGTTGATTCTTTCAGGGAGCGGATCACAAGTGAGGCAGAAGACTTGGT GGCAAATTTTTTCCCAAAGAAGTTATTAGAACTTGATAGTTTTCTGAAGGAACCAATCTTAAACATCCATGACCTAACTCAGATCCACTC TGACATGAATCTCCCAGTCCCTGACCCCATTCTTCTCACCAATAGCCATGATGGACTGGATGGTCCCACTTATAAGAAGCGAAGGTTGGA TGAGTGTGAAGAAGCCTTCCAAGGAACCAAGGTGTTTGTGATGCCCAATGGGATGCTGAAAAGCAACCAGCAGCTGGTGGACATTATTGA GAAAGTGAAACCTGAGATCCGGCTGTTGATTGAGAAATGTAACACGGTCAAAATGTGGGTACAGCTCCTGATTCCCAGGATAGAAGATGG AAACAACTTTGGGGTGTCCATTCAGGAGGAAACAGTTGCAGAGCTAAGAACTGTTGAGAGTGAAGCTGCATCTTATCTGGACCAGATTTC TAGATATTATATTACAAGAGCCAAATTGGTTTCTAAAATAGCTAAATATCCCCATGTGGAGGACTATCGCCGCACCGTGACAGAGATTGA TGAGAAAGAATATATCAGCCTTCGGCTCATCATATCAGAGCTGAGGAATCAATATGAACAATTGGCTTCTTTGCATGCTTTTGGTTTGTT ACCAAAATATACAGTGTGGTGAAGGTTGACTGAAGAAGTCCAGTGTGTCCAGTTAAAACAGAAATAAATTAAACTCTTCATCAACAAAGA CCTGTTTTTGTGACTGCCTTGAGTTTTATCAGAATTATTGGCCTAGTAATCCTTCAGAAACACCGTAATTCTAAATAAACCTCTTCCCAT ACACCTTTCCCCCATAAGATGTGTCTTCAACACTATAAAGCATTTGTATTGTGATTTGATTAAGTATATATTTGGTTGTTCTCAATGAAG AGCAAATTTAAATATTATGTGCATTTGTAAATACAGTAGCTATAAAATTTTCCATACTTCTAATGGCAGAATAGAGGAGGCCATATTAAA TAATACTGATGAAAGGCAGGACACTGCATTGTAAATAGGATTTTCTAGGCTCGGTAGGCAGAAAGAATTATTTTTCTTTGAAGGAAATAA CTTTTTATCATGGTAATTTTGAAGGATGATTCCTATGATGTGTTCACCAGGGGAATGTGGCTTTTAAAGAAAATCTTCTATTGGTTGTAA CTGTTCATATCTTCTTACTTTTCTGTGTTGACTTCATTATTCCCATGGTATTGGCCTTTTAAACTATGTGCCTCTGAGTCTTTCAATTTA TAAATTTGTTATCTTAATAAATATTATAAAAATGTCTTCATTGCATCATTACCTATTATATATTCAAGGAGGTTCTGTAAATACAGATCT ATTTACCAAGGTGTTTTGAAATGATTTATACTATTTTATCAGATTTTACTGGATTTTGTTAATGTAACACTTTCTGAGTTTTGATCTGAA TGGAATTTCACTGTGTAAGGCATTGTCTAAAATGTTCACTGTGAGGCAGTAGAGGAGGTTGAAGTCATGCATATCTAGACAGATGAATTA TCTGCAAGTACTTAAGGTGGAGCCTTACTACATTGTCATTAAGGAGTTTCTATTTTTGACTTGTTTTTCAAAATACAGTGTTTTAAGTCA GGTTTTATTACCTGGACATTAGCAATCACTAGCAAGTCTTCATCTTTATGATTTCATTGTTTTAATTTAGATTTCACAAACAGTTTTACT TCTAAGAGTTAAGAAATGCACTGTCTTTCACATTCATCTGACTAATTGAAATCCATTTATGTAGAATATAGGAATGGGGGGTGATTCTGA AGTATCGGTCTGCTTGTATCTGTGAGCATATTCATTTGTATTTTAGGGTCTCTCTGCTTGCTGCTTAGGAGAGCAAATCAGATACTGTCA >69753_69753_2_PSME3-TM9SF2_PSME3_chr17_40991397_ENST00000441946_TM9SF2_chr13_100214945_ENST00000376387_length(amino acids)=262AA_BP=244 MRSSEMEKWILKKIKYLQSGGLSASYYSYKVDSFRERITSEAEDLVANFFPKKLLELDSFLKEPILNIHDLTQIHSDMNLPVPDPILLTN