
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:PTCH1-IPPK (FusionGDB2 ID:69866) |
Fusion Gene Summary for PTCH1-IPPK |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: PTCH1-IPPK | Fusion gene ID: 69866 | Hgene | Tgene | Gene symbol | PTCH1 | IPPK | Gene ID | 5727 | 64768 |
| Gene name | patched 1 | inositol-pentakisphosphate 2-kinase | |
| Synonyms | BCNS|NBCCS|PTC|PTC1|PTCH | C9orf12|INSP5K2|IP5K|IPK1|bA476B13.1 | |
| Cytomap | 9q22.32 | 9q22.31 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein patched homolog 1 | inositol-pentakisphosphate 2-kinaseIPK1 homologbA476B13.1 (novel protein)inositol 1,3,4,5,6-pentakisphosphate 2-kinaseins(1,3,4,5,6)P5 2-kinaseinsP5 2-kinase | |
| Modification date | 20200314 | 20200313 | |
| UniProtAcc | . | Q9H8X2 | |
| Ensembl transtripts involved in fusion gene | ENST00000331920, ENST00000375274, ENST00000430669, ENST00000437951, ENST00000468211, ENST00000418258, ENST00000421141, ENST00000429896, ENST00000548379, | ENST00000375522, ENST00000486841, ENST00000287996, | |
| Fusion gene scores | * DoF score | 6 X 7 X 4=168 | 9 X 9 X 5=405 |
| # samples | 7 | 9 | |
| ** MAII score | log2(7/168*10)=-1.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/405*10)=-2.16992500144231 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: PTCH1 [Title/Abstract] AND IPPK [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | PTCH1(98268689)-IPPK(95381847), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | PTCH1-IPPK seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. PTCH1-IPPK seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. PTCH1-IPPK seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | PTCH1 | GO:0010875 | positive regulation of cholesterol efflux | 21931618 |
| Hgene | PTCH1 | GO:0072659 | protein localization to plasma membrane | 11278759 |
| Tgene | IPPK | GO:0052746 | inositol phosphorylation | 12084730 |
 Fusion gene breakpoints across PTCH1 (5'-gene) Fusion gene breakpoints across PTCH1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
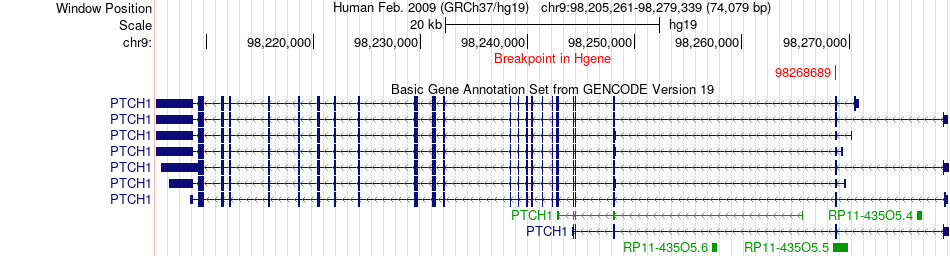 |
 Fusion gene breakpoints across IPPK (3'-gene) Fusion gene breakpoints across IPPK (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
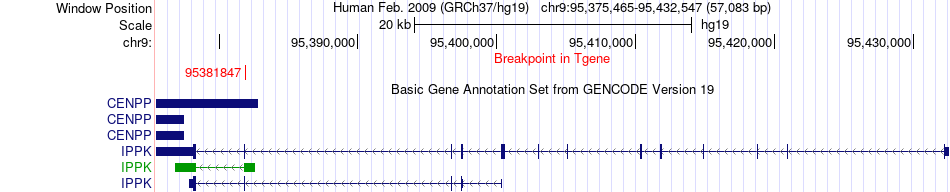 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-DX-A1KZ-01A | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| ChimerDB4 | SARC | TCGA-RN-AAAQ-01A | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
Top |
Fusion Gene ORF analysis for PTCH1-IPPK |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000331920 | ENST00000375522 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| 5CDS-5UTR | ENST00000331920 | ENST00000486841 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| 5CDS-5UTR | ENST00000375274 | ENST00000375522 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| 5CDS-5UTR | ENST00000375274 | ENST00000486841 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| 5CDS-5UTR | ENST00000430669 | ENST00000375522 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| 5CDS-5UTR | ENST00000430669 | ENST00000486841 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| 5CDS-5UTR | ENST00000437951 | ENST00000375522 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| 5CDS-5UTR | ENST00000437951 | ENST00000486841 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| 5CDS-5UTR | ENST00000468211 | ENST00000375522 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| 5CDS-5UTR | ENST00000468211 | ENST00000486841 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| 5UTR-3CDS | ENST00000418258 | ENST00000287996 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| 5UTR-3CDS | ENST00000421141 | ENST00000287996 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| 5UTR-3CDS | ENST00000429896 | ENST00000287996 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| 5UTR-5UTR | ENST00000418258 | ENST00000375522 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| 5UTR-5UTR | ENST00000418258 | ENST00000486841 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| 5UTR-5UTR | ENST00000421141 | ENST00000375522 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| 5UTR-5UTR | ENST00000421141 | ENST00000486841 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| 5UTR-5UTR | ENST00000429896 | ENST00000375522 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| 5UTR-5UTR | ENST00000429896 | ENST00000486841 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| Frame-shift | ENST00000430669 | ENST00000287996 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| Frame-shift | ENST00000468211 | ENST00000287996 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| In-frame | ENST00000331920 | ENST00000287996 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| In-frame | ENST00000375274 | ENST00000287996 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| In-frame | ENST00000437951 | ENST00000287996 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| intron-3CDS | ENST00000548379 | ENST00000287996 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| intron-5UTR | ENST00000548379 | ENST00000375522 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
