
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:PTK2-AGO2 (FusionGDB2 ID:70019) |
Fusion Gene Summary for PTK2-AGO2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: PTK2-AGO2 | Fusion gene ID: 70019 | Hgene | Tgene | Gene symbol | PTK2 | AGO2 | Gene ID | 5747 | 27161 |
| Gene name | protein tyrosine kinase 2 | argonaute RISC catalytic component 2 | |
| Synonyms | FADK|FAK|FAK1|FRNK|PPP1R71|p125FAK|pp125FAK | CASC7|EIF2C2|LINC00980|PPD|Q10 | |
| Cytomap | 8q24.3 | 8q24.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | focal adhesion kinase 1FADK 1FAK-related non-kinase polypeptidePTK2 protein tyrosine kinase 2focal adhesion kinase isoform FAK-Del33focal adhesion kinase-related nonkinaseprotein phosphatase 1 regulatory subunit 71 | protein argonaute-2PAZ Piwi domain proteinargonaute 2, RISC catalytic componentcancer susceptibility candidate 7cancer susceptibility candidate 7 (non-protein coding)eukaryotic translation initiation factor 2C, 2long intergenic non-protein coding RN | |
| Modification date | 20200327 | 20200327 | |
| UniProtAcc | . | Q9UKV8 | |
| Ensembl transtripts involved in fusion gene | ENST00000340930, ENST00000395218, ENST00000517887, ENST00000519419, ENST00000521059, ENST00000522684, ENST00000535192, ENST00000519881, ENST00000430260, ENST00000517712, ENST00000519465, ENST00000519635, ENST00000520151, ENST00000520892, ENST00000522950, ENST00000538769, | ENST00000517293, ENST00000519980, ENST00000220592, | |
| Fusion gene scores | * DoF score | 39 X 32 X 15=18720 | 5 X 6 X 4=120 |
| # samples | 57 | 6 | |
| ** MAII score | log2(57/18720*10)=-5.03747470541866 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/120*10)=-1 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: PTK2 [Title/Abstract] AND AGO2 [Title/Abstract] AND fusion [Title/Abstract] Germline AGO2 mutations impair RNA interference and human neurological development (pmid: 33199684) | ||
| Most frequent breakpoint | AGO2(141645584)-PTK2(141685598), # samples:3 PTK2(142011224)-AGO2(141595410), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | AGO2-PTK2 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. AGO2-PTK2 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. AGO2-PTK2 seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. PTK2-AGO2 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. PTK2-AGO2 seems lost the major protein functional domain in Hgene partner, which is a kinase due to the frame-shifted ORF. PTK2-AGO2 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | PTK2 | GO:0007179 | transforming growth factor beta receptor signaling pathway | 24036928 |
| Hgene | PTK2 | GO:0007229 | integrin-mediated signaling pathway | 24036928 |
| Hgene | PTK2 | GO:0010763 | positive regulation of fibroblast migration | 26763945 |
| Hgene | PTK2 | GO:0018108 | peptidyl-tyrosine phosphorylation | 10655584|11331870 |
| Hgene | PTK2 | GO:0022408 | negative regulation of cell-cell adhesion | 21703394 |
| Hgene | PTK2 | GO:0030335 | positive regulation of cell migration | 11331870|21703394 |
| Hgene | PTK2 | GO:0033628 | regulation of cell adhesion mediated by integrin | 10655584 |
| Hgene | PTK2 | GO:0046777 | protein autophosphorylation | 10655584|11331870 |
| Hgene | PTK2 | GO:0048013 | ephrin receptor signaling pathway | 10655584 |
| Hgene | PTK2 | GO:0060396 | growth hormone receptor signaling pathway | 10925297 |
| Hgene | PTK2 | GO:0090303 | positive regulation of wound healing | 26763945 |
| Tgene | AGO2 | GO:0010501 | RNA secondary structure unwinding | 19966796 |
| Tgene | AGO2 | GO:0031054 | pre-miRNA processing | 16424907|17671087|18178619|19966796 |
| Tgene | AGO2 | GO:0035087 | siRNA loading onto RISC involved in RNA interference | 19966796 |
| Tgene | AGO2 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA | 19966796|23661684 |
| Tgene | AGO2 | GO:0035278 | miRNA mediated inhibition of translation | 17671087|19801630 |
| Tgene | AGO2 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA | 15260970|17524464 |
| Tgene | AGO2 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA | 18178619|19966796 |
| Tgene | AGO2 | GO:0045766 | positive regulation of angiogenesis | 27208409 |
| Tgene | AGO2 | GO:0045947 | negative regulation of translational initiation | 17524464|19801630 |