SHDGLDGPTYKKRRLDECEEAFQGTKVFVMPNGMLKSNQQLVDIIEKVKPEIRLLIEKCNTVKMWVQLLIPRIEDGNNFGVSIQEETVAE -------------------------------------------------------------- >69753_69753_3_PSME3-TM9SF2_PSME3_chr17_40991397_ENST00000545225_TM9SF2_chr13_100214945_ENST00000376387_length(transcript)=2656nt_BP=1340nt AAGTGTAGGAGCCAAGGCCGCAGAGAATGGGGAGGCTCCGCTATTCTCTAAATTCAGGGGGCGGCGGGCGCGGTGGCTCACGCCTGTAAT CTCAGCACTTGGGGAGGCCGAGGAGGGCGGATCACGTGAGGTCAGGAGTTCGAGACCAGCCTGGCCAACACAGTGAAACCCCGTCTCTAC TAAAAATACAAAAACTAGCCGGGCGTGGTGGGGCACGCCTATAATCCCAGCTTAATCCCAGCTACTCGGGAGGCTGAGGCAGGAGAATCG CTTGAACCTGGGAGGTGGAGGTTGCAGTGAGCCGAGATCGGGCCACTGCACTCCAGCCTGGGCGACAGGGCAAGGCTCCATCTCAGAACA ACAACAGCAACAACAAAAGGCCGGGCATGGTGGCTCACGCTTGTAATTCCAGCACTTTGGGAGGCCGAGGCGGGCGGATCACAAGGCTGG AGTGAAGCGGAACAAACAAACATGGCTCACTGCAGATTTGACTTCCTGGCTCAAGCAATCCTCTTGCTTCAACCTCCAGTGTAGCAGGGA CCACAGCATGAAAGCCCCTTTAGGGGAGGTGACTGGAATCATCCCCAAAGAATTCTCACCTAGTCCTGGTGTGGCTGCTTCATAGTCCTG CGTTTCTTCCTGCTTGGCTGTCCCTAACATCTCAGAAACGCATAGAGAAGTAACGGGCTTCTAGAGAGGTTGATTCTTTCAGGGAGCGGA TCACAAGTGAGGCAGAAGACTTGGTGGCAAATTTTTTCCCAAAGAAGTTATTAGAACTTGATAGTTTTCTGAAGGAACCAATCTTAAACA TCCATGACCTAACTCAGATCCACTCTGACATGAATCTCCCAGTCCCTGACCCCATTCTTCTCACCAATAGCCATGATGGACTGGATGGTC CCACTTATAAGAAGCGAAGGTTGGATGAGTGTGAAGAAGCCTTCCAAGGAACCAAGGTGTTTGTGATGCCCAATGGGATGCTGAAAAGCA ACCAGCAGCTGGTGGACATTATTGAGAAAGTGAAACCTGAGATCCGGCTGTTGATTGAGAAATGTAACACGGTCAAAATGTGGGTACAGC TCCTGATTCCCAGGATAGAAGATGGAAACAACTTTGGGGTGTCCATTCAGGAGGAAACAGTTGCAGAGCTAAGAACTGTTGAGAGTGAAG CTGCATCTTATCTGGACCAGATTTCTAGATATTATATTACAAGAGCCAAATTGGTTTCTAAAATAGCTAAATATCCCCATGTGGAGGACT ATCGCCGCACCGTGACAGAGATTGATGAGAAAGAATATATCAGCCTTCGGCTCATCATATCAGAGCTGAGGAATCAATATGAACAATTGG CTTCTTTGCATGCTTTTGGTTTGTTACCAAAATATACAGTGTGGTGAAGGTTGACTGAAGAAGTCCAGTGTGTCCAGTTAAAACAGAAAT AAATTAAACTCTTCATCAACAAAGACCTGTTTTTGTGACTGCCTTGAGTTTTATCAGAATTATTGGCCTAGTAATCCTTCAGAAACACCG TAATTCTAAATAAACCTCTTCCCATACACCTTTCCCCCATAAGATGTGTCTTCAACACTATAAAGCATTTGTATTGTGATTTGATTAAGT ATATATTTGGTTGTTCTCAATGAAGAGCAAATTTAAATATTATGTGCATTTGTAAATACAGTAGCTATAAAATTTTCCATACTTCTAATG GCAGAATAGAGGAGGCCATATTAAATAATACTGATGAAAGGCAGGACACTGCATTGTAAATAGGATTTTCTAGGCTCGGTAGGCAGAAAG