| intron-5UTR | ENST00000548379 | ENST00000486841 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000331920 | PTCH1 | chr9 | 98268689 | - | ENST00000287996 | IPPK | chr9 | 95381847 | - | 3648 | 694 | 174 | 779 | 201 |
| ENST00000437951 | PTCH1 | chr9 | 98268689 | - | ENST00000287996 | IPPK | chr9 | 95381847 | - | 3644 | 690 | 547 | 155 | 130 |
| ENST00000375274 | PTCH1 | chr9 | 98268689 | - | ENST00000287996 | IPPK | chr9 | 95381847 | - | 3490 | 536 | 145 | 621 | 158 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000331920 | ENST00000287996 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - | 0.2075116 | 0.79248834 |
| ENST00000437951 | ENST00000287996 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - | 0.08590324 | 0.9140967 |
| ENST00000375274 | ENST00000287996 | PTCH1 | chr9 | 98268689 | - | IPPK | chr9 | 95381847 | - | 0.009071453 | 0.9909286 |
Top |
Fusion Genomic Features for PTCH1-IPPK |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
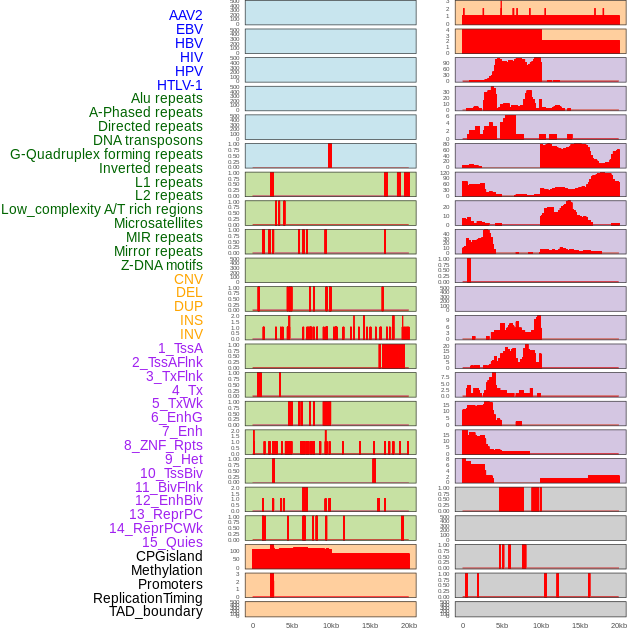 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for PTCH1-IPPK |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:98268689/chr9:95381847) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | IPPK |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Phosphorylates Ins(1,3,4,5,6)P5 at position 2 to form Ins(1,2,3,4,5,6)P6 (InsP6 or phytate). InsP6 is involved in many processes such as mRNA export, non-homologous end-joining, endocytosis, ion channel regulation. It also protects cells from TNF-alpha-induced apoptosis. {ECO:0000269|PubMed:12084730, ECO:0000269|PubMed:15967797}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 14_31 | 131 | 2534.3333333333335 | Compositional bias | Note=Gly-rich |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 14_31 | 130 | 1526.3333333333333 | Compositional bias | Note=Gly-rich |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 14_31 | 65 | 1382.0 | Compositional bias | Note=Gly-rich |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 14_31 | 65 | 2468.3333333333335 | Compositional bias | Note=Gly-rich |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 14_31 | 65 | 184.0 | Compositional bias | Note=Gly-rich |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 1_100 | 131 | 2534.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 1_100 | 130 | 1526.3333333333333 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 101_121 | 131 | 2534.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 101_121 | 130 | 1526.3333333333333 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 14_31 | 0 | 2461.3333333333335 | Compositional bias | Note=Gly-rich |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 14_31 | 0 | 2513.3333333333335 | Compositional bias | Note=Gly-rich |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 14_31 | 0 | 2093.3333333333335 | Compositional bias | Note=Gly-rich |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 438_598 | 131 | 2534.3333333333335 | Domain | SSD |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 438_598 | 130 | 1526.3333333333333 | Domain | SSD |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 438_598 | 0 | 2461.3333333333335 | Domain | SSD |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 438_598 | 0 | 2513.3333333333335 | Domain | SSD |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 438_598 | 0 | 2093.3333333333335 | Domain | SSD |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 438_598 | 65 | 1382.0 | Domain | SSD |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 438_598 | 65 | 2468.3333333333335 | Domain | SSD |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 438_598 | 65 | 184.0 | Domain | SSD |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 1049_1055 | 131 | 2534.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 1077_1083 | 131 | 2534.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 1105_1121 | 131 | 2534.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 1142_1154 | 131 | 2534.