| Tgene | AGO2 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA | 15260970 |
| Tgene | AGO2 | GO:1905618 | positive regulation of miRNA mediated inhibition of translation | 23409027 |
 Fusion gene breakpoints across PTK2 (5'-gene) Fusion gene breakpoints across PTK2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
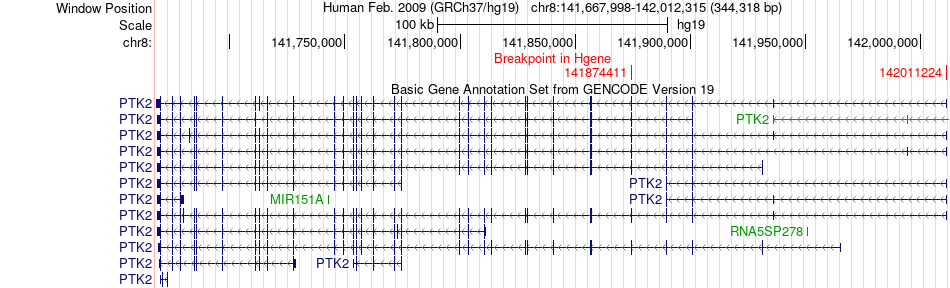 |
 Fusion gene breakpoints across AGO2 (3'-gene) Fusion gene breakpoints across AGO2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
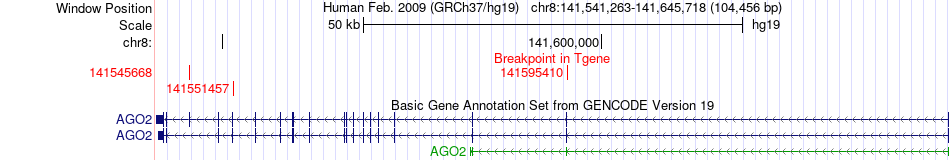 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-A1-A0SN-01A | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| ChimerDB4 | BRCA | TCGA-A2-A3XY-01A | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| ChimerDB4 | BRCA | TCGA-A2-A3XY-01A | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| ChimerDB4 | CESC | TCGA-VS-A94Y-01A | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| ChimerDB4 | STAD | TCGA-HU-A4GH-01A | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
Top |
Fusion Gene ORF analysis for PTK2-AGO2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000340930 | ENST00000517293 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| 5CDS-intron | ENST00000340930 | ENST00000519980 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| 5CDS-intron | ENST00000395218 | ENST00000517293 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| 5CDS-intron | ENST00000395218 | ENST00000519980 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| 5CDS-intron | ENST00000517887 | ENST00000517293 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| 5CDS-intron | ENST00000517887 | ENST00000519980 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| 5CDS-intron | ENST00000519419 | ENST00000517293 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| 5CDS-intron | ENST00000519419 | ENST00000519980 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| 5CDS-intron | ENST00000521059 | ENST00000517293 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| 5CDS-intron | ENST00000521059 | ENST00000519980 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| 5CDS-intron | ENST00000522684 | ENST00000517293 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| 5CDS-intron | ENST00000522684 | ENST00000519980 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| 5CDS-intron | ENST00000535192 | ENST00000517293 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| 5CDS-intron | ENST00000535192 | ENST00000519980 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| 5UTR-3CDS | ENST00000340930 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| 5UTR-3CDS | ENST00000340930 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| 5UTR-3CDS | ENST00000340930 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| 5UTR-3CDS | ENST00000340930 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| 5UTR-3CDS | ENST00000395218 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| 5UTR-3CDS | ENST00000395218 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| 5UTR-3CDS | ENST00000395218 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| 5UTR-3CDS | ENST00000395218 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| 5UTR-3CDS | ENST00000519881 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| 5UTR-3CDS | ENST00000519881 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| 5UTR-3CDS | ENST00000519881 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| 5UTR-3CDS | ENST00000519881 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| 5UTR-3CDS | ENST00000521059 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| 5UTR-3CDS | ENST00000521059 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| 5UTR-3CDS | ENST00000521059 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| 5UTR-3CDS | ENST00000521059 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| 5UTR-3CDS | ENST00000522684 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| 5UTR-3CDS | ENST00000522684 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| 5UTR-3CDS | ENST00000522684 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| 5UTR-3CDS | ENST00000522684 