AATTATTTTTCTTTGAAGGAAATAACTTTTTATCATGGTAATTTTGAAGGATGATTCCTATGATGTGTTCACCAGGGGAATGTGGCTTTT AAAGAAAATCTTCTATTGGTTGTAACTGTTCATATCTTCTTACTTTTCTGTGTTGACTTCATTATTCCCATGGTATTGGCCTTTTAAACT ATGTGCCTCTGAGTCTTTCAATTTATAAATTTGTTATCTTAATAAATATTATAAAAATGTCTTCATTGCATCATTACCTATTATATATTC AAGGAGGTTCTGTAAATACAGATCTATTTACCAAGGTGTTTTGAAATGATTTATACTATTTTATCAGATTTTACTGGATTTTGTTAATGT AACACTTTCTGAGTTTTGATCTGAATGGAATTTCACTGTGTAAGGCATTGTCTAAAATGTTCACTGTGAGGCAGTAGAGGAGGTTGAAGT CATGCATATCTAGACAGATGAATTATCTGCAAGTACTTAAGGTGGAGCCTTACTACATTGTCATTAAGGAGTTTCTATTTTTGACTTGTT TTTCAAAATACAGTGTTTTAAGTCAGGTTTTATTACCTGGACATTAGCAATCACTAGCAAGTCTTCATCTTTATGATTTCATTGTTTTAA TTTAGATTTCACAAACAGTTTTACTTCTAAGAGTTAAGAAATGCACTGTCTTTCACATTCATCTGACTAATTGAAATCCATTTATGTAGA ATATAGGAATGGGGGGTGATTCTGAAGTATCGGTCTGCTTGTATCTGTGAGCATATTCATTTGTATTTTAGGGTCTCTCTGCTTGCTGCT >69753_69753_3_PSME3-TM9SF2_PSME3_chr17_40991397_ENST00000545225_TM9SF2_chr13_100214945_ENST00000376387_length(amino acids)=218AA_BP=200 MVANFFPKKLLELDSFLKEPILNIHDLTQIHSDMNLPVPDPILLTNSHDGLDGPTYKKRRLDECEEAFQGTKVFVMPNGMLKSNQQLVDI IEKVKPEIRLLIEKCNTVKMWVQLLIPRIEDGNNFGVSIQEETVAELRTVESEAASYLDQISRYYITRAKLVSKIAKYPHVEDYRRTVTE -------------------------------------------------------------- >69753_69753_4_PSME3-TM9SF2_PSME3_chr17_40991397_ENST00000590720_TM9SF2_chr13_100214945_ENST00000376387_length(transcript)=2233nt_BP=917nt AGTTTCCGGCGTGAGCGGCGAAAGCCGGGAGGGCGAGCGAGAGAGCAAGCAGGCAGCAGGCTGCCGGCGGGCGGGCGGACGGCACAGAGG GAGGGAGCGAGCGAGCAGTGAGTAAGCCAGCAAGGGCGGTCGGGTCCCGAGGTCAGCCGAGATTTCTCAGGTCCCTCCGGCCCCCTCCCT GGAGTCCACAGCGCCTCCGGTGTCCAGAGGATCGGACACGGCCCGGCCCGGCCATGGCCTCGTTGCTGAAGGTGGATCAGGAAGTGAAGC TCAAGGTTGATTCTTTCAGGGAGCGGATCACAAGTGAGGCAGAAGACTTGGTGGCAAATTTTTTCCCAAAGAAGTTATTAGAACTTGATA GTTTTCTGAAGGAACCAATCTTAAACATCCATGACCTAACTCAGATCCACTCTGACATGAATCTCCCAGTCCCTGACCCCATTCTTCTCA CCAATAGCCATGATGGACTGGATGGTCCCACTTATAAGAAGCGAAGGTTGGATGAGTGTGAAGAAGCCTTCCAAGGAACCAAGGTGTTTG TGATGCCCAATGGGATGCTGAAAAGCAACCAGCAGCTGGTGGACATTATTGAGAAAGTGAAACCTGAGATCCGGCTGTTGATTGAGAAAT GTAACACGGTCAAAATGTGGGTACAGCTCCTGATTCCCAGGATAGAAGATGGAAACAACTTTGGGGTGTCCATTCAGGAGGAAACAGTTG CAGAGCTAAGAACTGTTGAGAGTGAAGCTGCATCTTATCTGGACCAGATTTCTAGATATTATATTACAAGAGCCAAATTGGTTTCTAAAA