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 1176_1447 | 131 | 2534.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 122_436 | 131 | 2534.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 458_472 | 131 | 2534.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 494_501 | 131 | 2534.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 523_547 | 131 | 2534.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 569_577 | 131 | 2534.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 599_748 | 131 | 2534.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 770_1027 | 131 | 2534.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 1049_1055 | 130 | 1526.3333333333333 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 1077_1083 | 130 | 1526.3333333333333 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 1105_1121 | 130 | 1526.3333333333333 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 1142_1154 | 130 | 1526.3333333333333 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 1176_1447 | 130 | 1526.3333333333333 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 122_436 | 130 | 1526.3333333333333 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 458_472 | 130 | 1526.3333333333333 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 494_501 | 130 | 1526.3333333333333 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 523_547 | 130 | 1526.3333333333333 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 569_577 | 130 | 1526.3333333333333 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 599_748 | 130 | 1526.3333333333333 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 770_1027 | 130 | 1526.3333333333333 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 1049_1055 | 0 | 2461.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 1077_1083 | 0 | 2461.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 1105_1121 | 0 | 2461.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 1142_1154 | 0 | 2461.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 1176_1447 | 0 | 2461.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 122_436 | 0 | 2461.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 1_100 | 0 | 2461.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 458_472 | 0 | 2461.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 494_501 | 0 | 2461.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 523_547 | 0 | 2461.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 569_577 | 0 | 2461.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 599_748 | 0 | 2461.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 770_1027 | 0 | 2461.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 1049_1055 | 0 | 2513.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 1077_1083 | 0 | 2513.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 1105_1121 | 0 | 2513.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 1142_1154 | 0 | 2513.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 1176_1447 | 0 | 2513.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 122_436 | 0 | 2513.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 1_100 | 0 | 2513.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 458_472 | 0 | 2513.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 494_501 | 0 | 2513.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 523_547 | 0 | 2513.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 569_577 | 0 | 2513.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 599_748 | 0 | 2513.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 770_1027 | 0 | 2513.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 1049_1055 | 0 | 2093.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 1077_1083 | 0 | 2093.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 1105_1121 | 0 | 2093.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 1142_1154 | 0 | 2093.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 1176_1447 | 0 | 2093.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 122_436 | 0 | 2093.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 1_100 | 0 | 2093.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 458_472 | 0 | 2093.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 494_501 | 0 | 2093.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 523_547 | 0 | 2093.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 569_577 | 0 | 2093.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 599_748 | 0 | 2093.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 770_1027 | 0 | 2093.