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| 5UTR-5UTR | ENST00000340930 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| 5UTR-5UTR | ENST00000395218 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| 5UTR-5UTR | ENST00000519881 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| 5UTR-5UTR | ENST00000521059 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| 5UTR-5UTR | ENST00000522684 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| 5UTR-intron | ENST00000340930 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| 5UTR-intron | ENST00000395218 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| 5UTR-intron | ENST00000519881 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| 5UTR-intron | ENST00000521059 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| 5UTR-intron | ENST00000522684 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| Frame-shift | ENST00000340930 | ENST00000220592 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| Frame-shift | ENST00000395218 | ENST00000220592 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| Frame-shift | ENST00000519419 | ENST00000220592 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| Frame-shift | ENST00000522684 | ENST00000220592 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| In-frame | ENST00000517887 | ENST00000220592 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| In-frame | ENST00000521059 | ENST00000220592 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| In-frame | ENST00000535192 | ENST00000220592 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-3CDS | ENST00000430260 | ENST00000220592 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-3CDS | ENST00000430260 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000430260 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000430260 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000430260 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000517712 | ENST00000220592 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-3CDS | ENST00000517712 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000517712 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000517712 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000517712 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000517887 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000517887 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000517887 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000517887 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000519419 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000519419 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000519419 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000519419 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000519465 | ENST00000220592 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-3CDS | ENST00000519465 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000519465 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000519465 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000519465 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000519635 | ENST00000220592 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-3CDS | ENST00000519635 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000519635 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000519635 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000519635 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000519881 | ENST00000220592 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-3CDS | ENST00000520151 | ENST00000220592 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-3CDS | ENST00000520151 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000520151 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000520151 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000520151 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000520892 | ENST00000220592 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-3CDS | ENST00000520892 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000520892 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000520892 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000520892 