TAGCTAAATATCCCCATGTGGAGGACTATCGCCGCACCGTGACAGAGATTGATGAGAAAGAATATATCAGCCTTCGGCTCATCATATCAG AGCTGAGGAATCAATATGAACAATTGGCTTCTTTGCATGCTTTTGGTTTGTTACCAAAATATACAGTGTGGTGAAGGTTGACTGAAGAAG TCCAGTGTGTCCAGTTAAAACAGAAATAAATTAAACTCTTCATCAACAAAGACCTGTTTTTGTGACTGCCTTGAGTTTTATCAGAATTAT TGGCCTAGTAATCCTTCAGAAACACCGTAATTCTAAATAAACCTCTTCCCATACACCTTTCCCCCATAAGATGTGTCTTCAACACTATAA AGCATTTGTATTGTGATTTGATTAAGTATATATTTGGTTGTTCTCAATGAAGAGCAAATTTAAATATTATGTGCATTTGTAAATACAGTA GCTATAAAATTTTCCATACTTCTAATGGCAGAATAGAGGAGGCCATATTAAATAATACTGATGAAAGGCAGGACACTGCATTGTAAATAG GATTTTCTAGGCTCGGTAGGCAGAAAGAATTATTTTTCTTTGAAGGAAATAACTTTTTATCATGGTAATTTTGAAGGATGATTCCTATGA TGTGTTCACCAGGGGAATGTGGCTTTTAAAGAAAATCTTCTATTGGTTGTAACTGTTCATATCTTCTTACTTTTCTGTGTTGACTTCATT ATTCCCATGGTATTGGCCTTTTAAACTATGTGCCTCTGAGTCTTTCAATTTATAAATTTGTTATCTTAATAAATATTATAAAAATGTCTT CATTGCATCATTACCTATTATATATTCAAGGAGGTTCTGTAAATACAGATCTATTTACCAAGGTGTTTTGAAATGATTTATACTATTTTA TCAGATTTTACTGGATTTTGTTAATGTAACACTTTCTGAGTTTTGATCTGAATGGAATTTCACTGTGTAAGGCATTGTCTAAAATGTTCA CTGTGAGGCAGTAGAGGAGGTTGAAGTCATGCATATCTAGACAGATGAATTATCTGCAAGTACTTAAGGTGGAGCCTTACTACATTGTCA TTAAGGAGTTTCTATTTTTGACTTGTTTTTCAAAATACAGTGTTTTAAGTCAGGTTTTATTACCTGGACATTAGCAATCACTAGCAAGTC TTCATCTTTATGATTTCATTGTTTTAATTTAGATTTCACAAACAGTTTTACTTCTAAGAGTTAAGAAATGCACTGTCTTTCACATTCATC TGACTAATTGAAATCCATTTATGTAGAATATAGGAATGGGGGGTGATTCTGAAGTATCGGTCTGCTTGTATCTGTGAGCATATTCATTTG >69753_69753_4_PSME3-TM9SF2_PSME3_chr17_40991397_ENST00000590720_TM9SF2_chr13_100214945_ENST00000376387_length(amino acids)=246AA_BP=228 MASLLKVDQEVKLKVDSFRERITSEAEDLVANFFPKKLLELDSFLKEPILNIHDLTQIHSDMNLPVPDPILLTNSHDGLDGPTYKKRRLD ECEEAFQGTKVFVMPNGMLKSNQQLVDIIEKVKPEIRLLIEKCNTVKMWVQLLIPRIEDGNNFGVSIQEETVAELRTVESEAASYLDQIS -------------------------------------------------------------- >69753_69753_5_PSME3-TM9SF2_PSME3_chr17_40991397_ENST00000592169_TM9SF2_chr13_100214945_ENST00000376387_length(transcript)=1968nt_BP=652nt CGAGCGAGCAGTGAGTAAGCCAGCAAGGGCGGTCGGGTCCCGAGGTCAGCCGAGATTTCTCAGGTCCCTCCGGCCCCCTCCCTGGAGTCC ACAGCGCCTCCGGTGTCCAGAGGATCGGACACGGCCCGGCCCGGCCATGGCCTCGTTGCTGAAGGTGGATCAGGAAGTGAAGCTCAAGGT TGATTCTTTCAGGGAGCGGATCACAAGTGAGCCCACTTATAAGAAGCGAAGGTTGGATGAGTGTGAAGAAGCCTTCCAAGGAACCAAGGT GTTTGTGATGCCCAATGGGATGCTGAAAAGCAACCAGCAGCTGGTGGACATTATTGAGAAAGTGAAACCTGAGATCCGGCTGTTGATTGA