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 1049_1055 | 65 | 1382.0 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 1077_1083 | 65 | 1382.0 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 1105_1121 | 65 | 1382.0 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 1142_1154 | 65 | 1382.0 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 1176_1447 | 65 | 1382.0 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 122_436 | 65 | 1382.0 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 1_100 | 65 | 1382.0 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 458_472 | 65 | 1382.0 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 494_501 | 65 | 1382.0 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 523_547 | 65 | 1382.0 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 569_577 | 65 | 1382.0 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 599_748 | 65 | 1382.0 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 770_1027 | 65 | 1382.0 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 1049_1055 | 65 | 2468.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 1077_1083 | 65 | 2468.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 1105_1121 | 65 | 2468.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 1142_1154 | 65 | 2468.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 1176_1447 | 65 | 2468.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 122_436 | 65 | 2468.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 1_100 | 65 | 2468.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 458_472 | 65 | 2468.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 494_501 | 65 | 2468.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 523_547 | 65 | 2468.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 569_577 | 65 | 2468.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 599_748 | 65 | 2468.3333333333335 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 770_1027 | 65 | 2468.3333333333335 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 1049_1055 | 65 | 184.0 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 1077_1083 | 65 | 184.0 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 1105_1121 | 65 | 184.0 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 1142_1154 | 65 | 184.0 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 1176_1447 | 65 | 184.0 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 122_436 | 65 | 184.0 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 1_100 | 65 | 184.0 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 458_472 | 65 | 184.0 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 494_501 | 65 | 184.0 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 523_547 | 65 | 184.0 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 569_577 | 65 | 184.0 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 599_748 | 65 | 184.0 | Topological domain | Cytoplasmic |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 770_1027 | 65 | 184.0 | Topological domain | Extracellular |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 1028_1048 | 131 | 2534.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 1056_1076 | 131 | 2534.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 1084_1104 | 131 | 2534.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 1122_1141 | 131 | 2534.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 1155_1175 | 131 | 2534.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 437_457 | 131 | 2534.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 473_493 | 131 | 2534.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 502_522 | 131 | 2534.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 548_568 | 131 | 2534.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 578_598 | 131 | 2534.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000331920 | - | 2 | 24 | 749_769 | 131 | 2534.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 1028_1048 | 130 | 1526.3333333333333 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 1056_1076 | 130 | 1526.3333333333333 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 1084_1104 | 130 | 1526.3333333333333 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 1122_1141 | 130 | 1526.3333333333333 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 1155_1175 | 130 | 1526.3333333333333 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 437_457 | 130 | 1526.3333333333333 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 473_493 | 130 | 1526.3333333333333 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 502_522 | 130 | 1526.3333333333333 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 548_568 | 130 | 1526.3333333333333 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 578_598 | 130 | 1526.3333333333333 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000375274 | - | 2 | 24 | 749_769 | 130 | 1526.3333333333333 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 101_121 | 0 | 2461.