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000522950 | ENST00000220592 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-3CDS | ENST00000522950 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000522950 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000522950 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000522950 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000535192 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000535192 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000535192 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000535192 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000538769 | ENST00000220592 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-3CDS | ENST00000538769 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000538769 | ENST00000220592 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-3CDS | ENST00000538769 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-3CDS | ENST00000538769 | ENST00000519980 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-5UTR | ENST00000430260 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-5UTR | ENST00000517712 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-5UTR | ENST00000517887 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-5UTR | ENST00000519419 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-5UTR | ENST00000519465 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-5UTR | ENST00000519635 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-5UTR | ENST00000520151 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-5UTR | ENST00000520892 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-5UTR | ENST00000522950 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-5UTR | ENST00000535192 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-5UTR | ENST00000538769 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141595410 | - |
| intron-intron | ENST00000430260 | ENST00000517293 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-intron | ENST00000430260 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-intron | ENST00000430260 | ENST00000519980 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-intron | ENST00000517712 | ENST00000517293 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-intron | ENST00000517712 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-intron | ENST00000517712 | ENST00000519980 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-intron | ENST00000517887 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-intron | ENST00000519419 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-intron | ENST00000519465 | ENST00000517293 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-intron | ENST00000519465 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-intron | ENST00000519465 | ENST00000519980 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-intron | ENST00000519635 | ENST00000517293 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-intron | ENST00000519635 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-intron | ENST00000519635 | ENST00000519980 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-intron | ENST00000519881 | ENST00000517293 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-intron | ENST00000519881 | ENST00000519980 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-intron | ENST00000520151 | ENST00000517293 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-intron | ENST00000520151 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-intron | ENST00000520151 | ENST00000519980 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-intron | ENST00000520892 | ENST00000517293 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-intron | ENST00000520892 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-intron | ENST00000520892 | ENST00000519980 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-intron | ENST00000522950 | ENST00000517293 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-intron | ENST00000522950 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-intron | ENST00000522950 | ENST00000519980 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-intron | ENST00000535192 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-intron | ENST00000538769 | ENST00000517293 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