GAAATGTAACACGGTCAAAATGTGGGTACAGCTCCTGATTCCCAGGATAGAAGATGGAAACAACTTTGGGGTGTCCATTCAGGAGGAAAC AGTTGCAGAGCTAAGAACTGTTGAGAGTGAAGCTGCATCTTATCTGGACCAGATTTCTAGATATTATATTACAAGAGCCAAATTGGTTTC TAAAATAGCTAAATATCCCCATGTGGAGGACTATCGCCGCACCGTGACAGAGATTGATGAGAAAGAATATATCAGCCTTCGGCTCATCAT ATCAGAGCTGAGGAATCAATATGAACAATTGGCTTCTTTGCATGCTTTTGGTTTGTTACCAAAATATACAGTGTGGTGAAGGTTGACTGA AGAAGTCCAGTGTGTCCAGTTAAAACAGAAATAAATTAAACTCTTCATCAACAAAGACCTGTTTTTGTGACTGCCTTGAGTTTTATCAGA ATTATTGGCCTAGTAATCCTTCAGAAACACCGTAATTCTAAATAAACCTCTTCCCATACACCTTTCCCCCATAAGATGTGTCTTCAACAC TATAAAGCATTTGTATTGTGATTTGATTAAGTATATATTTGGTTGTTCTCAATGAAGAGCAAATTTAAATATTATGTGCATTTGTAAATA CAGTAGCTATAAAATTTTCCATACTTCTAATGGCAGAATAGAGGAGGCCATATTAAATAATACTGATGAAAGGCAGGACACTGCATTGTA AATAGGATTTTCTAGGCTCGGTAGGCAGAAAGAATTATTTTTCTTTGAAGGAAATAACTTTTTATCATGGTAATTTTGAAGGATGATTCC TATGATGTGTTCACCAGGGGAATGTGGCTTTTAAAGAAAATCTTCTATTGGTTGTAACTGTTCATATCTTCTTACTTTTCTGTGTTGACT TCATTATTCCCATGGTATTGGCCTTTTAAACTATGTGCCTCTGAGTCTTTCAATTTATAAATTTGTTATCTTAATAAATATTATAAAAAT GTCTTCATTGCATCATTACCTATTATATATTCAAGGAGGTTCTGTAAATACAGATCTATTTACCAAGGTGTTTTGAAATGATTTATACTA TTTTATCAGATTTTACTGGATTTTGTTAATGTAACACTTTCTGAGTTTTGATCTGAATGGAATTTCACTGTGTAAGGCATTGTCTAAAAT GTTCACTGTGAGGCAGTAGAGGAGGTTGAAGTCATGCATATCTAGACAGATGAATTATCTGCAAGTACTTAAGGTGGAGCCTTACTACAT TGTCATTAAGGAGTTTCTATTTTTGACTTGTTTTTCAAAATACAGTGTTTTAAGTCAGGTTTTATTACCTGGACATTAGCAATCACTAGC AAGTCTTCATCTTTATGATTTCATTGTTTTAATTTAGATTTCACAAACAGTTTTACTTCTAAGAGTTAAGAAATGCACTGTCTTTCACAT TCATCTGACTAATTGAAATCCATTTATGTAGAATATAGGAATGGGGGGTGATTCTGAAGTATCGGTCTGCTTGTATCTGTGAGCATATTC >69753_69753_5_PSME3-TM9SF2_PSME3_chr17_40991397_ENST00000592169_TM9SF2_chr13_100214945_ENST00000376387_length(amino acids)=190AA_BP=172 MASLLKVDQEVKLKVDSFRERITSEPTYKKRRLDECEEAFQGTKVFVMPNGMLKSNQQLVDIIEKVKPEIRLLIEKCNTVKMWVQLLIPR IEDGNNFGVSIQEETVAELRTVESEAASYLDQISRYYITRAKLVSKIAKYPHVEDYRRTVTEIDEKEYISLRLIISELRNQYEQLASLHA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for PSME3-TM9SF2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for PSME3-TM9SF2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for PSME3-TM9SF2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