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 1028_1048 | 0 | 2461.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 1056_1076 | 0 | 2461.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 1084_1104 | 0 | 2461.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 1122_1141 | 0 | 2461.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 1155_1175 | 0 | 2461.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 437_457 | 0 | 2461.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 473_493 | 0 | 2461.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 502_522 | 0 | 2461.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 548_568 | 0 | 2461.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 578_598 | 0 | 2461.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000418258 | - | 2 | 24 | 749_769 | 0 | 2461.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 101_121 | 0 | 2513.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 1028_1048 | 0 | 2513.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 1056_1076 | 0 | 2513.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 1084_1104 | 0 | 2513.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 1122_1141 | 0 | 2513.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 1155_1175 | 0 | 2513.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 437_457 | 0 | 2513.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 473_493 | 0 | 2513.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 502_522 | 0 | 2513.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 548_568 | 0 | 2513.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 578_598 | 0 | 2513.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000421141 | - | 2 | 24 | 749_769 | 0 | 2513.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 101_121 | 0 | 2093.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 1028_1048 | 0 | 2093.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 1056_1076 | 0 | 2093.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 1084_1104 | 0 | 2093.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 1122_1141 | 0 | 2093.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 1155_1175 | 0 | 2093.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 437_457 | 0 | 2093.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 473_493 | 0 | 2093.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 502_522 | 0 | 2093.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 548_568 | 0 | 2093.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 578_598 | 0 | 2093.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000429896 | - | 2 | 24 | 749_769 | 0 | 2093.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 101_121 | 65 | 1382.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 1028_1048 | 65 | 1382.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 1056_1076 | 65 | 1382.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 1084_1104 | 65 | 1382.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 1122_1141 | 65 | 1382.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 1155_1175 | 65 | 1382.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 437_457 | 65 | 1382.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 473_493 | 65 | 1382.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 502_522 | 65 | 1382.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 548_568 | 65 | 1382.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 578_598 | 65 | 1382.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000430669 | - | 2 | 23 | 749_769 | 65 | 1382.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 101_121 | 65 | 2468.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 1028_1048 | 65 | 2468.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 1056_1076 | 65 | 2468.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 1084_1104 | 65 | 2468.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 1122_1141 | 65 | 2468.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 1155_1175 | 65 | 2468.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 437_457 | 65 | 2468.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 473_493 | 65 | 2468.