| intron-intron | ENST00000538769 | ENST00000517293 | PTK2 | chr8 | 142011224 | - | AGO2 | chr8 | 141551457 | - |
| intron-intron | ENST00000538769 | ENST00000519980 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000535192 | PTK2 | chr8 | 141874411 | - | ENST00000220592 | AGO2 | chr8 | 141545668 | - | 1771 | 481 | 31 | 891 | 286 |
| ENST00000517887 | PTK2 | chr8 | 141874411 | - | ENST00000220592 | AGO2 | chr8 | 141545668 | - | 2178 | 888 | 264 | 1298 | 344 |
| ENST00000521059 | PTK2 | chr8 | 141874411 | - | ENST00000220592 | AGO2 | chr8 | 141545668 | - | 1922 | 632 | 101 | 1042 | 313 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000535192 | ENST00000220592 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - | 0.004498199 | 0.9955018 |
| ENST00000517887 | ENST00000220592 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - | 0.001420452 | 0.9985795 |
| ENST00000521059 | ENST00000220592 | PTK2 | chr8 | 141874411 | - | AGO2 | chr8 | 141545668 | - | 0.001170414 | 0.99882954 |
Top |
Fusion Genomic Features for PTK2-AGO2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
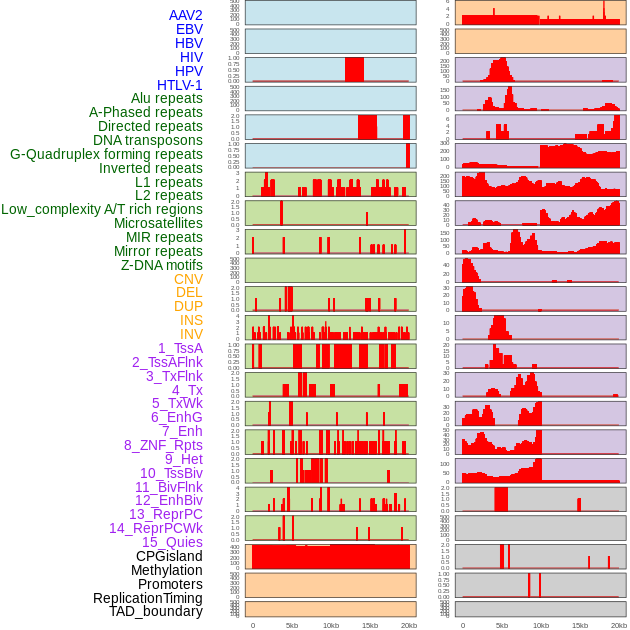 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for PTK2-AGO2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:141645584/chr8:141685598) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | AGO2 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Required for RNA-mediated gene silencing (RNAi) by the RNA-induced silencing complex (RISC). The 'minimal RISC' appears to include AGO2 bound to a short guide RNA such as a microRNA (miRNA) or short interfering RNA (siRNA). These guide RNAs direct RISC to complementary mRNAs that are targets for RISC-mediated gene silencing. The precise mechanism of gene silencing depends on the degree of complementarity between the miRNA or siRNA and its target. Binding of RISC to a perfectly complementary mRNA generally results in silencing due to endonucleolytic cleavage of the mRNA specifically by AGO2. Binding of RISC to a partially complementary mRNA results in silencing through inhibition of translation, and this is independent of endonuclease activity. May inhibit translation initiation by binding to the 7-methylguanosine cap, thereby preventing the recruitment of the translation initiation factor eIF4-E. May also inhibit translation initiation via interaction with EIF6, which itself binds to the 60S ribosomal subunit and prevents its association with the 40S ribosomal subunit. The inhibition of translational initiation leads to the accumulation of the affected mRNA in cytoplasmic processing bodies (P-bodies), where mRNA degradation may subsequently occur. In some cases RISC-mediated translational repression is also observed for miRNAs that perfectly match the 3' untranslated region (3'-UTR). Can also up-regulate the translation of specific mRNAs under certain growth conditions. Binds to the AU element of the 3'-UTR of the TNF (TNF-alpha) mRNA and up-regulates translation under conditions of serum starvation. Also required for transcriptional gene silencing (TGS), in which short RNAs known as antigene RNAs or agRNAs direct the transcriptional repression of complementary promoter regions. {ECO:0000250|UniProtKB:Q8CJG0, ECO:0000255|HAMAP-Rule:MF_03031, ECO:0000269|PubMed:15105377, ECO:0000269|PubMed:15260970, ECO:0000269|PubMed:15284456, ECO:0000269|PubMed:15337849, ECO:0000269|PubMed:15800637, ECO:0000269|PubMed:16081698, ECO:0000269|PubMed:16142218, ECO:0000269|PubMed:16271387, ECO:0000269|PubMed:16289642, ECO:0000269|PubMed:16357216, ECO:0000269|PubMed:16756390, ECO:0000269|PubMed:16936728, ECO:0000269|PubMed:17382880, ECO:0000269|PubMed:17507929, ECO:0000269|PubMed:17524464, ECO:0000269|PubMed:17531811, ECO:0000269|PubMed:17932509, ECO:0000269|PubMed:18048652, ECO:0000269|PubMed:18178619, ECO:0000269|PubMed:18690212, ECO:0000269|PubMed:18771919, ECO:0000269|PubMed:19167051, ECO:0000269|PubMed:23746446}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | AGO2 | chr8:141874411 | chr8:141545668 | ENST00000519980 | 0 | 18 | 235_348 | 0 | 826.0 | Domain | PAZ | |
| Tgene | AGO2 | chr8:141874411 | chr8:141545668 | ENST00000519980 | 0 | 18 | 517_818 | 0 | 826.0 | Domain | Piwi |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000340930 | - | 5 | 33 | 712_733 | 150 | 1066.0 | Compositional bias | Note=Pro-rich |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000340930 | - | 5 | 33 | 863_913 | 150 | 1066.0 | Compositional bias | Note=Pro-rich |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000521059 | - | 5 | 32 | 712_733 | 150 | 1053.0 | Compositional bias | Note=Pro-rich |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000521059 | - | 5 | 32 | 863_913 | 150 | 1053.0 | Compositional bias | Note=Pro-rich |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000522684 | - | 5 | 32 | 712_733 | 150 | 1053.0 | Compositional bias | Note=Pro-rich |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000522684 | - | 5 | 32 | 863_913 | 150 | 1053.0 | Compositional bias | Note=Pro-rich |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000340930 | - | 5 | 33 | 35_355 | 150 | 1066.0 | Domain | FERM |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000340930 | - | 5 | 33 | 422_680 | 150 | 1066.0 | Domain | Protein kinase |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000521059 | - | 5 | 32 | 35_355 | 150 | 1053.0 | Domain | FERM |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000521059 | - | 5 | 32 | 422_680 | 150 | 1053.0 | Domain | Protein kinase |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000522684 | - | 5 | 32 | 35_355 | 150 | 1053.0 | Domain | FERM |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000522684 | - | 5 | 32 | 422_680 | 150 | 1053.0 | Domain | Protein kinase |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000340930 | - | 5 | 33 | 428_434 | 150 | 1066.0 | Nucleotide binding | ATP |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000340930 | - | 5 | 33 | 500_502 | 150 | 1066.0 | Nucleotide binding | ATP |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000521059 | - | 5 | 32 | 428_434 | 150 | 1053.0 | Nucleotide binding | ATP |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000521059 | - | 5 | 32 | 500_502 | 150 | 1053.0 | Nucleotide binding | ATP |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000522684 | - | 5 | 32 | 428_434 | 150 | 1053.0 | Nucleotide binding | ATP |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000522684 | - | 5 | 32 | 500_502 | 150 | 1053.0 | Nucleotide binding | ATP |
| Tgene | AGO2 | chr8:141874411 | chr8:141545668 | ENST00000220592 | 15 | 19 | 235_348 | 723 | 860.0 | Domain | PAZ | |
| Tgene | AGO2 | chr8:141874411 | chr8:141545668 | ENST00000220592 | 15 | 19 | 517_818 | 723 | 860.0 | Domain | Piwi |
Top |
Fusion Gene Sequence for PTK2-AGO2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >70019_70019_1_PTK2-AGO2_PTK2_chr8_141874411_ENST00000517887_AGO2_chr8_141545668_ENST00000220592_length(transcript)=2178nt_BP=888nt TAGGAACACAGAGCCATGGGGTCACATTACTAAGCTGGTTTCATTCTGTTCAGTAGCTTGATCTAGCCAGGATGGTCTGTGTTTCAGATT GCGGAAGCATGAGAAGATTCTGTAACTGAGAGGCATTCTTCAAGCTGCTGGAAAATATTACAGAAGGAACACCTTCCAAATTTTGCTTCA AGCTCTGAAAAAAGGAGCACACAGGGGAACCTTAGTCTTGCTTGTGGATCGAAGCAAATACTATCACAGCGAGGAACTGGGACTTTGTGT GCAGACTCCTGGCTTGATCTTTATAGGATTGGGTGTATGTTGTTGGAACTGGCAGGCCAAGAGGCCCTGAAACCAGCAGGTGCCATCTAT ATGGAGAAAAGCGGATGTAGTCCATTTCCAGTGTGTTGGGCTAAAGAATATGACAGATACCTAGCATCTAGCAAAATAATGGCAGCTGCT TACCTTGACCCCAACTTGAATCACACACCAAATTCGAGTACTAAGACTCACCTGGGTACTGGTATGGAACGTTCTCCTGGTGCAATGGAG CGAGTATTAAAGGTCTTTCATTATTTTGAAAGCAATAGTGAGCCAACCACCTGGGCCAGTATTATCAGGCATGGAGATGCTACTGATGTC AGGGGCATCATTCAGAAGATAGTGGACAGTCACAAAGTAAAGCATGTGGCCTGCTATGGATTCCGCCTCAGTCACCTGCGGTCAGAGGAG GTTCACTGGCTTCACGTGGATATGGGCGTCTCCAGTGTGAGGGAGAAGTATGAGCTTGCTCACCCACCAGAGGAGTGGAAATATGAATTG AGAATTCGTTATTTGCCAAAAGGATTTCTAAACCAGTTTACTGAAGATAAGCCAACTTTGAATTTCTTCTATCAACAGGTTGGGAAAAGT GGAAACATTCCAGCAGGCACGACTGTGGACACGAAAATCACCCACCCCACCGAGTTCGACTTCTACCTGTGTAGTCACGCTGGCATCCAG GGGACAAGCAGGCCTTCGCACTATCACGTCCTCTGGGACGACAATCGTTTCTCCTCTGATGAGCTGCAGATCCTAACCTACCAGCTGTGT CACACCTACGTGCGCTGCACACGCTCCGTGTCCATCCCAGCGCCAGCATACTACGCTCACCTGGTGGCCTTCCGGGCCAGGTACCACCTG GTGGATAAGGAACATGACAGTGCTGAAGGAAGCCATACCTCTGGGCAGAGTAACGGGCGAGACCACCAAGCACTGGCCAAGGCGGTCCAG