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 502_522 | 65 | 2468.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 548_568 | 65 | 2468.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 578_598 | 65 | 2468.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000437951 | - | 2 | 24 | 749_769 | 65 | 2468.3333333333335 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 101_121 | 65 | 184.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 1028_1048 | 65 | 184.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 1056_1076 | 65 | 184.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 1084_1104 | 65 | 184.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 1122_1141 | 65 | 184.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 1155_1175 | 65 | 184.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 437_457 | 65 | 184.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 473_493 | 65 | 184.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 502_522 | 65 | 184.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 548_568 | 65 | 184.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 578_598 | 65 | 184.0 | Transmembrane | Helical |
| Hgene | PTCH1 | chr9:98268689 | chr9:95381847 | ENST00000468211 | - | 2 | 5 | 749_769 | 65 | 184.0 | Transmembrane | Helical |
| Tgene | IPPK | chr9:98268689 | chr9:95381847 | ENST00000287996 | 10 | 13 | 136_140 | 390 | 492.0 | Motif | Note=EXKPK motif |
Top |
Fusion Gene Sequence for PTCH1-IPPK |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >69866_69866_1_PTCH1-IPPK_PTCH1_chr9_98268689_ENST00000331920_IPPK_chr9_95381847_ENST00000287996_length(transcript)=3648nt_BP=694nt GAACTGGATGTGGGCAGCGGCGGCCGCAGAGACCTCGGGACCCCCGCGCAATGTGGCAATGGAAGGCGCAGGGTCTGACTCCCCGGCAGC GGCCGCGGCCGCAGCGGCAGCAGCGCCCGCCGTGTGAGCAGCAGCAGCGGCTGGTCTGTCAACCGGAGCCCGAGCCCGAGCAGCCTGCGG CCAGCAGCGTCCTCGCAAGCCGAGCGCCCAGGCGCGCCAGGAGCCCGCAGCAGCGGCAGCAGCGCGCCGGGCCGCCCGGGAAGCCTCCGT CCCCGCGGCGGCGGCGGCGGCGGCGGCAACATGGCCTCGGCTGGTAACGCCGCCGAGCCCCAGGACCGCGGCGGCGGCGGCAGCGGCTGT ATCGGTGCCCCGGGACGGCCGGCTGGAGGCGGGAGGCGCAGACGGACGGGGGGGCTGCGCCGTGCTGCCGCGCCGGACCGGGACTATCTG CACCGGCCCAGCTACTGCGACGCCGCCTTCGCTCTGGAGCAGATTTCCAAGGGGAAGGCTACTGGCCGGAAAGCGCCGCTGTGGCTGAGA GCGAAGTTTCAGAGACTCTTATTTAAACTGGGTTGTTACATTCAAAAAAACTGCGGCAAGTTCTTGGTTGTGGGCCTCCTCATATTTGGG GCCTTCGCGGTGGGATTAAAAGCAGCGAACCTCGAGACCAACGTGGAGGAGCTGTGGGTGGAAGGTGCAGCAGTACCGCGTCGCCATGAC TGCCAAGGACTGCTCCATCATGATTGCACTGTCTCCCTGTCTGCAGGATGCCAGCTCTGATCAAAGGCCTGTCGTCCCTTCATCGAGGTC CAGGTTTGCCTTTTCCGTGTCTGTGCTGGACCTTGACCTCAAGCCCTACGAGAGCATTCCCCATCAGTATAAACTGGACGGCAAGATCGT CAACTATTATTCAAAGACTGTACGTGCCAAAGACAACGCCGTGATGTCGACTCGGTTCAAGGAAAGCGAAGATTGCACATTAGTTCTCCA CAAGGTCTAACTCTTTCCCTGCAGTGTCTTTGAAACTTGAACATAATGTGAAGGCTGAATGATAGAGATATTTTCTGTTGTGTTGGGTGA CCTTTGGTTGTGAATGTTTTTGCTTTTAACCCCTTTTGAGGTGGGATTGCCTCTTGGAGACATGGAATTGAAGAGCACTAGAAACAACTT CCTGGACAAGGAATGTAGGAAGTGAGTGCTGTGTCCCAGGAAGCTGCTCACACTCTTAAAATGGAAGTGTCCGTTAAGCCCTGGGAAGAC GTTCTGGATAGTTCTTCTTTCCCAACCAGGGCTCATGTCTGATTCTCTAATGCGAAAAGCCTTATTCTAAGACCCAAGGTTTGGATCTGC TACCACCAGACTCCTAACATAGAAAACTTGAATTGTCACATACATTTTACAGTTTGGACTTTTAAGAAAACATGGATACTACTGGAACTT CCCCCAGCTGAGTTACATGGTCACTTTTTCAGTGCAAGCCACATATCAACACAGGTTTTTAGGTGGTTCCCTGCTGCACAGCGTGACCTC GTGGTCGCGCAGATGCCGATTCTGAGCAGTGTAGACTCAGGGTCCCTGTGGAGGTGCTGAAGCCCACAGTTCCACACAGGGCAGCAGGGA GCGCAAGGCTCAGGTCAGGATGGACACCACTCATGCGAGCATTGACCTTTTTTTTTTTGCTTTACAATTTAATCTTTCACATGAGGGGGA GATGAGCTGTTCTTTCCCACTAAAAATTGCTGCTGTAGAAGTTGGAGGTGGAGCTGTGACTGGCTGAGCTGCTGTGGCCGGGCTGGGCAG TGTGCCCCAACAGCTCAGTGCTTTCCTGACACTCCAGTGTCTGGGGTGGTTGAGGAGCGAGTTCTCTCTTCCTCCCAGACCAAGTTCCTC CCTCGGGTTTGCCTTGAGACGTGTTGCGTTTTTGGGCCCCGTGGCCTCTCCCTGTTAGGCTGCCACAGGCCCTGCTTCTGGAAGGTGAAC AGCTCCTGGCTGCTGCCGAGAGGGTTCTCGTTGGGGTCACCAAAGTGTGCCCGGCTGCTATGAAAAACGTTGGGAATCTTGGTTTCAGTT TTTTATTCTATGCTAGGTTGTACAGACTTATTTATATCATCGTTTTGAGGGACTAATGGAGGCTTATTGTAACATATAATATTAGTGAAA CCATGGAATTATATGAAAATGATACATGAGAAATAAGGAAACTATTTTGCTGATTGTAAATTTTTGTGGGAAATTTTGTGATAACTTGAG AATTATACTTGTTTGAATCAAGGCAACTCTTCTAGAATTTATTGTTCAATTCTGTCATATTTACTTCTAATTCTACTCTCAGAGGTGCAA AAAGATACTTATCTAACTGGACACTTTTACTGTTTTCTTCTCACTTCCAATGACACCGTCTCATACACACGGGTCCTGTGTGCTGGGATG GGGGCTTTGGTGAAGTGCACGGAGCTCACGGACAAGCCGCAGGCTCTGGGAAGCGTGGAGGTAGTGGCCGTTGAGTACTGTCTTGACCCA AGGGAGTTCACTGTTGCTGGGCTGGGGCTGGGGCAGCCTGGTTTGGGATGATTTTGCCCATTGTCACGGTGTGTGGAGGATGCTGGGTCC TGGGTTCAAGTGCACATGTGAGGGAACCCCAGTCCGTGGGAAGGTGGGCTGGGCTCCCTAGGTCCTTCAGGGCTGCAAGAGTTATTGCCT AGCACAGGGCCCAGACAGGGCTGGACTAGACACGATGAAAACAGCCTGTATTTGAGGGAGAAGAGAAAACAGATGGAGACCCCCACTCTG TCTGGTCTTGGCTCCCACCAGCCCGGGCTGTGAGGAGCCCCTGAGCCGCGGGTGCCACTGCTGTGGATGCGTTTCTGGCCCGTCTTGGAA