GTTCACCAAGACACTCTGCGCACCATGTACTTTGCTTGACATGTTTTAGTGTTTAGCGATTGTGTACCGAGTGGGATTCACGAGACCAGC TACACTCAGACCAACAGATGGCCAGCCCTTCCGTGACAGCCAGCATCGAACATGAGACGTCATTGATTTTATTAGATTCTCCGTTTTCCA GAATGCCTTCCGTCCCAGATTTCAAACTTGGATTTTGAACTGCAGACCTGTATGAGAACCCAATGTCATAGGAAATATGGTTTGCTAAAA TCTATAAGCTGCTTATTAAAACAGAGTCCCGTGTGTCCTAAAAATCTCCTAAAACCAGTCTATGAACTCAGGGCTTTAAAACATTTTTAA TTTATTTGGTCATTCAATTTACTTGTTTTTAATACATGATTCTCTATGAAATTGATGGGCTCAAACTAGCTGTGAATCTTCTGAGAGTGA AAGCAACACAAAACACAAGTGTGGTTTTAAAGCCTTGAACATTCTGATGTTGTCACTAAAGTTGATTTCCAGGCGATGCTCGTGTGCCCC TGGCGTGGCTCACCCAAGTTCCTCGACTGAGGGCCGGTGGCCATGCAGAGGCGTCCGCAGGTGCCGTGTTCTGCCAGCACCGCCCTTCAC CCGGCCTGAACTAAGGAGCAGTGGCAGAAGGTGGGCCCCGTGTGTTTACAGCATTTCCAGGTCCAGAGAGGTTGGCAGACAAGTGCCATT TTAATAAAAAATGTATTTAAAAAAAGAAAAAAGACAATATACTGACAGTCTAATCATCAGATTTGTCCTATAGGACTGTGACATTTATTT TCCAAAGTTTTTAAAGAATACGGGTCTGTGGTGATAAATACAGTACAATCCTTTTTCACTGTTATGTCTTTAATGTAAGAGAAGAATATA >70019_70019_1_PTK2-AGO2_PTK2_chr8_141874411_ENST00000517887_AGO2_chr8_141545668_ENST00000220592_length(amino acids)=344AA_BP=1 MCADSWLDLYRIGCMLLELAGQEALKPAGAIYMEKSGCSPFPVCWAKEYDRYLASSKIMAAAYLDPNLNHTPNSSTKTHLGTGMERSPGA MERVLKVFHYFESNSEPTTWASIIRHGDATDVRGIIQKIVDSHKVKHVACYGFRLSHLRSEEVHWLHVDMGVSSVREKYELAHPPEEWKY ELRIRYLPKGFLNQFTEDKPTLNFFYQQVGKSGNIPAGTTVDTKITHPTEFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQ -------------------------------------------------------------- >70019_70019_2_PTK2-AGO2_PTK2_chr8_141874411_ENST00000521059_AGO2_chr8_141545668_ENST00000220592_length(transcript)=1922nt_BP=632nt GCGCGGCTGCTGTCGCCCAGCGCCGCCCCGTCGTCGTCTGCCTTCGCTTCACGGCGCCGAGCCGCGGTCCGAAGTCTTGCTGTGTCACCC AGGCTGCCAGGCTGGAGTGGAGTGGCATGATCTCGGCTGACTGCAACCTCTGCCTCCCAGAATATGACAGATACCTAGCATCTAGCAAAA TAATGGCAGCTGCTTACCTTGACCCCAACTTGAATCACACACCAAATTCGAGTACTAAGACTCACCTGGGTACTGGTATGGAACGTTCTC CTGGTGCAATGGAGCGAGTATTAAAGGTCTTTCATTATTTTGAAAGCAATAGTGAGCCAACCACCTGGGCCAGTATTATCAGGCATGGAG ATGCTACTGATGTCAGGGGCATCATTCAGAAGATAGTGGACAGTCACAAAGTAAAGCATGTGGCCTGCTATGGATTCCGCCTCAGTCACC TGCGGTCAGAGGAGGTTCACTGGCTTCACGTGGATATGGGCGTCTCCAGTGTGAGGGAGAAGTATGAGCTTGCTCACCCACCAGAGGAGT GGAAATATGAATTGAGAATTCGTTATTTGCCAAAAGGATTTCTAAACCAGTTTACTGAAGATAAGCCAACTTTGAATTTCTTCTATCAAC AGGTTGGGAAAAGTGGAAACATTCCAGCAGGCACGACTGTGGACACGAAAATCACCCACCCCACCGAGTTCGACTTCTACCTGTGTAGTC ACGCTGGCATCCAGGGGACAAGCAGGCCTTCGCACTATCACGTCCTCTGGGACGACAATCGTTTCTCCTCTGATGAGCTGCAGATCCTAA CCTACCAGCTGTGTCACACCTACGTGCGCTGCACACGCTCCGTGTCCATCCCAGCGCCAGCATACTACGCTCACCTGGTGGCCTTCCGGG CCAGGTACCACCTGGTGGATAAGGAACATGACAGTGCTGAAGGAAGCCATACCTCTGGGCAGAGTAACGGGCGAGACCACCAAGCACTGG CCAAGGCGGTCCAGGTTCACCAAGACACTCTGCGCACCATGTACTTTGCTTGACATGTTTTAGTGTTTAGCGATTGTGTACCGAGTGGGA TTCACGAGACCAGCTACACTCAGACCAACAGATGGCCAGCCCTTCCGTGACAGCCAGCATCGAACATGAGACGTCATTGATTTTATTAGA TTCTCCGTTTTCCAGAATGCCTTCCGTCCCAGATTTCAAACTTGGATTTTGAACTGCAGACCTGTATGAGAACCCAATGTCATAGGAAAT ATGGTTTGCTAAAATCTATAAGCTGCTTATTAAAACAGAGTCCCGTGTGTCCTAAAAATCTCCTAAAACCAGTCTATGAACTCAGGGCTT TAAAACATTTTTAATTTATTTGGTCATTCAATTTACTTGTTTTTAATACATGATTCTCTATGAAATTGATGGGCTCAAACTAGCTGTGAA TCTTCTGAGAGTGAAAGCAACACAAAACACAAGTGTGGTTTTAAAGCCTTGAACATTCTGATGTTGTCACTAAAGTTGATTTCCAGGCGA TGCTCGTGTGCCCCTGGCGTGGCTCACCCAAGTTCCTCGACTGAGGGCCGGTGGCCATGCAGAGGCGTCCGCAGGTGCCGTGTTCTGCCA GCACCGCCCTTCACCCGGCCTGAACTAAGGAGCAGTGGCAGAAGGTGGGCCCCGTGTGTTTACAGCATTTCCAGGTCCAGAGAGGTTGGC AGACAAGTGCCATTTTAATAAAAAATGTATTTAAAAAAAGAAAAAAGACAATATACTGACAGTCTAATCATCAGATTTGTCCTATAGGAC TGTGACATTTATTTTCCAAAGTTTTTAAAGAATACGGGTCTGTGGTGATAAATACAGTACAATCCTTTTTCACTGTTATGTCTTTAATGT >70019_70019_2_PTK2-AGO2_PTK2_chr8_141874411_ENST00000521059_AGO2_chr8_141545668_ENST00000220592_length(amino acids)=313AA_BP=1 MEWSGMISADCNLCLPEYDRYLASSKIMAAAYLDPNLNHTPNSSTKTHLGTGMERSPGAMERVLKVFHYFESNSEPTTWASIIRHGDATD VRGIIQKIVDSHKVKHVACYGFRLSHLRSEEVHWLHVDMGVSSVREKYELAHPPEEWKYELRIRYLPKGFLNQFTEDKPTLNFFYQQVGK SGNIPAGTTVDTKITHPTEFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAYYAHLVAFRARYH -------------------------------------------------------------- >70019_70019_3_PTK2-AGO2_PTK2_chr8_141874411_ENST00000535192_AGO2_chr8_141545668_ENST00000220592_length(transcript)=1771nt_BP=481nt ATATGACAGATACCTAGCATCTAGCAAAATAATGGCAGCTGCTTACCTTGACCCCAACTTGAATCACACACCAAATTCGAGTACTAAGAC TCACCTGGGTACTGGTATGGAACGTTCTCCTGGTGCAATGGAGCGAGTATTAAAGGTCTTTCATTATTTTGAAAGCAATAGTGAGCCAAC CACCTGGGCCAGTATTATCAGGCATGGAGATGCTACTGATGTCAGGGGCATCATTCAGAAGATAGTGGACAGTCACAAAGTAAAGCATGT GGCCTGCTATGGATTCCGCCTCAGTCACCTGCGGTCAGAGGAGGTTCACTGGCTTCACGTGGATATGGGCGTCTCCAGTGTGAGGGAGAA GTATGAGCTTGCTCACCCACCAGAGGAGTGGAAATATGAATTGAGAATTCGTTATTTGCCAAAAGGATTTCTAAACCAGTTTACTGAAGA TAAGCCAACTTTGAATTTCTTCTATCAACAGGTTGGGAAAAGTGGAAACATTCCAGCAGGCACGACTGTGGACACGAAAATCACCCACCC CACCGAGTTCGACTTCTACCTGTGTAGTCACGCTGGCATCCAGGGGACAAGCAGGCCTTCGCACTATCACGTCCTCTGGGACGACAATCG TTTCTCCTCTGATGAGCTGCAGATCCTAACCTACCAGCTGTGTCACACCTACGTGCGCTGCACACGCTCCGTGTCCATCCCAGCGCCAGC ATACTACGCTCACCTGGTGGCCTTCCGGGCCAGGTACCACCTGGTGGATAAGGAACATGACAGTGCTGAAGGAAGCCATACCTCTGGGCA GAGTAACGGGCGAGACCACCAAGCACTGGCCAAGGCGGTCCAGGTTCACCAAGACACTCTGCGCACCATGTACTTTGCTTGACATGTTTT AGTGTTTAGCGATTGTGTACCGAGTGGGATTCACGAGACCAGCTACACTCAGACCAACAGATGGCCAGCCCTTCCGTGACAGCCAGCATC GAACATGAGACGTCATTGATTTTATTAGATTCTCCGTTTTCCAGAATGCCTTCCGTCCCAGATTTCAAACTTGGATTTTGAACTGCAGAC CTGTATGAGAACCCAATGTCATAGGAAATATGGTTTGCTAAAATCTATAAGCTGCTTATTAAAACAGAGTCCCGTGTGTCCTAAAAATCT CCTAAAACCAGTCTATGAACTCAGGGCTTTAAAACATTTTTAATTTATTTGGTCATTCAATTTACTTGTTTTTAATACATGATTCTCTAT GAAATTGATGGGCTCAAACTAGCTGTGAATCTTCTGAGAGTGAAAGCAACACAAAACACAAGTGTGGTTTTAAAGCCTTGAACATTCTGA TGTTGTCACTAAAGTTGATTTCCAGGCGATGCTCGTGTGCCCCTGGCGTGGCTCACCCAAGTTCCTCGACTGAGGGCCGGTGGCCATGCA GAGGCGTCCGCAGGTGCCGTGTTCTGCCAGCACCGCCCTTCACCCGGCCTGAACTAAGGAGCAGTGGCAGAAGGTGGGCCCCGTGTGTTT ACAGCATTTCCAGGTCCAGAGAGGTTGGCAGACAAGTGCCATTTTAATAAAAAATGTATTTAAAAAAAGAAAAAAGACAATATACTGACA GTCTAATCATCAGATTTGTCCTATAGGACTGTGACATTTATTTTCCAAAGTTTTTAAAGAATACGGGTCTGTGGTGATAAATACAGTACA >70019_70019_3_PTK2-AGO2_PTK2_chr8_141874411_ENST00000535192_AGO2_chr8_141545668_ENST00000220592_length(amino acids)=286AA_BP=0 MAAAYLDPNLNHTPNSSTKTHLGTGMERSPGAMERVLKVFHYFESNSEPTTWASIIRHGDATDVRGIIQKIVDSHKVKHVACYGFRLSHL RSEEVHWLHVDMGVSSVREKYELAHPPEEWKYELRIRYLPKGFLNQFTEDKPTLNFFYQQVGKSGNIPAGTTVDTKITHPTEFDFYLCSH AGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAYYAHLVAFRARYHLVDKEHDSAEGSHTSGQSNGRDHQALA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for PTK2-AGO2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | AGO2 | chr8:141874411 | chr8:141545668 | ENST00000220592 | 15 | 19 | 753_761 | 723.0 | 860.0 | guide RNA | |
| Tgene | AGO2 | chr8:141874411 | chr8:141545668 | ENST00000220592 | 15 | 19 | 790_812 | 723.0 | 860.0 | guide RNA | |
| Tgene | AGO2 | chr8:141874411 | chr8:141545668 | ENST00000519980 | 0 | 18 | 311_316 | 0 | 826.0 | guide RNA | |
| Tgene | AGO2 | chr8:141874411 | chr8:141545668 | ENST00000519980 | 0 | 18 | 524_566 | 0 | 826.0 | guide RNA | |
| Tgene | AGO2 | chr8:141874411 | chr8:141545668 | ENST00000519980 | 0 | 18 | 709_710 | 0 | 826.0 | guide RNA | |
| Tgene | AGO2 | chr8:141874411 | chr8:141545668 | ENST00000519980 | 0 | 18 | 753_761 | 0 | 826.0 | guide RNA | |
| Tgene | AGO2 | chr8:141874411 | chr8:141545668 | ENST00000519980 | 0 | 18 | 790_812 | 0 | 826.0 | guide RNA | |
| Tgene | AGO2 | chr8:141874411 | chr8:141545668 | ENST00000519980 | 0 | 18 | 587_590 | 0 | 826.0 | GW182 family members | |
| Tgene | AGO2 | chr8:141874411 | chr8:141545668 | ENST00000519980 | 0 | 18 | 650_660 | 0 | 826.0 | GW182 family members |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000340930 | - | 5 | 33 | 912_1052 | 150.0 | 1066.0 | ARHGEF28 |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000521059 | - | 5 | 32 | 912_1052 | 150.0 | 1053.0 | ARHGEF28 |
| Hgene | PTK2 | chr8:141874411 | chr8:141545668 | ENST00000522684 | - | 5 | 32 | 912_1052 | 150.0 | 1053.0 | ARHGEF28 |
| Tgene | AGO2 | chr8:141874411 | chr8:141545668 | ENST00000220592 | 15 | 19 | 311_316 | 723.0 | 860.0 | guide RNA | |
| Tgene | AGO2 | chr8:141874411 | chr8:141545668 | ENST00000220592 | 15 | 19 | 524_566 | 723.0 | 860.0 | guide RNA | |
| Tgene | AGO2 | chr8:141874411 | chr8:141545668 | ENST00000220592 | 15 | 19 | 709_710 | 723.0 | 860.0 | guide RNA | |
| Tgene | AGO2 | chr8:141874411 | chr8:141545668 | ENST00000220592 | 15 | 19 | 587_590 | 723.0 | 860.0 | GW182 family members | |
| Tgene | AGO2 | chr8:141874411 | chr8:141545668 | ENST00000220592 | 15 | 19 | 650_660 | 723.0 | 860.0 | GW182 family members |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for PTK2-AGO2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for PTK2-AGO2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