ACTGCTGCTGCTGCGTAGTCATGACGGCCCTGACTGGTTTGAGACAACACGCGTTCACAGATCTTAACAATGCTAAATGGGCGACTGATG AGGTCACGGTTCTTAAGGCACATCCGGGACGATCGATATAGCACTTAGGCTATGCAGGGGGGTCTTTCTTTCATAAAGGACCTCATTCCT TGCGTCCCAGCCCAGGAACAGTGCTGTCTTTGCTGTCTTAGGCCATTCTTGTTATTAAGCTGACAGCTTCACTTTCTTGTGGAAGACCAA GAACAGCATGAGGACTTGCTGAGACTGCCTCACGGTGCCACCATGCGGACCGGGGGGAGGCAGGGAGTGAGGGTGTGGCTCCCCTGGGCT ACTAAGGACAGTTTAAAGATGAGGCGCCATAGGCATCCGTCTCCAGAGCCCCCAGGAGGCAGGAACCAGGATAAGGGTGGCTTCTCCCAG CTGAGTAATGTGGCCACCATGTCCAAGGTGGACTCGGTGTGAGGCTGCAGTGGGTGGGGGATGGATCCTCCCCACACATCTGAGGACACA CAACCACTTGGTACATTTTCTCAGATGTTAAGCTTATTTTTATAAACCACATTGTACGTGTCACTTTTATTTGGCATTTTATACTTAACT AGTTGTATTTATTTTTCATGTCATGAGAGTTTCCATATAATGTGCCATAACTGCATAGACAGCACGTCTTCAGGGTGTGGTTTCTAAGCA >69866_69866_1_PTCH1-IPPK_PTCH1_chr9_98268689_ENST00000331920_IPPK_chr9_95381847_ENST00000287996_length(amino acids)=201AA_BP=173 MRPAASSQAERPGAPGARSSGSSAPGRPGSLRPRGGGGGGGNMASAGNAAEPQDRGGGGSGCIGAPGRPAGGGRRRRTGGLRRAAAPDRD YLHRPSYCDAAFALEQISKGKATGRKAPLWLRAKFQRLLFKLGCYIQKNCGKFLVVGLLIFGAFAVGLKAANLETNVEELWVEGAAVPRR -------------------------------------------------------------- >69866_69866_2_PTCH1-IPPK_PTCH1_chr9_98268689_ENST00000375274_IPPK_chr9_95381847_ENST00000287996_length(transcript)=3490nt_BP=536nt ACCCAGGAGGGCTGTGCTGATAGCACGTTCCTCGAGTTTACTTTGCTTTCCTTGAGTTTATTGTAAAGGGGTAAAGTTTTCGGATCCGTC ACGTGACCCTGACAGGTCCTGCCTATGGCGCGGCAGACCACCCACGCCGAGGGCCATGGAACTGCTTAATAGAAACAGGCTTGTAATTGT GAGTCCGCGCTGCACTCCGCCGAAAGCCTCCGGCGGCCCAGCGCGCCGGGGTTTTTACACTTTCCGTTCCTTTTGTAAAGACGGAGGAGG AGGAGAAGAAGAAGAAGAAAACGGAGGAGAAGAAAAAGACGACAGGGGAGACAAAGAGACCCGCAGCGACAAGGGGAAGGCTACTGGCCG GAAAGCGCCGCTGTGGCTGAGAGCGAAGTTTCAGAGACTCTTATTTAAACTGGGTTGTTACATTCAAAAAAACTGCGGCAAGTTCTTGGT TGTGGGCCTCCTCATATTTGGGGCCTTCGCGGTGGGATTAAAAGCAGCGAACCTCGAGACCAACGTGGAGGAGCTGTGGGTGGAAGGTGC AGCAGTACCGCGTCGCCATGACTGCCAAGGACTGCTCCATCATGATTGCACTGTCTCCCTGTCTGCAGGATGCCAGCTCTGATCAAAGGC CTGTCGTCCCTTCATCGAGGTCCAGGTTTGCCTTTTCCGTGTCTGTGCTGGACCTTGACCTCAAGCCCTACGAGAGCATTCCCCATCAGT ATAAACTGGACGGCAAGATCGTCAACTATTATTCAAAGACTGTACGTGCCAAAGACAACGCCGTGATGTCGACTCGGTTCAAGGAAAGCG AAGATTGCACATTAGTTCTCCACAAGGTCTAACTCTTTCCCTGCAGTGTCTTTGAAACTTGAACATAATGTGAAGGCTGAATGATAGAGA TATTTTCTGTTGTGTTGGGTGACCTTTGGTTGTGAATGTTTTTGCTTTTAACCCCTTTTGAGGTGGGATTGCCTCTTGGAGACATGGAAT TGAAGAGCACTAGAAACAACTTCCTGGACAAGGAATGTAGGAAGTGAGTGCTGTGTCCCAGGAAGCTGCTCACACTCTTAAAATGGAAGT GTCCGTTAAGCCCTGGGAAGACGTTCTGGATAGTTCTTCTTTCCCAACCAGGGCTCATGTCTGATTCTCTAATGCGAAAAGCCTTATTCT AAGACCCAAGGTTTGGATCTGCTACCACCAGACTCCTAACATAGAAAACTTGAATTGTCACATACATTTTACAGTTTGGACTTTTAAGAA AACATGGATACTACTGGAACTTCCCCCAGCTGAGTTACATGGTCACTTTTTCAGTGCAAGCCACATATCAACACAGGTTTTTAGGTGGTT CCCTGCTGCACAGCGTGACCTCGTGGTCGCGCAGATGCCGATTCTGAGCAGTGTAGACTCAGGGTCCCTGTGGAGGTGCTGAAGCCCACA GTTCCACACAGGGCAGCAGGGAGCGCAAGGCTCAGGTCAGGATGGACACCACTCATGCGAGCATTGACCTTTTTTTTTTTGCTTTACAAT TTAATCTTTCACATGAGGGGGAGATGAGCTGTTCTTTCCCACTAAAAATTGCTGCTGTAGAAGTTGGAGGTGGAGCTGTGACTGGCTGAG CTGCTGTGGCCGGGCTGGGCAGTGTGCCCCAACAGCTCAGTGCTTTCCTGACACTCCAGTGTCTGGGGTGGTTGAGGAGCGAGTTCTCTC TTCCTCCCAGACCAAGTTCCTCCCTCGGGTTTGCCTTGAGACGTGTTGCGTTTTTGGGCCCCGTGGCCTCTCCCTGTTAGGCTGCCACAG GCCCTGCTTCTGGAAGGTGAACAGCTCCTGGCTGCTGCCGAGAGGGTTCTCGTTGGGGTCACCAAAGTGTGCCCGGCTGCTATGAAAAAC GTTGGGAATCTTGGTTTCAGTTTTTTATTCTATGCTAGGTTGTACAGACTTATTTATATCATCGTTTTGAGGGACTAATGGAGGCTTATT GTAACATATAATATTAGTGAAACCATGGAATTATATGAAAATGATACATGAGAAATAAGGAAACTATTTTGCTGATTGTAAATTTTTGTG GGAAATTTTGTGATAACTTGAGAATTATACTTGTTTGAATCAAGGCAACTCTTCTAGAATTTATTGTTCAATTCTGTCATATTTACTTCT AATTCTACTCTCAGAGGTGCAAAAAGATACTTATCTAACTGGACACTTTTACTGTTTTCTTCTCACTTCCAATGACACCGTCTCATACAC ACGGGTCCTGTGTGCTGGGATGGGGGCTTTGGTGAAGTGCACGGAGCTCACGGACAAGCCGCAGGCTCTGGGAAGCGTGGAGGTAGTGGC CGTTGAGTACTGTCTTGACCCAAGGGAGTTCACTGTTGCTGGGCTGGGGCTGGGGCAGCCTGGTTTGGGATGATTTTGCCCATTGTCACG GTGTGTGGAGGATGCTGGGTCCTGGGTTCAAGTGCACATGTGAGGGAACCCCAGTCCGTGGGAAGGTGGGCTGGGCTCCCTAGGTCCTTC AGGGCTGCAAGAGTTATTGCCTAGCACAGGGCCCAGACAGGGCTGGACTAGACACGATGAAAACAGCCTGTATTTGAGGGAGAAGAGAAA ACAGATGGAGACCCCCACTCTGTCTGGTCTTGGCTCCCACCAGCCCGGGCTGTGAGGAGCCCCTGAGCCGCGGGTGCCACTGCTGTGGAT GCGTTTCTGGCCCGTCTTGGAAACTGCTGCTGCTGCGTAGTCATGACGGCCCTGACTGGTTTGAGACAACACGCGTTCACAGATCTTAAC AATGCTAAATGGGCGACTGATGAGGTCACGGTTCTTAAGGCACATCCGGGACGATCGATATAGCACTTAGGCTATGCAGGGGGGTCTTTC TTTCATAAAGGACCTCATTCCTTGCGTCCCAGCCCAGGAACAGTGCTGTCTTTGCTGTCTTAGGCCATTCTTGTTATTAAGCTGACAGCT TCACTTTCTTGTGGAAGACCAAGAACAGCATGAGGACTTGCTGAGACTGCCTCACGGTGCCACCATGCGGACCGGGGGGAGGCAGGGAGT GAGGGTGTGGCTCCCCTGGGCTACTAAGGACAGTTTAAAGATGAGGCGCCATAGGCATCCGTCTCCAGAGCCCCCAGGAGGCAGGAACCA GGATAAGGGTGGCTTCTCCCAGCTGAGTAATGTGGCCACCATGTCCAAGGTGGACTCGGTGTGAGGCTGCAGTGGGTGGGGGATGGATCC TCCCCACACATCTGAGGACACACAACCACTTGGTACATTTTCTCAGATGTTAAGCTTATTTTTATAAACCACATTGTACGTGTCACTTTT ATTTGGCATTTTATACTTAACTAGTTGTATTTATTTTTCATGTCATGAGAGTTTCCATATAATGTGCCATAACTGCATAGACAGCACGTC >69866_69866_2_PTCH1-IPPK_PTCH1_chr9_98268689_ENST00000375274_IPPK_chr9_95381847_ENST00000287996_length(amino acids)=158AA_BP=130 MELLNRNRLVIVSPRCTPPKASGGPARRGFYTFRSFCKDGGGGEEEEENGGEEKDDRGDKETRSDKGKATGRKAPLWLRAKFQRLLFKLG -------------------------------------------------------------- >69866_69866_3_PTCH1-IPPK_PTCH1_chr9_98268689_ENST00000437951_IPPK_chr9_95381847_ENST00000287996_length(transcript)=3644nt_BP=690nt ACCCAGGAGGGCTGTGCTGATAGCACGTTCCTCGAGTTTACTTTGCTTTCCTTGAGTTTATTGTAAAGGGGTAAAGTTTTCGGATCCGTC ACGTGACCCTGACAGGTCCTGCCTATGGCGCGGCAGACCACCCACGCCGAGGGCCATGGAACTGCTTAATAGAAACAGGCTTGTAATTGT GAGTCCGCGCTGCACTCCGCCGAAAGCCTCCGGCGGCCCAGCGCGCCGGGGTTTTTACACTTTCCGTTCCTTTTGTAAAGACGGAGGAGG AGGAGAAGAAGAAGAAGAAAACGGAGGAGAAGAAAAAGACGACAGGGGAGACAAAGAGACCCGCAGCGACAAGGCAAGGGGGAGACGAGG GAAGACTGGGAGAAGACGGAGGAGCGGAGGACGAGGAAAGGGGGGCCAGGGAAAAAAAAGGAATTGATGTGAAATCCAAGCCCAGCGTCC GCGCCATCGGCACCCGCGCTCCGAGCAGGGGTTGACGGCCGGCTATGGGGAAGGCTACTGGCCGGAAAGCGCCGCTGTGGCTGAGAGCGA AGTTTCAGAGACTCTTATTTAAACTGGGTTGTTACATTCAAAAAAACTGCGGCAAGTTCTTGGTTGTGGGCCTCCTCATATTTGGGGCCT TCGCGGTGGGATTAAAAGCAGCGAACCTCGAGACCAACGTGGAGGAGCTGTGGGTGGAAGGTGCAGCAGTACCGCGTCGCCATGACTGCC AAGGACTGCTCCATCATGATTGCACTGTCTCCCTGTCTGCAGGATGCCAGCTCTGATCAAAGGCCTGTCGTCCCTTCATCGAGGTCCAGG TTTGCCTTTTCCGTGTCTGTGCTGGACCTTGACCTCAAGCCCTACGAGAGCATTCCCCATCAGTATAAACTGGACGGCAAGATCGTCAAC TATTATTCAAAGACTGTACGTGCCAAAGACAACGCCGTGATGTCGACTCGGTTCAAGGAAAGCGAAGATTGCACATTAGTTCTCCACAAG GTCTAACTCTTTCCCTGCAGTGTCTTTGAAACTTGAACATAATGTGAAGGCTGAATGATAGAGATATTTTCTGTTGTGTTGGGTGACCTT TGGTTGTGAATGTTTTTGCTTTTAACCCCTTTTGAGGTGGGATTGCCTCTTGGAGACATGGAATTGAAGAGCACTAGAAACAACTTCCTG GACAAGGAATGTAGGAAGTGAGTGCTGTGTCCCAGGAAGCTGCTCACACTCTTAAAATGGAAGTGTCCGTTAAGCCCTGGGAAGACGTTC TGGATAGTTCTTCTTTCCCAACCAGGGCTCATGTCTGATTCTCTAATGCGAAAAGCCTTATTCTAAGACCCAAGGTTTGGATCTGCTACC ACCAGACTCCTAACATAGAAAACTTGAATTGTCACATACATTTTACAGTTTGGACTTTTAAGAAAACATGGATACTACTGGAACTTCCCC CAGCTGAGTTACATGGTCACTTTTTCAGTGCAAGCCACATATCAACACAGGTTTTTAGGTGGTTCCCTGCTGCACAGCGTGACCTCGTGG TCGCGCAGATGCCGATTCTGAGCAGTGTAGACTCAGGGTCCCTGTGGAGGTGCTGAAGCCCACAGTTCCACACAGGGCAGCAGGGAGCGC AAGGCTCAGGTCAGGATGGACACCACTCATGCGAGCATTGACCTTTTTTTTTTTGCTTTACAATTTAATCTTTCACATGAGGGGGAGATG AGCTGTTCTTTCCCACTAAAAATTGCTGCTGTAGAAGTTGGAGGTGGAGCTGTGACTGGCTGAGCTGCTGTGGCCGGGCTGGGCAGTGTG CCCCAACAGCTCAGTGCTTTCCTGACACTCCAGTGTCTGGGGTGGTTGAGGAGCGAGTTCTCTCTTCCTCCCAGACCAAGTTCCTCCCTC GGGTTTGCCTTGAGACGTGTTGCGTTTTTGGGCCCCGTGGCCTCTCCCTGTTAGGCTGCCACAGGCCCTGCTTCTGGAAGGTGAACAGCT CCTGGCTGCTGCCGAGAGGGTTCTCGTTGGGGTCACCAAAGTGTGCCCGGCTGCTATGAAAAACGTTGGGAATCTTGGTTTCAGTTTTTT ATTCTATGCTAGGTTGTACAGACTTATTTATATCATCGTTTTGAGGGACTAATGGAGGCTTATTGTAACATATAATATTAGTGAAACCAT GGAATTATATGAAAATGATACATGAGAAATAAGGAAACTATTTTGCTGATTGTAAATTTTTGTGGGAAATTTTGTGATAACTTGAGAATT ATACTTGTTTGAATCAAGGCAACTCTTCTAGAATTTATTGTTCAATTCTGTCATATTTACTTCTAATTCTACTCTCAGAGGTGCAAAAAG ATACTTATCTAACTGGACACTTTTACTGTTTTCTTCTCACTTCCAATGACACCGTCTCATACACACGGGTCCTGTGTGCTGGGATGGGGG CTTTGGTGAAGTGCACGGAGCTCACGGACAAGCCGCAGGCTCTGGGAAGCGTGGAGGTAGTGGCCGTTGAGTACTGTCTTGACCCAAGGG AGTTCACTGTTGCTGGGCTGGGGCTGGGGCAGCCTGGTTTGGGATGATTTTGCCCATTGTCACGGTGTGTGGAGGATGCTGGGTCCTGGG TTCAAGTGCACATGTGAGGGAACCCCAGTCCGTGGGAAGGTGGGCTGGGCTCCCTAGGTCCTTCAGGGCTGCAAGAGTTATTGCCTAGCA CAGGGCCCAGACAGGGCTGGACTAGACACGATGAAAACAGCCTGTATTTGAGGGAGAAGAGAAAACAGATGGAGACCCCCACTCTGTCTG GTCTTGGCTCCCACCAGCCCGGGCTGTGAGGAGCCCCTGAGCCGCGGGTGCCACTGCTGTGGATGCGTTTCTGGCCCGTCTTGGAAACTG CTGCTGCTGCGTAGTCATGACGGCCCTGACTGGTTTGAGACAACACGCGTTCACAGATCTTAACAATGCTAAATGGGCGACTGATGAGGT CACGGTTCTTAAGGCACATCCGGGACGATCGATATAGCACTTAGGCTATGCAGGGGGGTCTTTCTTTCATAAAGGACCTCATTCCTTGCG TCCCAGCCCAGGAACAGTGCTGTCTTTGCTGTCTTAGGCCATTCTTGTTATTAAGCTGACAGCTTCACTTTCTTGTGGAAGACCAAGAAC AGCATGAGGACTTGCTGAGACTGCCTCACGGTGCCACCATGCGGACCGGGGGGAGGCAGGGAGTGAGGGTGTGGCTCCCCTGGGCTACTA AGGACAGTTTAAAGATGAGGCGCCATAGGCATCCGTCTCCAGAGCCCCCAGGAGGCAGGAACCAGGATAAGGGTGGCTTCTCCCAGCTGA GTAATGTGGCCACCATGTCCAAGGTGGACTCGGTGTGAGGCTGCAGTGGGTGGGGGATGGATCCTCCCCACACATCTGAGGACACACAAC CACTTGGTACATTTTCTCAGATGTTAAGCTTATTTTTATAAACCACATTGTACGTGTCACTTTTATTTGGCATTTTATACTTAACTAGTT GTATTTATTTTTCATGTCATGAGAGTTTCCATATAATGTGCCATAACTGCATAGACAGCACGTCTTCAGGGTGTGGTTTCTAAGCAGAAA >69866_69866_3_PTCH1-IPPK_PTCH1_chr9_98268689_ENST00000437951_IPPK_chr9_95381847_ENST00000287996_length(amino acids)=130AA_BP= MKLRSQPQRRFPASSLPHSRPSTPARSAGADGADAGLGFHINSFFFPGPPFLVLRSSVFSQSSLVSPLPCRCGSLCLPCRLFLLLRFLLL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for PTCH1-IPPK |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for PTCH1-IPPK |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for PTCH1-IPPK |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
